Transcriptome Response of Liver and Muscle in Heat-Stressed Laying Hens
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Tissue Collection
2.2. Total RNA Isolation
2.3. RNA-Seq Design and cDNA Library Construction
2.4. Sequence Read Quality Control, Mapping, and Counting
2.5. Differential Expression Analysis
2.6. Additional Analyses on DEGs
2.7. Correlation of RNA-Seq and qPCR Results
3. Results
3.1. Transcriptome Alignment, Mapping, and PCA Analysis
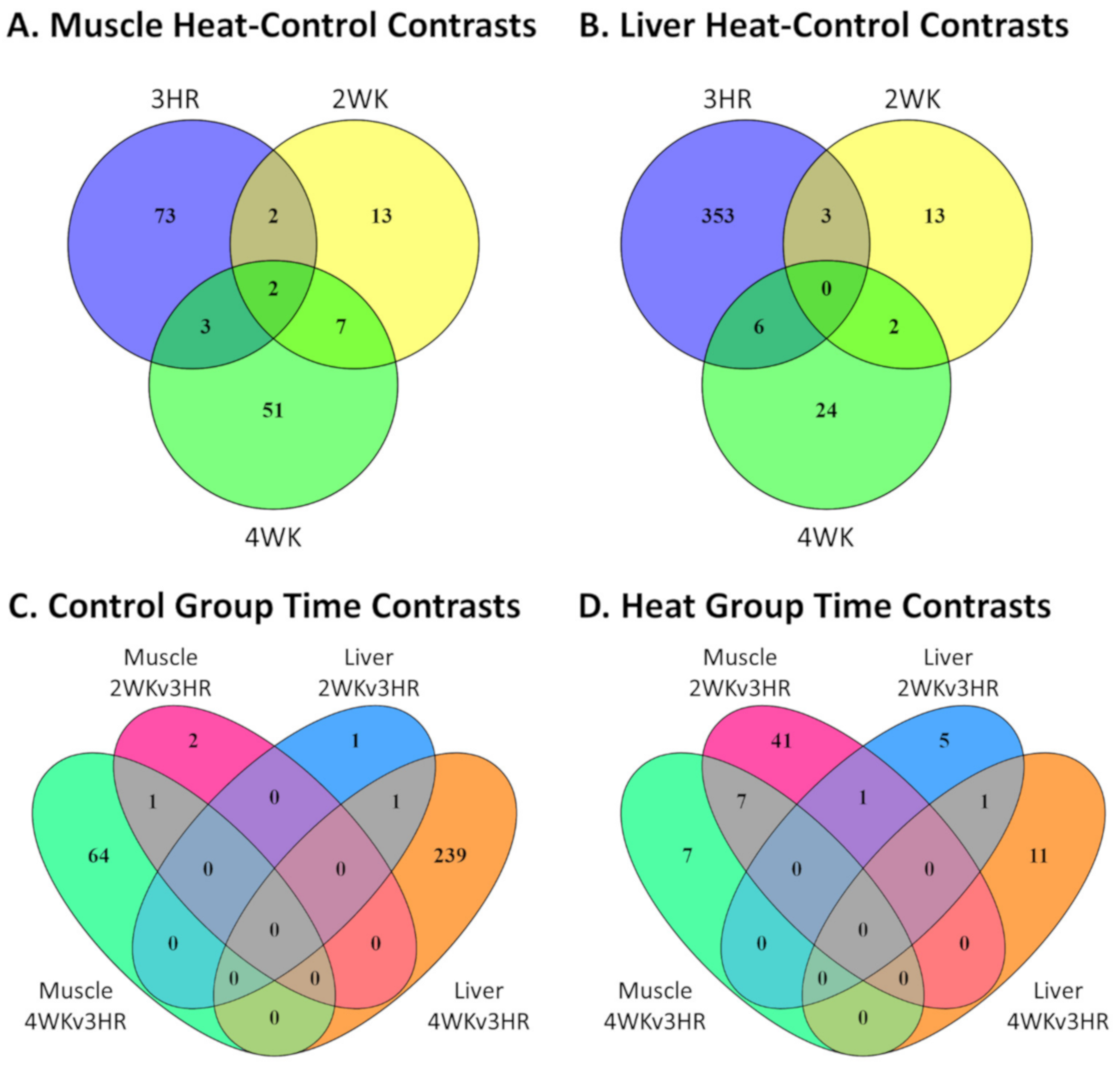
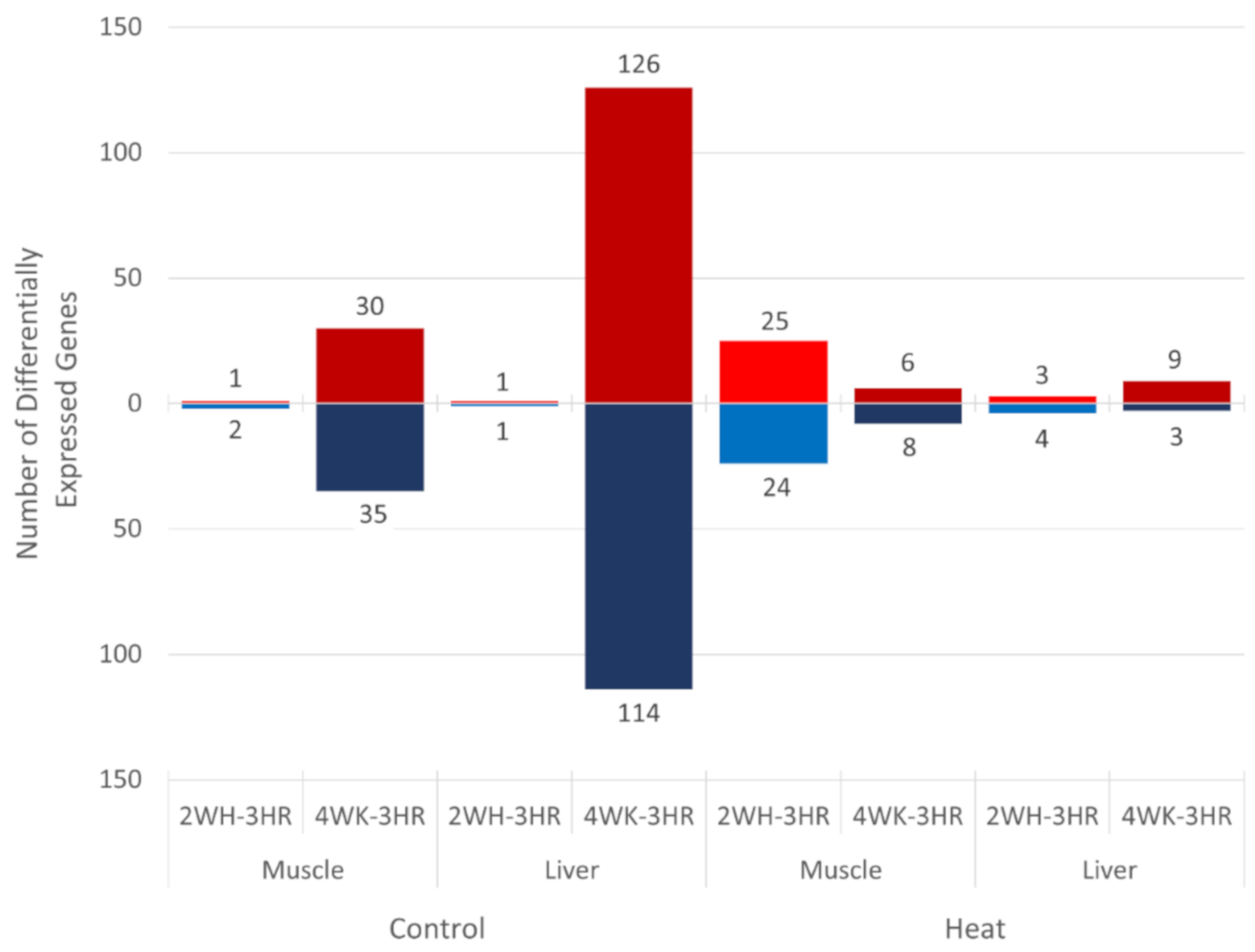
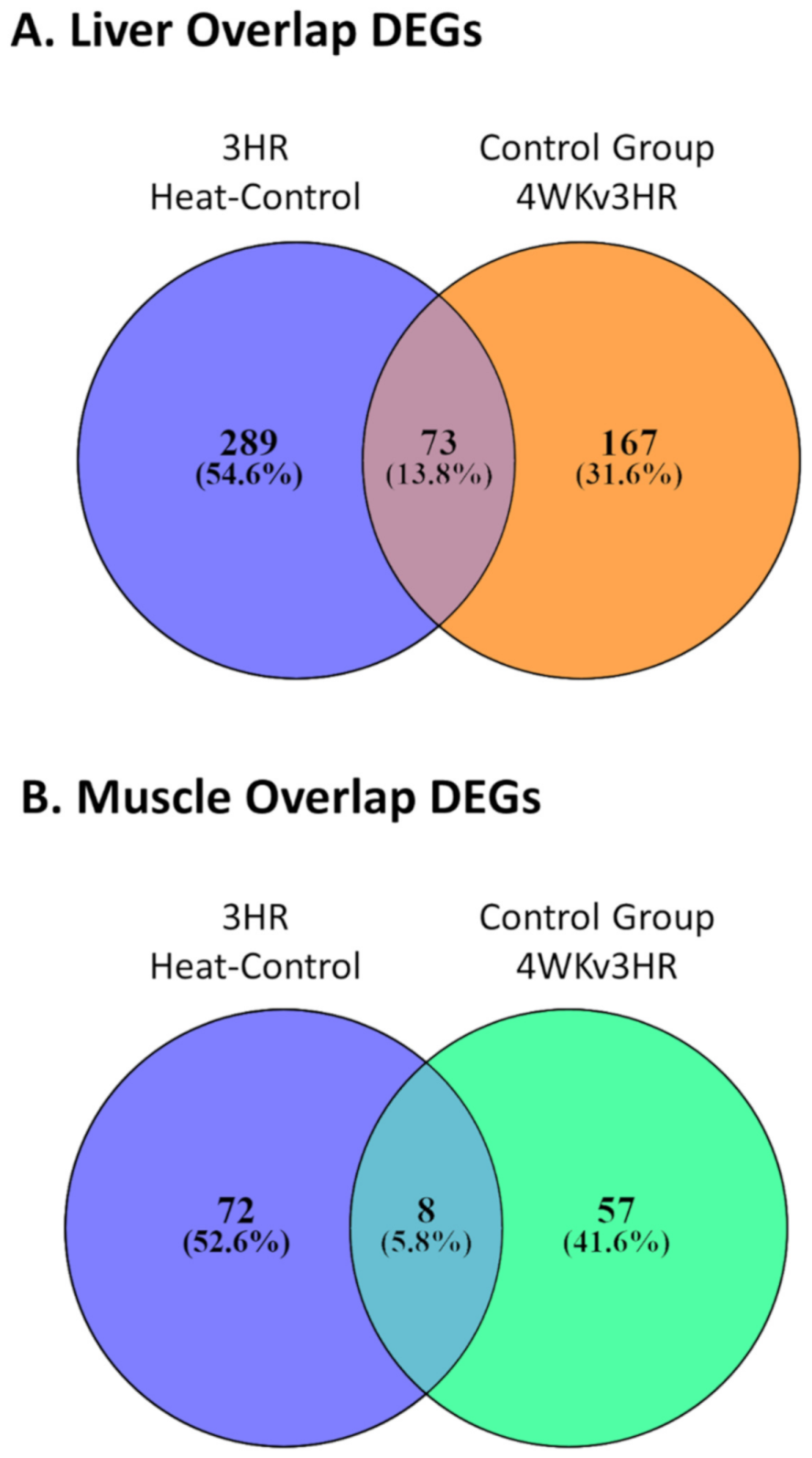
3.2. Effect of Heat Stress on Gene Expression in Egg-Producing Layers
3.3. Gene Expression Changes in Layers during Early Production
3.4. IPA Functional Predictions from Heat-Control and Control Group Time Contrasts and Corresponding Overlapping Genes
3.5. Correlation of RNA-Seq and qPCR Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Babinszky, L.; Halas, V.; Verstegen, M.W.A. Impacts of climate change on animal production and quality of animal food products. In Climate Change-Socioeconomic Effects; InTech: London, UK, 2011. [Google Scholar]
- Settar, P.; Yalçin, S.; Türkmut, L.; Ozkan, S.; Cahanar, A. Season by genotype interaction related to broiler growth rate and heat tolerance. Poult. Sci. 1999, 78, 1353–1358. [Google Scholar] [CrossRef]
- Deeb, N.; Cahaner, A. Genotype-by-environment interaction with broiler genotypes differing in growth rate. 3. Growth rate and water consumption of broiler progeny from weight-selected versus nonselected parents under normal and high ambient temperatures. Poult. Sci. 2002, 81, 293–301. [Google Scholar] [CrossRef]
- Lara, L.; Rostagno, M. Impact of heat stress on poultry production. Animals 2013, 3, 356–369. [Google Scholar] [CrossRef]
- Daghir, N.J. Present status and future of the poultry industry in hot regions. In Poultry Production in Hot Climates; Daghir, N.J., Ed.; CAB International: Wallingford, UK, 2008. [Google Scholar]
- Attia, Y.A.; Abd El-Hamid, A.E.H.E.; Abedalla, A.A.; Berika, M.A.; Al-Harthi, M.A.; Kucuk, O.; Sahin, K.; Abou-Shehema, B.M. Laying performance, digestibility and plasma hormones in laying hens exposed to chronic heat stress as affected by betaine, vitamin C, and/or vitamin E supplementation. Springerplus 2016, 5, 1619. [Google Scholar] [CrossRef] [PubMed]
- Rowland, K.; Ashwell, C.M.; Persia, M.E.; Rothschild, M.F.; Schmidt, C.; Lamont, S.J. Genetic analysis of production, physiological, and egg quality traits in heat-challenged commercial white egg-laying hens using 600k SNP array data. Genet. Sel. Evol. 2019, 51, 31. [Google Scholar] [CrossRef]
- Barrett, N.W.; Rowland, K.; Schmidt, C.J.; Lamont, S.J.; Rothschild, M.F.; Ashwell, C.M.; Persia, M.E. Effects of acute and chronic heat stress on the performance, egg quality, body temperature, and blood gas parameters of laying hens. Poult. Sci. 2019, 98, 6684–6692. [Google Scholar] [CrossRef]
- Wilson, H.R.; Wilcox, C.J.; Voitle, R.A.; Baird, C.D.; Dorminey, R.W. Characteristics of white leghorn chickens selected for heat tolerance. Poult. Sci. 1975, 54, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Tang, L.; Lu, L.; Zhang, L.; Lin, X.; Liu, H.C.; Odle, J.; Luo, X. Effects of acute and chronic heat stress on plasma metabolites, hormones and oxidant status in restrictedly fed broiler breeders. Poult. Sci. 2015, 94, 1635–1644. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Hsieh, J.C.F.; Schmidt, C.J.; Zhu, Q.; Lamont, S.J. Liver transcriptome response to hyperthermic stress in three distinct chicken lines. BMC Genom. 2016, 17, 955. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Tang, L.; Lu, L.; Zhang, L.; Xi, L.; Liu, H.C.; Odle, J.; Luo, X. Differential expression of heat shock transcription factors and heat shock proteins after acute and chronic heat stress in laying chickens (Gallus gallus). PLoS ONE 2014, 9, e102204. [Google Scholar] [CrossRef]
- Azad, K.M.A.; Kikusato, M.; Hoque, A.M.; Toyomizu, M. Effect of chronic heat stress on performance and oxidative damage in different strains of chickens. J. Poult. Sci. 2010, 47, 333–337. [Google Scholar] [CrossRef][Green Version]
- Sandercock, D.A.; Hunter, R.R.; Mitchell, M.A.; Hocking, P.M. Thermoregulatory capacity and muscle membrane integrity are compromised in broilers compared with layers at the same age or body weight. Br. Poult. Sci. 2006, 47, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Zahoor, I.; De Koning, D.J.; Hocking, P.M. Transcriptional profile of breast muscle in heat stressed layers is similar to that of broiler chickens at control temperature. Genet. Sel. Evol. 2017, 49, 69. [Google Scholar] [CrossRef]
- Lin, H.; Decuypere, E.; Buyse, J. Acute heat stress induces oxidative stress in broiler chickens. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2006, 144, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Tan, G.Y.; Fu, Y.Q.; Feng, J.H.; Zhang, M.H. Effects of acute heat stress and subsequent stress removal on function of hepatic mitochondrial respiration, ROS production and lipid peroxidation in broiler chickens. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2010, 151, 204–208. [Google Scholar] [CrossRef]
- Wallimann, T.; Turner, D.C.; Eppenberger, H.M. Localization of creatine kinase isoenzymes in myofibrils: I. chicken skeletal muscle. J. Cell Biol. 1977, 75, 297–317. [Google Scholar] [CrossRef]
- Melesse, A.; Maak, S.; Schmidt, R.; Von Lengerken, G. Effect of long-term heat stress on key enzyme activities and T 3 levels in commercial layer hens. Int. J. Livest. Prod. 2011, 2, 107–116. [Google Scholar]
- Luo, Q.B.; Song, X.Y.; Ji, C.L.; Zhang, X.Q.; Zhang, D.X. Exploring the molecular mechanism of acute heat stress exposure in broiler chickens using gene expression profiling. Gene 2014, 546, 200–205. [Google Scholar] [CrossRef]
- Désert, C.; Duclos, M.J.; Blavy, P.; Lecerf, F.; Moreews, F.; Klopp, C.; Aubry, M.; Herault, F.; Le Roy, P.; Berri, C.; et al. Transcriptome profiling of the feeding-to-fasting transition in chicken liver. BMC Genom. 2008, 9, 611. [Google Scholar] [CrossRef] [PubMed]
- Mashaly, M.M.; Hendricks, G.L.; Kalama, M.A.; Gehad, A.E.; Abbas, A.O.; Patterson, P.H. Effect of heat stress on production parameters and immune responses of commercial laying hens. Poult. Sci. 2004, 83, 889–894. [Google Scholar] [CrossRef]
- Rozenboim, I.; Tako, E.; Gal-Garber, O.; Proudman, J.A.; Uni, Z. The effect of heat stress on ovarian function of laying hens. Poult. Sci. 2007, 86, 1760–1765. [Google Scholar] [CrossRef]
- Jastrebski, S.F.; Lamont, S.J.; Schmidt, C.J. Chicken hepatic response to chronic heat stress using integrated transcriptome and metabolome analysis. PLoS ONE 2017, 12, e0181900. [Google Scholar] [CrossRef]
- Wang, Y.; Saelao, P.; Kern, C.; Jin, S.; Gallardo, R.A.; Kelly, T.; Dekkers, J.M.; Lamont, S.J.; Zhou, H. Liver Transcriptome Responses to Heat Stress and Newcastle Disease Virus Infection in Genetically Distinct Chicken Inbred Lines. Genes 2020, 11, 1067. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Dobin, A.; Gingeras, T.R. Mapping RNA-seq reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11.14.1–11.14.19. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2014, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Marini, F.; Binder, H. Development of applications for interactive and reproducible research: A case study. Genom. Comput. Biol. 2016, 3, 39. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 2018, 57, 289–300. [Google Scholar] [CrossRef]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, D183–D189. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J.; Tugendreich, S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Coble, D.J.; Fleming, D.; Persia, M.E.; Ashwell, C.M.; Rothschild, M.F.; Schmidt, C.J.; Lamont, S.J. RNA-seq analysis of broiler liver transcriptome reveals novel responses to high ambient temperature. BMC Genom. 2014, 15, 1084. [Google Scholar] [CrossRef]
- Wang, S.H.; Cheng, C.Y.; Tang, P.C.; Chen, C.F.; Chen, H.H.; Lee, Y.P.; Huang, S.Y. Differential gene expressions in testes of L2 strain Taiwan country chicken in response to acute heat stress. Theriogenology 2013, 79, 374–382.e7. [Google Scholar] [CrossRef]
- Maxfield, F.R.; Iaea, D.B.; Pipalia, N.H. Role of STARD4 and NPC1 in intracellular sterol transport1. Biochem. Cell Biol. 2016, 94, 499–506. [Google Scholar] [CrossRef]
- Abdalla, B.A.; Chen, J.; Nie, Q.; Zhang, X. Genomic insights into the multiple factors controlling abdominal fat deposition in a chicken model. Front. Genet. 2018, 9, 262. [Google Scholar] [CrossRef] [PubMed]
- Claire D’Andre, H.; Paul, W.; Shen, X.; Jia, X.; Zhang, R.; Sun, L.; Zhang, X. Identification and characterization of genes that control fat deposition in chickens. J. Anim. Sci. Biotechnol. 2013, 4, 43. [Google Scholar] [CrossRef]
- Zheng, Q.; Zhang, Y.; Chen, Y.; Yang, N.; Wang, X.J.; Zhu, D. Systematic identification of genes involved in divergent skeletal muscle growth rates of broiler and layer chickens. BMC Genom. 2009, 10, 87. [Google Scholar] [CrossRef]
- Furqon, A.; Gunawan, A.; Ulupi, N.; Suryati, T.; Sumantri, C. Expression and association of SCD gene polymorphisms and fatty acid compositions in chicken cross. Media Peternak. 2017, 40, 151–157. [Google Scholar] [CrossRef]
- Zhuo, Z.; Lamont, S.J.; Lee, W.R.; Abasht, B. RNA-Seq Analysis of Abdominal Fat Reveals Differences between Modern Commercial Broiler Chickens with High and Low Feed Efficiencies. PLoS ONE 2015, 10, e0135810. [Google Scholar] [CrossRef]
- Wang, S.H.; Cheng, C.Y.; Chen, C.J.; Chan, H.L.; Chen, H.H.; Tang, P.C.; Chen, C.F.; Lee, Y.P.; Huang, S.Y. Acute Heat Stress Changes Protein Expression in the Testes of a Broiler-Type Strain of Taiwan Country Chickens. Anim. Biotechnol. 2019, 30, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Yebra-Pimentel, E.S.; Reis, B.; Gessner, J.; Wuertz, S.; Dirks, R.P.H. Temperature training improves transcriptional homeostasis after heat shock in juvenile Atlantic sturgeon (Acipenser oxyrinchus). Fish Physiol. Biochem. 2020, 46, 1653–1664. [Google Scholar] [CrossRef] [PubMed]
- Cogburn, L.A.; Smarsh, D.N.; Wang, X.; Trakooljul, N.; Carré, W.; White, H.B. Transcriptional profiling of liver in riboflavin-deficient chicken embryos explains impaired lipid utilization, energy depletion, massive hemorrhaging, and delayed feathering. BMC Genom. 2018, 19, 1–30. [Google Scholar] [CrossRef]
- Emslie, R.G.; Henry, K.M. Glycogen formation in the fowl. Biochem. J. 1933, 27, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Bigland, C.H.; Triantaphyllopoulos, D.C. Chicken prothrombin, thrombin, and fibrinogen. Am. J. Physiol. 1961, 200, 1013–1017. [Google Scholar] [CrossRef]
- Mutryn, M.F.; Brannick, E.M.; Fu, W.; Lee, W.R.; Abasht, B. Characterization of a novel chicken muscle disorder through differential gene expression and pathway analysis using RNA-sequencing. BMC Genom. 2015, 16, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, H.-W.; Ku, W.-L.; Lin, C.-J.; Chang, C.-F.; Wu, G.-C. Two distinct vitellogenin genes are similar in function and expression in the bigfin reef squid Sepioteuthis lessoniana. Biol. Reprod. 2018, 99, 1034–1044. [Google Scholar] [CrossRef]
- Shao, F.; Bao, H.; Li, H.; Duan, J.; Li, J.; Ling, Y.; Wu, C. Ovary removal modifies liver message RNA profiles in single Comb White Leghorn chickens. Poult. Sci. 2020, 99, 1813–1821. [Google Scholar] [CrossRef]
- Diaz, R.; Hurwitz, S.; Chattopadhyay, N.; Pines, M.; Yang, Y.; Kifor, O.; Einat, M.S.; Butters, R.; Hebert, S.C.; Brown, E.M. Cloning, expression, and tissue localization of the calcium-sensing receptor in chicken (Gallus domesticus). Am. J. Physiol.-Regul. Integr. Comp. Physiol. 1997, 273, R1008–R1016. [Google Scholar] [CrossRef]
- Ahmed, T.A.E.; Suso, H.P.; Hincke, M.T. In-depth comparative analysis of the chicken eggshell membrane proteome. J. Proteom. 2017, 155, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Smith-Mungo, L.I.; Kagan, H.M. Lysyl oxidase: Properties, regulation and multiple functions in biology. Matrix Biol. 1998, 16, 387–398. [Google Scholar] [CrossRef]
- Bae, S.K.; Bessho, Y.; Hojo, M.; Kageyama, R. The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development 2000, 127, 2933–2943. [Google Scholar]
- Chabes, A.; Thelander, L. Controlled protein degradation regulates ribonucleotide reductase activity in proliferating mammalian cells during the normal cell cycle and in response to DNA damage and replication blocks. J. Biol. Chem. 2000, 275, 17747–17753. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.P.; Wang, G.; Bermudez, V.; Hurwitz, J.; Chen, X.S. Crystal structure of the GINS complex and functional insights into its role in DNA replication. Proc. Natl. Acad. Sci. USA 2007, 104, 12685–12690. [Google Scholar] [CrossRef]
- Slawinska, A.; Hsieh, J.C.; Schmidt, C.J.; Lamont, S.J. Heat Stress and Lipopolysaccharide Stimulation of Chicken Macrophage-Like Cell Line Activates Expression of Distinct Sets of Genes. PLoS ONE 2016, 11, e0164575. [Google Scholar] [CrossRef]
- Srikanth, K.; Kumar, H.; Park, W.; Byun, M.; Lim, D.; Kemp, S.; te Pas, M.F.W.; Kim, J.M.; Park, J.E. Cardiac and Skeletal Muscle Transcriptome Response to Heat Stress in Kenyan Chicken Ecotypes Adapted to Low and High Altitudes Reveal Differences in Thermal Tolerance and Stress Response. Front. Genet. 2019, 10, 993. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, S.; Jiang, R.; Qu, L. Expression Analysis of SFRP1 and PDK4 Gene in Livers of Chicken under Heat Stress. China Poult. 2012, 14, 360–367. [Google Scholar]
- Zhang, J.; Schmidt, C.J.; Lamont, S.J. Transcriptome analysis reveals potential mechanisms underlying differential heart development in fast-and slow-growing broilers under heat stress. BMC Genom. 2017, 18, 295. [Google Scholar] [CrossRef]


| Ensembl ID | Gene ID | Contrast: HvC | Tissue | Full Name and Known Function |
| ENSGALG00000000241 | STARD4 | 3 HR, 2 WK, 4 WK | Muscle | StAR-Related Lipid Transfer Domain Containing 4, cholesterol homeostasis |
| ENSGALG00000001000 | HSPA5 | 3 HR, 4 WK | Liver | Heat Shock Protein Family A (Hsp70) Member 5, protein folding |
| ENSGALG00000001749 | ACSBG2 | 2 WK, 4 WK | Muscle | Acyl-CoA Synthetase Bubblegum Family Member 2, fatty acyl-CoA biosynthesis |
| ENSGALG00000005318 | OVCH2 | 2 WK, 4 WK | Liver | Ovochymase 2, serine-type endopeptidase activity |
| ENSGALG00000005739 | SCD | 3 HR, 4 WK | Muscle | Stearoyl-CoA Desaturase, fatty acid biosynthesis |
| ENSGALG00000008349 | LMCD1 | 2 WK, 4 WK | Muscle | LIM and Cysteine Rich Domains 1, transcription factor co-regulator |
| ENSGALG00000011376 | ANKRD9 | 3 HR, 2 WK | Liver | Ankyrin Repeat Domain 9, integral membrane protein |
| ENSGALG00000012610 | CTSV | 3 HR, 4 WK | Muscle | Cathepsin V, collagen chain trimerization |
| ENSGALG00000017394 | INSIG1 | 2 WK, 4 WK | Muscle | Insulin Induced Gene 1, cholesterol metabolism, lipogenesis, and glucose homeostasis |
| ENSGALG00000017804 | Y_RNA | 3 HR, 4 WK | Liver | Small non-coding RNA, component of the Ro60 ribonucleoprotein, target of autoimmune antibodies |
| ENSGALG00000029006 | SYPL1 | 2 WK, 4 WK | Muscle | Synaptophysin Like 1, transporter activity for synaptic vesicle |
| ENSGALG00000031306 | BHLHA15 | 3 HR, 4 WK | Liver | Basic Helix-Loop-Helix Family Member A15, controls transcriptional activity of myoblast differentiation |
| ENSGALG00000037156 | Novel | 2 WK, 4 WK | Muscle | Orthologue = zgc:136493, zinc-finger protein, predicted to be involved with oxidation-reduction process |
| ENSGALG00000037852 | HSD17B7 | 3 HR, 2 WK, 4 WK | Muscle | Hydroxysteroid 17-Beta Dehydrogenase 7, biosynthesis of sex steroids |
| ENSGALG00000040070 | PDIA2 | 3 HR, 2 WK | Liver | Protein Disulfide Isomerase Family A Member 2, protein folding |
| ENSGALG00000049504 | Novel | 2 WK, 4 WK | Liver | Orthologue = SLC2A4, Solute Carrier Family 2 Member 4, facilitated glucose transporter |
| ENSGALG00000050857 | PTPN20 | 3 HR, 4 WK | Liver | Protein Tyrosine Phosphatase, Non-Receptor Type 20, protein tyrosine phosphatase |
| ENSGALG00000051251 | Novel | 2 WK, 4 WK | Muscle | Orthologue = H2B, human histone cluster 1, class H2B, histone |
| ENSGALG00000051816 | Novel | 3 HR, 4 WK | Muscle | LncRNA |
| ENSGALG00000052045 | Novel | 2 WK, 4 WK | Muscle | LncRNA |
| ENSGALG00000053112 | Novel | 3 HR, 4 WK | Liver | Orthologue = FRMPD2, FERM and PDZ Domain Containing 2, cell polarization |
| ENSGALG00000053926 | Novel | 3 HR, 4 WK | Liver | LncRNA |
| ENSGALG00000054926 | Novel | 3 HR, 2 WK | Liver | Unknown |
| Ensembl ID | Gene ID | Contrast | Tissue | Full Name and Known Function |
| ENSGALG00000001749 | ACSBG2 | C_2 WKv3 HR, C_4 WKv3 HR | Muscle | Acyl-CoA Synthetase Bubblegum Family Member 2, fatty acyl-CoA biosynthesis |
| ENSGALG00000006009 | Novel | H_2 WKv3 HR, H_4 WKv3 HR | Muscle | Orthologue = PODXL2, Podocalyxin Like 2, CD34 family of cell surface transmembrane protein |
| ENSGALG00000008912 | ABCB1 | H_2 WKv3 HR, H_4 WKv3 HR | Liver | ATP Binding Cassette Subfamily B Member 1, multidrug resistance |
| ENSGALG00000009700 | PDK4 | H_2 WKv3 HR | Muscle, Liver | Pyruvate Dehydrogenase Kinase 4, mitochondrial protein in regulation of glucose and fatty acid metabolism |
| ENSGALG00000014554 | ATN1 | H_2 WKv3 HR, H_4 WKv3 HR | Muscle | Atrophin 1, transcriptional corepressor, promotes vascular smooth cell migration |
| ENSGALG00000023819 | Novel | H_2 WKv3 HR, H_4 WKv3 HR | Muscle | Orthologue = HSPB11, Heat Shock Protein Family B (Small) Member 11, SHH signaling |
| ENSGALG00000036850 | DNAJA4 | H_2 WKv3 HR, H_4 WKv3 HR | Muscle | DnaJ Heat Shock Protein Family (Hsp40) Member A4, unfolded protein binding |
| ENSGALG00000036956 | Novel | H_2 WKv3 HR, H_4 WKv3 HR | Muscle | Mitochondrial rRNA |
| ENSGALG00000041459 | DDIT4 | H_2 WKv3 HR, H_4 WKv3 HR | Muscle | DNA Damage Inducible Transcript 4, response to cellular energy levels and cellular stress from hypoxia and DNA damage |
| ENSGALG00000049873 | Novel | C_2 WKv3 HR, C_4 WKv3 HR | Liver | LncRNA |
| ENSGALG00000051251 | Novel | H_2 WKv3 HR, H_4 WKv3 HR | Muscle | Orthologue = H2B, human histone cluster 1, class H2B, histone |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Jia, X.; Hsieh, J.C.F.; Monson, M.S.; Zhang, J.; Shu, D.; Nie, Q.; Persia, M.E.; Rothschild, M.F.; Lamont, S.J. Transcriptome Response of Liver and Muscle in Heat-Stressed Laying Hens. Genes 2021, 12, 255. https://doi.org/10.3390/genes12020255
Wang Y, Jia X, Hsieh JCF, Monson MS, Zhang J, Shu D, Nie Q, Persia ME, Rothschild MF, Lamont SJ. Transcriptome Response of Liver and Muscle in Heat-Stressed Laying Hens. Genes. 2021; 12(2):255. https://doi.org/10.3390/genes12020255
Chicago/Turabian StyleWang, Yan, Xinzheng Jia, John C. F. Hsieh, Melissa S. Monson, Jibin Zhang, Dingming Shu, Qinghua Nie, Michael E. Persia, Max F. Rothschild, and Susan J. Lamont. 2021. "Transcriptome Response of Liver and Muscle in Heat-Stressed Laying Hens" Genes 12, no. 2: 255. https://doi.org/10.3390/genes12020255
APA StyleWang, Y., Jia, X., Hsieh, J. C. F., Monson, M. S., Zhang, J., Shu, D., Nie, Q., Persia, M. E., Rothschild, M. F., & Lamont, S. J. (2021). Transcriptome Response of Liver and Muscle in Heat-Stressed Laying Hens. Genes, 12(2), 255. https://doi.org/10.3390/genes12020255







