Formation of the Codon Degeneracy during Interdependent Development between Metabolism and Replication
Abstract
:1. Introduction
2. Materials and Methods
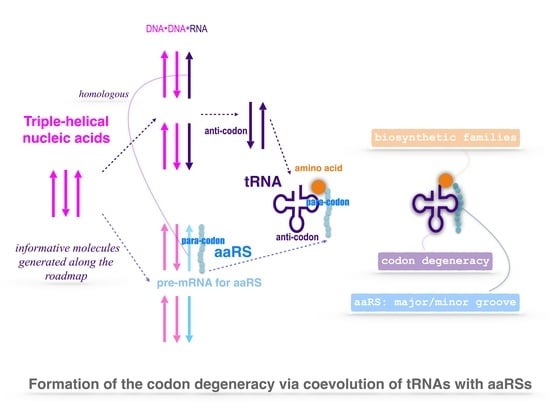
2.1. Triplex Picture
Nomenclature and Notation
- Notations for the 20 amino acids, 20 aaRSs, and the corresponding tRNAs (): amino acid ↔↔ tRNA , , , , , , where the amino acids from to are, respectively, as follows: , , , , , , , , , , , , , , , , , , , , and are, respectively, as follows: (namely ), (namely ), and so on.
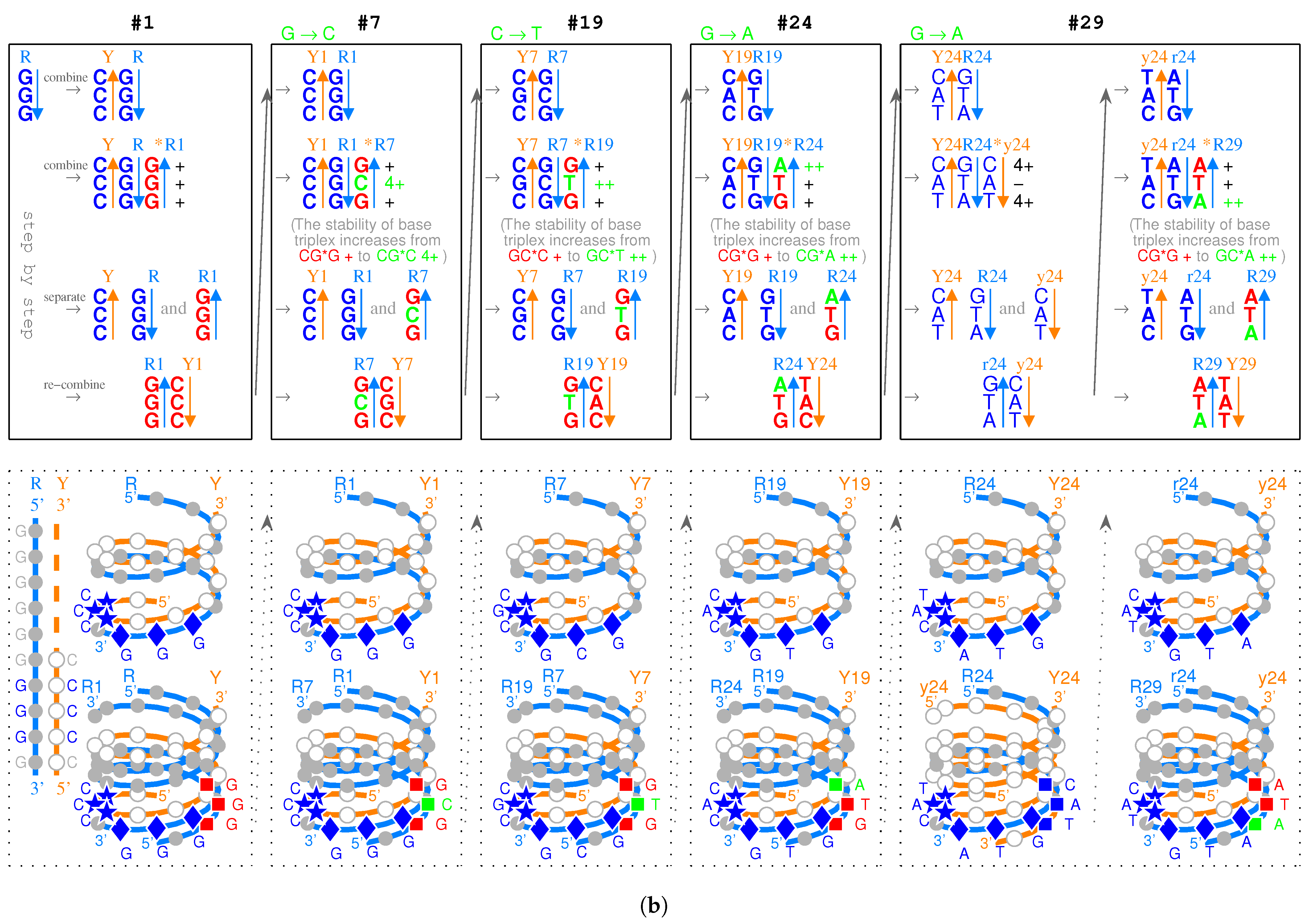
- Triplex DNAs (): , and the inverse triplex DNAs: , , where Y, y stands for pyrimidine strands, and R, r purine strands.
- Triplex DNA·DNA*RNA (): , , , , where two types of tRNAs can be generated by linking the RNA strands or , and aaRSs can approach tRNAs from major groove side (M) or minor groove side (m).
- Codon pairs: , etc.; pair connections: , etc.; route dualities: , etc., where the numbers () indicates the positions on the roadmap
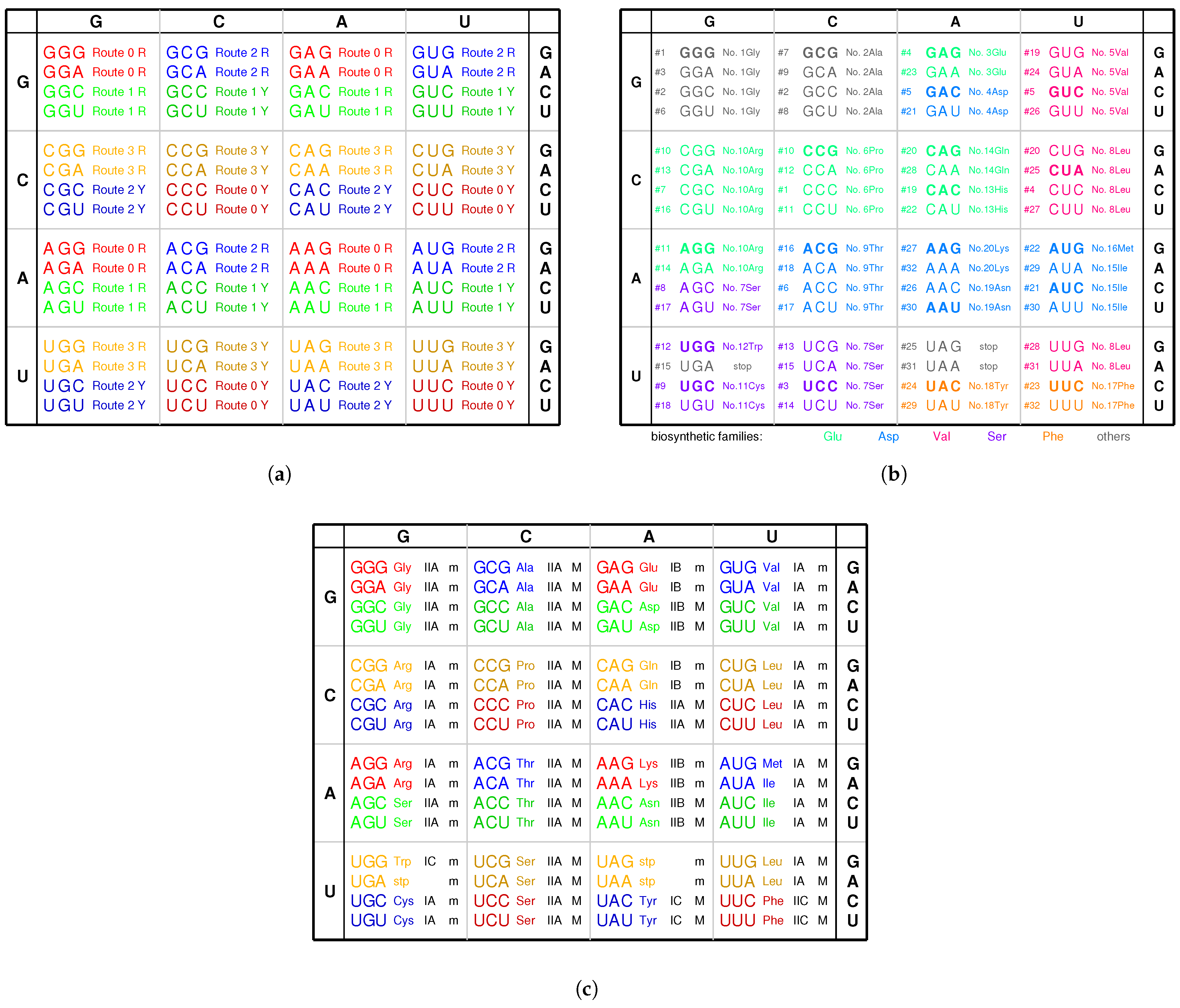
2.2. Origin of the Genetic Code
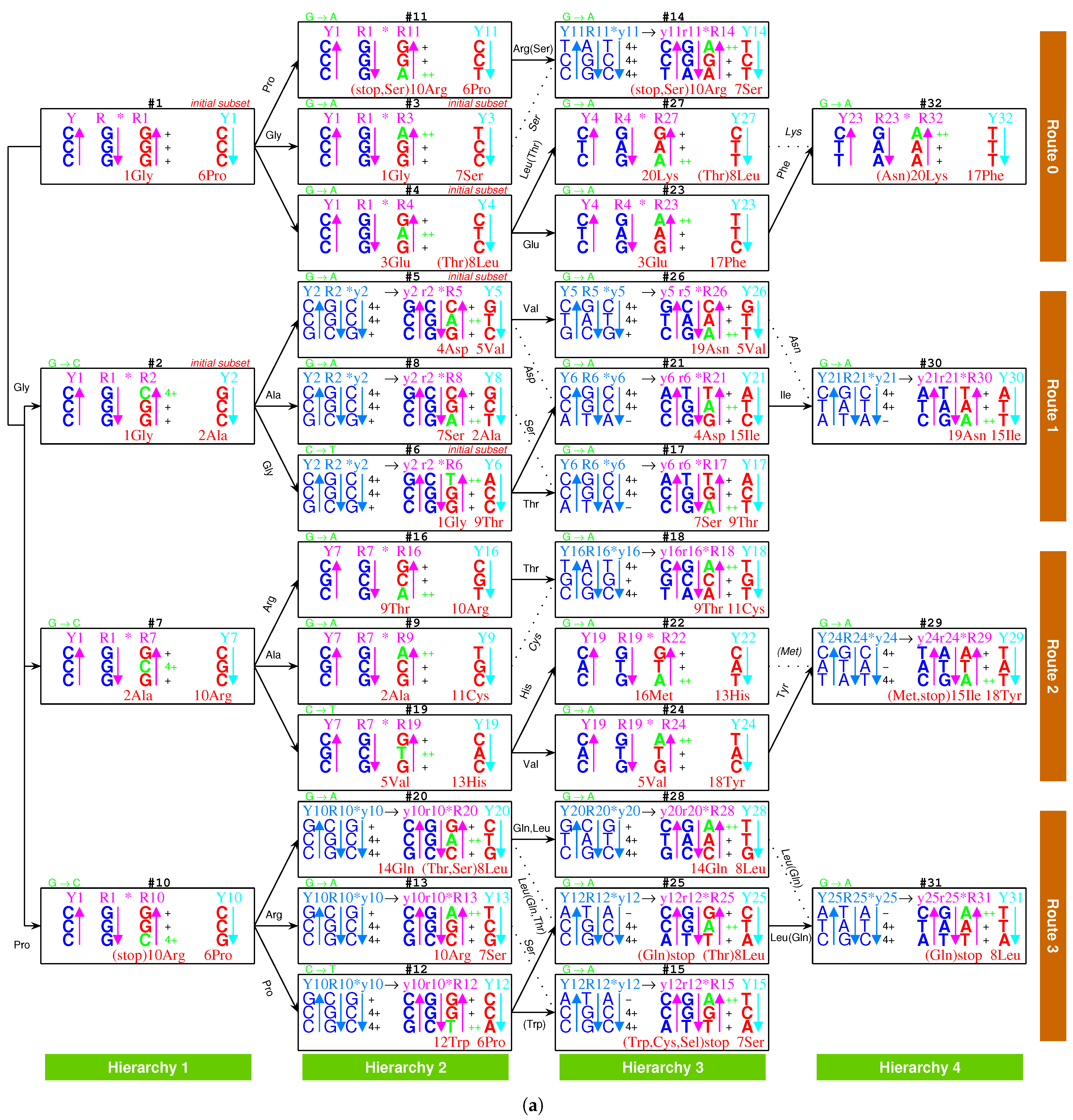
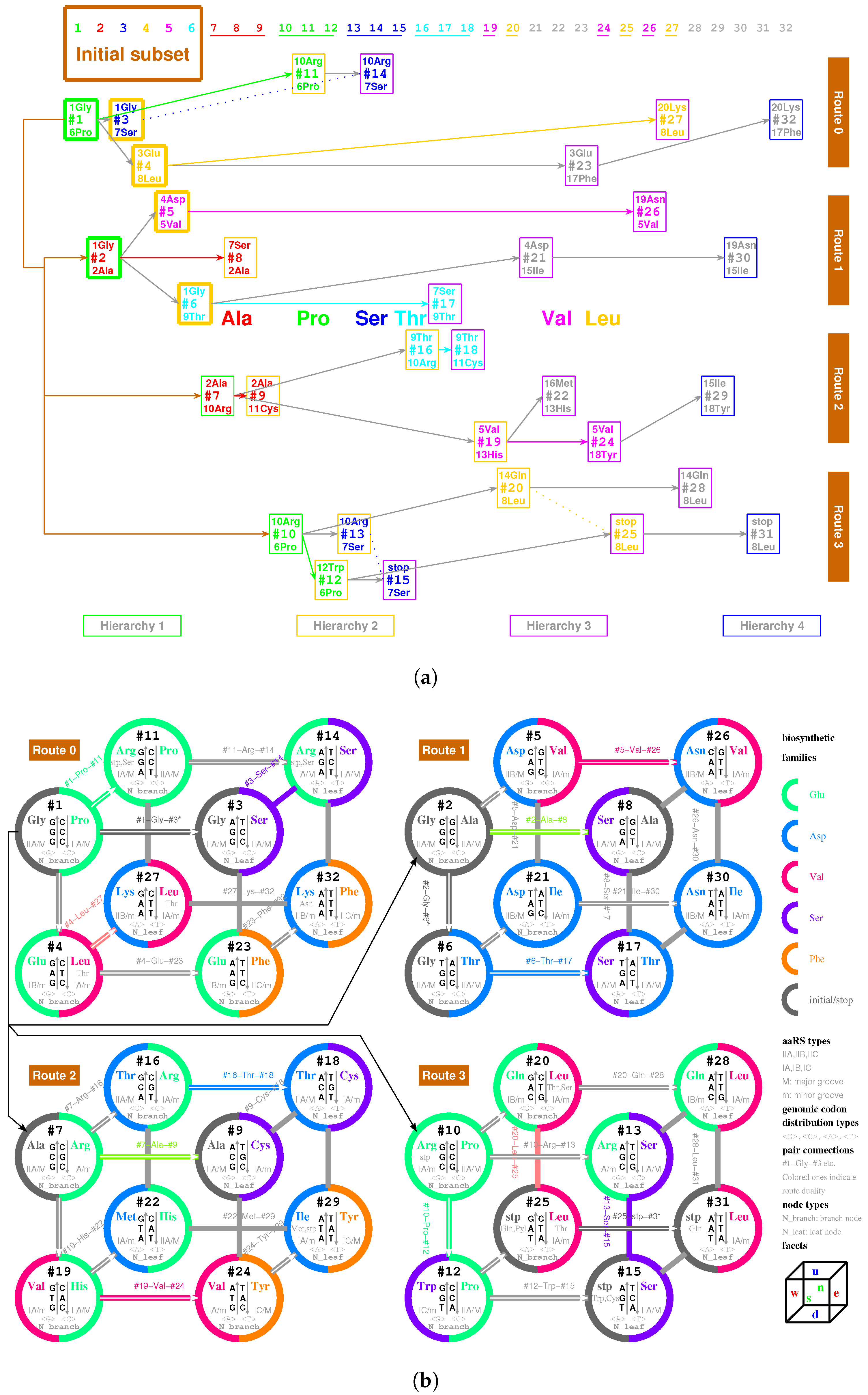
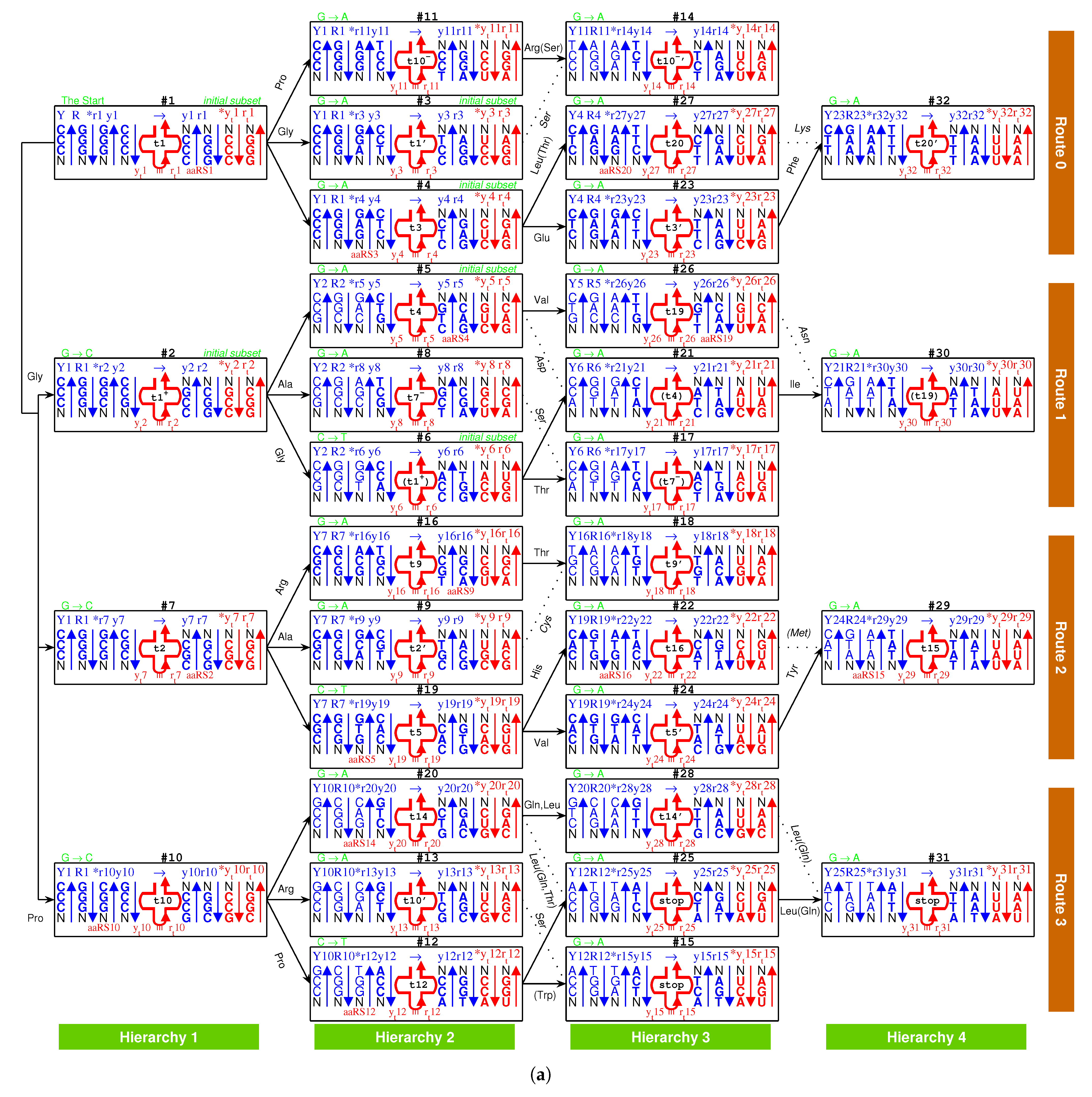
2.2.1. The Roadmap
, , , , , , , , , , , , , , , , , , , , , , , , , , , , , , , ;
, , , , , , , , , , , , , , , , , , , .
- Initiation
- step 1: 1GlyVacant #1
- step 2: 1Gly Vacant #1 1Gly Vacant #2
- step 3: 1Gly Vacant #1 1Gly 2Ala #2
- step 4: 1Gly Vacant #1 1Gly 2Ala #2 1Gly Vacant #3
- step 5: 1Gly Vacant #1 1Gly 2Ala #2 1Gly Vacant #3 3Glu Vacant #4
- step 6: 1Gly Vacant #1 1Gly 2Ala #2 1Gly Vacant #3 3Glu Vacant #4 4Asp Vacant #5
- step 7: 1Gly Vacant #1 1Gly 2Ala #2 1Gly Vacant #3 3Glu Vacant #4 4Asp 5Val #5
- step 8: 1Gly 6Pro #1 1Gly 2Ala #2 1Gly Vacant #3 3Glu Vacant #4 4Asp 5Val #5
- step 9: 1Gly 6Pro #1 1Gly 2Ala #2 1Gly 7Ser #3 3Glu Vacant #4 4Asp 5Val #5
- step 10: 1Gly 6Pro #1 1Gly 2Ala #2 1Gly 7Ser #3 3Glu 8Leu #4 4Asp 5Val #5
- step 11: 1Gly 6Pro #1 1Gly 2Ala #2 1Gly 7Ser #3 3Glu 8Leu #4 4Asp 5Val #5 1Gly Vacant #6
- step 12: 1Gly 6Pro #1 1Gly 2Ala #2 1Gly 7Ser #3 3Glu 8Leu #4 4Asp 5Val #5 1Gly 9Thr #6
- Midway & ending
- step 13: (#1 ∼ #6 are fully filled by 1Gly to 9Thr, the same below for the following steps) 2Ala 10Arg #7
- and the following steps (omitting the previously fully filled #1 ∼ #(n-1) codon pairs in step #n, from #8 to #32):
- 7Ser 2Ala #8; 2Ala 11Cys #9; 10Arg 6Pro #10; 10Arg 6Pro #11; 12Trp 6Pro #12; 10Arg 7Ser #13; 10Arg 7Ser #14; stop 7Ser #15; 9Thr 10Arg #16; 7Ser 9Thr #17; 9Thr 11Cys #18; 5Val 13His #19; 14Gln 8Leu #20; 4Asp 15Ile #21; 16Met 13His #22; 3Glu 17Phe #23; 5Val 18Tyr #24; stop 8Leu #25; 19Asn 5Val #26; 20Lys 8Leu #27; 14Gln 8Leu #28; 15Ile 18Tyr #29; 19Asn 15Ile #30; stop 8Leu #31; 20Lys 17Phe #32.
2.2.2. Initiation
, , ,
, , .
2.2.3. Midway
2.2.4. The Ending
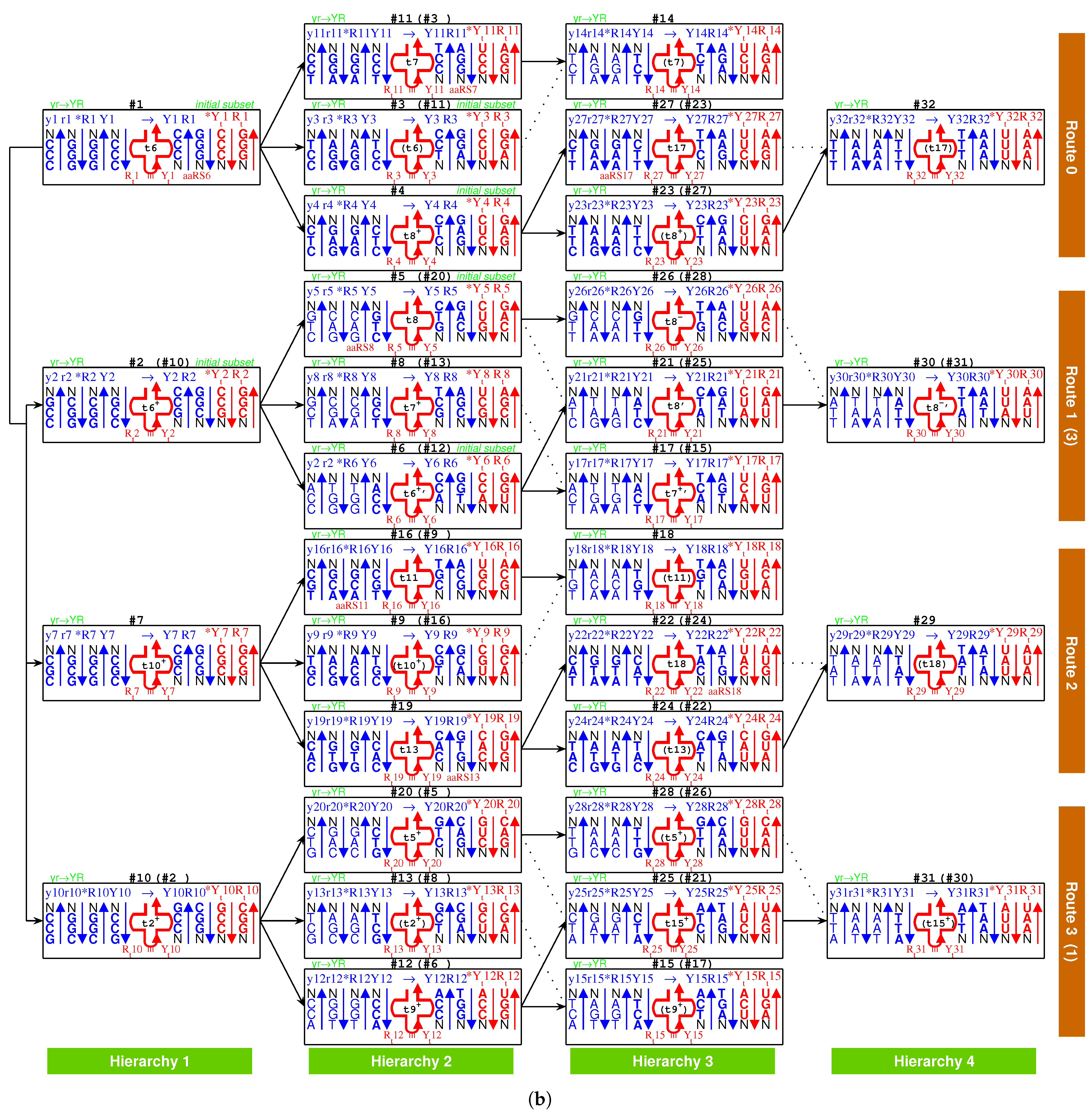
2.3. Origin of tRNA
2.3.1. Anti-Codon
2.3.2. Evolution of tRNA
| , , | ||
| , , | ||
| , | ||
| , , | ||
| , , | ||
| , , , | ||
| , , , , | ||
| , , | ||
| ,,,, | ||
| , | ||
| , | ||
| , |
2.3.3. Palindrome
2.4. Origin of aaRS
2.4.1. Para-Codon
2.4.2. Coevolution of tRNA with aaRS
2.5. Recruitment of Codons
2.5.1. Pair Connection
| 1Gly, aaRS1, t1→t1’: #1 R-Gly-#3 R | 2Ala, aaRS2, t2→t2’: #7 R-Ala-#9 R |
| 3Glu, aaRS3, t3→t3’: #4 R-Glu-#23 R | 4Asp, aaRS4, t4 wobbling: #5 R-Asp-#21 R |
| 5Val, aaRS5, t5→t5’: #19 R-Val-#24 R | 6Pro, aaRS6, t6 wobbling: #1 Y-Pro-#11 Y |
| 7Ser, aaRS7, t7 wobbling: #3 Y-Ser-#14 Y | 8Leu, aaRS8, t8→t8’: #20 Y-Leu-#25 Y |
| 9Thr, aaRS9, t9→t9’: #16 R-Thr-#18 R | 10Arg, aaRS10, t10→t10’: #10 R-Arg-#13 R |
| 11Cys, aaRS11, t11 wobbling: #9 Y-Cys-#18 Y | 12Trp, aaRS12, t12 wobbling: #12 R-Trp-#(15 R) |
| 13His, aaRS13, t13 wobbling: #19 Y-His-#22 Y | 14Gln, aaRS14, t14→t14’: #20 R-Gln-#28 R |
| 15Ile/16Met,aaRS15/16,t15/t16:#29R-Ile/Met-#22R | 17Phe, aaRS17, t17 wobbling: #23 Y-Phe-#32 Y |
| 18Tyr, aaRS18, t18 wobbling: #24 Y-Tyr-#29 Y | 19Asn, aaRS19, t19 wobbling: #26 R-Asn-#30 R |
| 20Lys, aaRS20, t20→t20’: #27 R-Lys-#32 R | stop, no aaRS, no tRNA: #25 R-stop-#31 R |
| 1Gly, aaRS1, wobbling: #2 R-Gly-#6 R | 2Ala, aaRS2, wobbling: #2 Y-Ala-#8 Y |
| 5Val, aaRS5, wobbling: #5 Y-Val-#26 Y | 6Pro, aaRS6, →: #10 Y-Pro-#12 Y |
| 7Ser, aaRS7, →: #13 Y-Ser-#15 Y | 8Leu, aaRS8, wobbling: #4 Y-Leu-#27 Y |
| 9Thr, aaRS9, wobbling: #6 Y-Thr-#17 Y | 10Arg, aaRS10, wobbling: #7 Y-Arg-#16 Y |
| 15Ile, aaRS15, wobbling: #21 Y-Ile-#30 Y |
| 7Ser, aaRS7, wobbling: #8 R-Ser-#17 R | 8Leu, aaRS8, → : #28 Y-Leu-#31 Y |
| 10Arg, aaRS10, → : #11 R-Arg-#14 R |
| 7Ser, aaRS7, → : #11 R-Ser-#14 R | stop, no aaRS, no tRNA: #11 R-Ser-#14 R |
| 9Thr, aaRS9, wobbling: #4 Y-Thr-#27 Y | 9Thr, aaRS9, →: #20 Y-Thr-#25 Y |
| 14Gln, aaRS14, → : #25 R-Gln-#31 R |
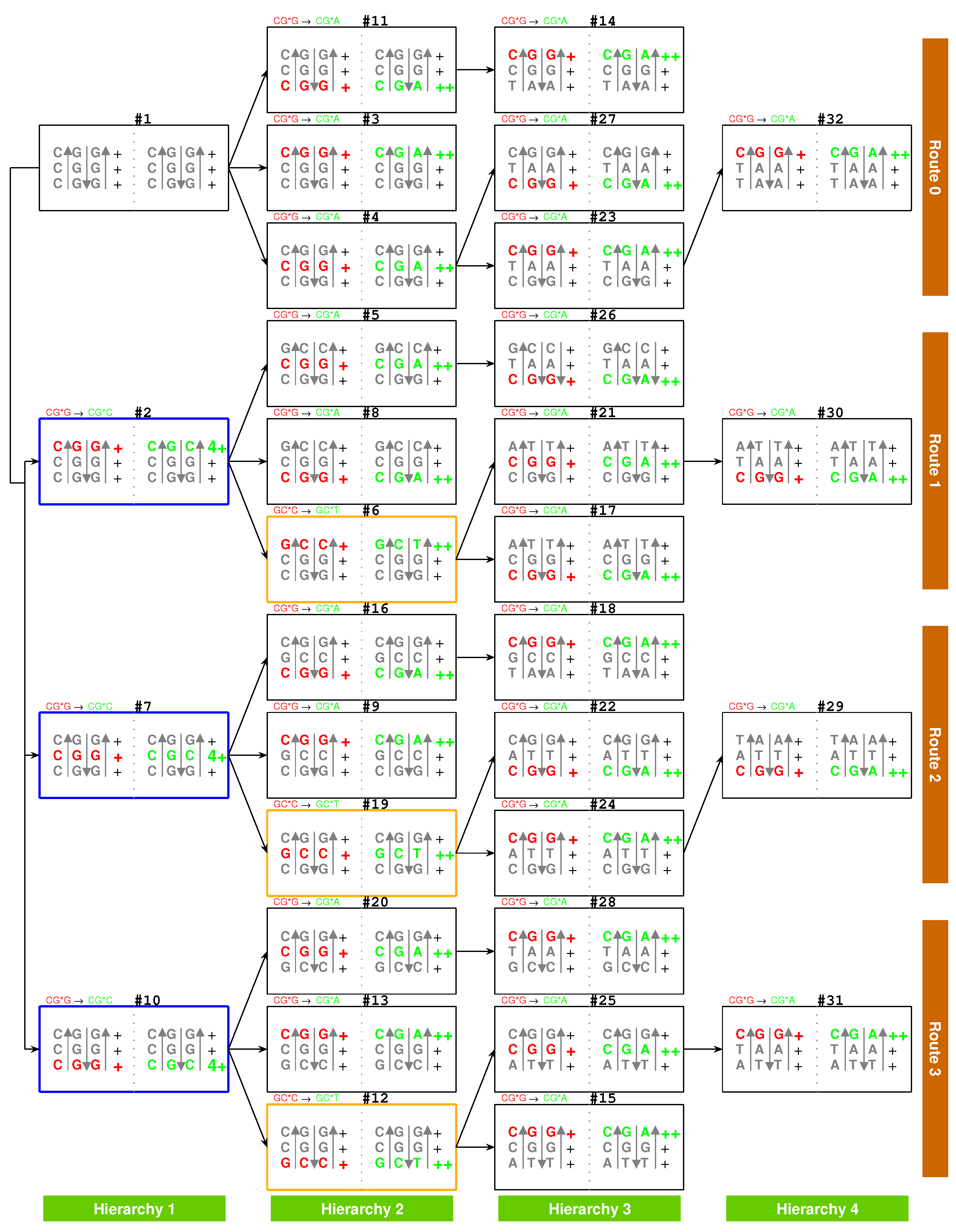
2.5.2. Route Duality
| 1Gly, aaRS1, t1 → | #1-Gly-#3 (Route 0) ∼ #2-Gly-#6 (Route 1) |
| 2Ala, aaRS2, t2 → | #7-Ala-#9 (Route 2) ∼ #2-Ala-#8 (Route 1) |
| 5Val, aaRS5, t5 → | #19-Val-#24 (Route 2) ∼ #5-Val-#26 (Route 1) |
| 6Pro, aaRS6, t6 → | #1-Pro-#11 (Route 0) ∼ #10-Pro-#12 (Route 3) |
| 7Ser, aaRS7, t7 → | #3-Ser-#14 (Route 0) ∼ #13-Ser-#15 (Route 3) |
| and t7 → | #3-Ser-#14 (Route 0) ∼ #8-Ser-#17 (Route 1) |
| 8Leu, aaRS8, t8 → | #20-Leu-#25 (Route 3) ∼ #4-Leu-#27 (Route 0) |
| and t8 → | #20-Leu-#25 (Route 3) ∼ #28-Leu-#31 (Route 3) |
| 9Thr, aaRS9, t9 → | #16-Thr-#18 (Route 2) ∼ #6-Thr-#17 (Route 1) |
| 10Arg, aaRS10, t10 → | #10-Arg-#13 (Route 3) ∼ #7-Arg-#16 (Route 2) |
| and t10 → | #10-Arg-#13 (Route 3) ∼ #11-Arg-#14 (Route 0) |
| 3Glu/4Asp, /, aaRS3 → aaRS4 | #4-Glu-#23 (Route 0) ∼#5-Asp-#21 (Route 1) |
| 7Ser/10Arg, /, aaRS7 / aaRS10 | #8-Ser-#17 (Route 1) ∼ #11-Arg-#14 (Route 0) |
| 11Cys/12Trp, /, aaRS11 → aaRS12 | #9-Cys-#18 (Route 2) ∼#12-Trp-(#15) (Route 3) |
| 13His/14Gln, /, aaRS13 → aaRS14 | #19-His-#22 (Route 2) ∼#20-Gln-#28 (Route 3) |
| 15Ile/16Met,,/,aaRS15→aaRS16 | #29-Ile/Met-#22 (Route 2) ∼ #21-Ile-#30 (Route 1) |
| 8Leu/17Phe, /, aaRS8 → aaRS17 | #28-Leu-#31 (Route 3) ∼ #23-Phe-#32 (Route 0) |
| 18Tyr/stop, t18, aaRS18 | #24-Tyr-#29 (Route 2) ∼#25-stop-#31 (Route 3) |
| 19Asn/20Lys, /, aaRS19 → aaRS20 | #26-Asn-#30 (Route 1) ∼#27-Lys-#32 (Route 0) |
| 7Ser, aaRS7, → | #8-Ser-#17 (Route 1) ∼ #11-(Ser)-#14 (Route 0) |
| 9Thr, aaRS9, → | #4-(Thr)-#27 (Route 0) ∼ #20-(Thr)-#25 (Route 3) |
| stop | #11-(stop)-#14 (Route 0) ∼ #15-stop-#31 (Route 3) |
2.6. Codon Degeneracy
3. Results
3.1. Driving Force in the Prebiotic Sequence Evolution
3.2. Explanation of Two Classes of aaRSs According to Coevolution of tRNAs with aaRSs
3.3. Explanation of the Codon Degeneracy on the Genetic Code Chart
4. Conclusions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Wong, J.T. A coevolution theory of the genetic code. Proc. Natl. Acad. Sci. USA 1975, 72, 1909–1912. [Google Scholar] [CrossRef] [Green Version]
- Wong, J.T.; Lazcano, A. Prebiotic Evolution and Astrobiology; Landes Bioscience: Austin, TX, USA, 2009. [Google Scholar]
- De Pouplana, L.R. (Ed.) The Genetic Code and the Origin of Life; Kluwer Academic: New York, NY, USA, 2004. [Google Scholar]
- De Pouplana, L.R.; Schimmel, P. Aminoacyl-tRNA synthetases: Potential markers of genetic code development. Trends Biochem. Sci. 2001, 26, 591–596. [Google Scholar] [CrossRef]
- Woese, C.R.; Kandler, O.; Wheelis, M.L. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 1990, 87, 4576–4579. [Google Scholar] [CrossRef] [Green Version]
- Gibson, D.G.; Glass, J.I.; Lartigue, C.; Noskov, V.N.; Chuang, R.Y.; Algire, M.A.; Benders, G.A.; Montague, M.G.; Ma, L.; Moodie, M.M.; et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 2010, 329, 52–56. [Google Scholar] [CrossRef] [Green Version]
- Wong, J.T. Coevolution theory of the genetic code at age thirty. BioEssays 2005, 27, 416–425. [Google Scholar] [CrossRef]
- Trifonov, E.N.; Gabdank, I.; Barash, D.; Sobolevsky, Y. Primordia vita. deconvolution from modern sequences. Orig. Life Evol. Biosph. 2006, 36, 559–565. [Google Scholar] [CrossRef]
- Reznick, J.S. Embracing the future as stewards of the past: Charting a course forward for historical medical libraries and archives. RBM 2014, 15, 111–123. [Google Scholar] [CrossRef] [Green Version]
- Soyfer, V.N.; Potaman, V.N. Triple-Helical Nucleic Acids; Springer: New York, NY, USA, 1996. [Google Scholar]
- Belotserkovskii, B.P.; Veselkov, A.G.; Filippov, S.A.; Dobrynin, V.N.; Mirkin, S.M.; Frank-Kamenetskii, M.D. Formation of intramolecular triplex in homopurine-homopyrimidine mirror repeats with point substitutions. Nucleic Acids Res. 1990, 18, 6621–6624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sklenář, V.; Felgon, J. Formation of a stable triplex from a single DNA strand. Nature 1990, 345, 836–838. [Google Scholar] [CrossRef] [PubMed]
- Frank-Kamenetskii, M.D. Triplex DNA structrutures. Annu. Rev. Biochem. 1995, 64, 65–95. [Google Scholar] [CrossRef] [PubMed]
- Robertus, J.D.; Ladner, J.E.; Finch, J.T.; Rhodes, D.; Brown, R.S.; Clark, B.F.C.; Klug, A. Structure of yeast phenylalanine tRNA at 3Å resolution. Nature 1974, 250, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Oro, J. Mechanism of synthesis of adenine from hydrogen cyanide under possible primitive Earth conditions. Nature 1961, 191, 1193–1194. [Google Scholar] [CrossRef] [PubMed]
- Orgel, L.E. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 2006, 39, 99–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyakawa, S.; Murasawa, K.; Kobayashi, K.; Sawaoka, A.B. Abiotic synthesis of guanine with high temperature plasma. Orig. Life Evol. Biosph. 2000, 30, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Ferris, J.P.; Sanchez, R.A.; Orgel, L.E. Studies in prebiotic synthesis. 3. Synthesis of pyrimidines from cyanoacetylene and cyanate. J. Mol. Biol. 1968, 33, 693–704. [Google Scholar] [CrossRef]
- Sanchez, R.; Ferris, J.P.; Orgel, L.E. Cyanoacetylene in prebiotic synthesis. Science 1966, 154, 784–785. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tsanakopoulou, M.; Magnani, C.J.; Szabla, R.; Šponer, J.E.; Šponer, J.; Góra, R.W.; Sutherland, J.D. A prebiotically plausible synthesis of pyrimidine β-ribonucleosides and their phosphate derivatives involving photoanomerization. Nat. Chem. 2017, 9, 303–309. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Prywes, N.; Tam, C.P.; O’flaherty, D.K.; Lelyveld, V.S.; Izgu, E.C.; Pal, A.; Szostak, J.W. Enhanced nonenzymatic RNA copying with 2-aminoimidazole activated nucleotides. J. Am. Chem. Soc. 2017, 139, 1810–1813. [Google Scholar] [CrossRef]
- Becker, S.; Feldmann, J.; Wiedemann, S.; Okamura, H.; Schneider, C.; Iwan, K.; Crisp, A.; Rossa, M.; Amatov, T.; Carell, T. Unified prebiotically plausible synthesis of pyrimidine and purine RNA ribonucleotides. Science 2019, 366, 76–82. [Google Scholar] [CrossRef] [Green Version]
- Powner, M.W.; Zheng, S.; Szostak, J.W. Multicomponent assembly of proposed DNA precursors in water. J. Am. Chem. Soc. 2012, 134, 13889–13895. [Google Scholar] [CrossRef]
- Trevinoa, S.G.; Zhanga, N.; Elenkoa, M.P.; Luptákb, A.; Szostak, J.W. Evolution of functional nucleic acids in the presence of nonheritable backbone heterogeneity. Proc. Natl. Acad. Sci. USA 2011, 108, 13492–13497. [Google Scholar] [CrossRef] [Green Version]
- Bhowmik, S.; Krishnamurthy, R. The role of sugar-backbone heterogeneity and chimeras in the simultaneous emergence of RNA and DNA. Nat. Chem. 2019, 11, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Green, N.J.; Gibard, C.; Krishnamurthy, R.; Sutherl, J.D. Prebiotic phosphorylation of 2-thiouridine provides either nucleotides or DNA building blocks via photoreduction. Nat. Chem. 2019, 11, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chmela, V.; Green, N.J.; Russell, D.A.; Janicki, M.J.; Góra, R.W.; Szabla, R.; Bond, A.D.; Sutherland, J.D. Selective prebiotic formation of RNA pyrimidine and DNA purine nucleosides. Nature 2020, 582, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.T.; Ng, S.; Mat, W.; Hu, T.; Xue, H. Coevolution theory of the genetic code at age forty: Pathway to translation and synthetic life. Life 2016, 6, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nirenberg, M.W.; Matthaei, J.H. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc. Natl. Acad. Sci. USA 1961, 47, 1588–1602. [Google Scholar] [CrossRef] [Green Version]
- Crick, F.H.C. Codon-anticodon pairing: The wobble hypothesis. J. Mol. Biol. 1966, 19, 548–555. [Google Scholar] [CrossRef]
- Eriani, G.; Delarue, M.; Poch, O.; Gangloff, J.; Moras, D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 1990, 347, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.L.; Urey, H.C. Organic compound synthes on the primitive earth. Science 1959, 130, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.H.; Macko, S.A. Isotopic evidence for extraterrestrial non-racemic amino acids in the Murchison meteorite. Nature 1997, 389, 265–268. [Google Scholar] [CrossRef]
- Wong, J.T. Coevolution of the genetic code and amino acid biosynthesis. Trends Biochem. Sci. 1981, 6, 33–36. [Google Scholar] [CrossRef]
- Kobayashi, K.; Kaneko, T.; Saito, T.; Oshima, T. Amino acid formation in gas mixtures by high energy particle irradiation. Orig. Life Evol. Biosph. 1998, 28, 155–165. [Google Scholar] [CrossRef]
- Li, D.J.; Zhang, S. Genetic code evolution as an initial driving force for molecular evolution. Phys. A 2009, 388, 3809–3825. [Google Scholar] [CrossRef] [Green Version]
- Muto, A.; Osawa, S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc. Natl. Acad. Sci. USA 1987, 84, 166–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woese, C.R.; Dugre, D.H.; Dugre, S.A.; Kondo, M.; Saxinger, W.C. On the fundamental nature and evolution of the genetic code. Cold Spring Harbour. Symp. Quant. Biol. 1966, 31, 723–736. [Google Scholar] [CrossRef]
- Crick, F.H.C. The origin of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Yarus, M. A specific amino acid binding site composed of RNA. Science 1988, 240, 1751–1758. [Google Scholar] [CrossRef]
- Di Giulio, M. The Extension Reached by the Minimization of the Polarity Distances during the Evolution of the Genetic Code. J. Mol. Evol. 1989, 29, 288–293. [Google Scholar] [CrossRef]
- Di Giulio, M. Some Aspects of the Organization and Evolution of the Genetic Code. J. Mol. Evol. 1989, 29, 191–201. [Google Scholar] [CrossRef]
- Osawa, S.; Jukes, T.H. Codon Reassignment (Codon Capture) in Evolution. J. Mol. Evol. 1989, 28, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Root-Bernstein, R. Simultaneous origin of homochirality, the genetic code and its directionality. Bioessays 2007, 29, 689–698. [Google Scholar] [CrossRef]
- Rodin, A.S.; Szathmáry, E.; Rodin, S.N. One ancestor for two codes viewed from the perspective of two complementary modes of tRNA aminoacylation. Biol. Direct 2009, 4, 4. [Google Scholar] [CrossRef] [Green Version]
- Knight, R.D.; Freel, S.J.; Landweber, L.F. Rewiring the keyboard: Evolvability of the genetic code. Nat. Rev. Genet. 2001, 2, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Higgs, P.G. Pathways of Genetic Code Evolution in Ancient and Modern Organisms. J. Mol. Evol. 2015, 80, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Yang, X.; Higgs, P.G. The Mechanisms of Codon Reassignments in Mitochondrial Genetic Codes. J. Mol. Evol. 2007, 64, 662–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escudé, C.; Francçois, J.C.; Sun, J.S.; Ott, G.; Sprinzl, M.; Garestier, T.; Heélene, J.C. Stability of triple helices containing RNA and DNA strands: Experimental and molecular modeling studies. Nucleic Acids Res. 1993, 21, 5547–5553. [Google Scholar] [CrossRef]
- Han, H.; Dervan, P.B. Sequence-specific recognition of double helical RNA and RNA·DNA by triple helix formation. Proc. Natl. Acad. Sci. USA 1993, 90, 3806–3810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Kool, E.T. Relative stabilities of triple helices composed of combinations of DNA, RNA and 2’-O-methyl-RNA backbones: Chimeric circular oligonucleotides as probes. Nucleic Acids Res. 1995, 23, 1157–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altwegg, K.; Balsiger, H.; Bar-Nun, A.; Berthelier, J.J.; Bieler, A.; Bochsler, P.; Briois, C.; Calmonte, U.; Combi, M.R.; Cottin, H.; et al. Prebiotic chemicals–amino acid and phosphorus–in the coma of comet 67P/Churyumov-Gerasimenko. Sci. Adv. 2016, 2, e1600285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.J. Concurrent origins of the genetic code and the homochirality of life, and the origin and evolution of biodiversity Part I: Observations and explanations. bioRxiv 2015. [Google Scholar] [CrossRef] [Green Version]
- Roberts, R.W.; Crothers, D.M. Stability and properties of double and triple helices: Dramatic effects of RNA or DNA backbone composition. Science 1992, 258, 1463–1466. [Google Scholar] [CrossRef]
- Di Giulio, M. On the origin of the transfer RNA molecule. J. Theor. Biol. 1992, 159, 199–214. [Google Scholar] [CrossRef]
- Di Giulio, M. Was it an ancient gene codifying for a hairpin RNA that, by means of direct duplication, gave rise to the primitive tRNA molecule? J. Theor. Biol. 1995, 177, 95–101. [Google Scholar] [CrossRef]
- Di Giulio, M. The nonmonophyletic origin of tRNA molecule. J. Theor. Biol. 1999, 197, 403–414. [Google Scholar] [CrossRef]
- Di Giulio, M. The origin of the tRNA molecule: Implications for the origin of protein synthesis. J. Theor. Biol. 2004, 226, 89–93. [Google Scholar] [CrossRef]
- Di Giulio, M. Nanoarchaeum equitans is a living fossil. J. Theor. Biol. 2006, 242, 257–260. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, B.J.; Holl, J.J. Denatured DNA as a Direct Template for in vitro Protein Synthesis. Proc. Natl. Acad. Sci. USA 1965, 54, 880–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uzawa, T.; Yamagishi, A.; Oshima, T. Polypeptide Synthesis Directed by DNA as a Messenger in Cell-Free Polypeptide Synthesis by Extreme Thermophiles, Thermus thermophilus HB27 and Sulfolobus tokodaii Strain 7. J. Biochem. 2002, 131, 849–853. [Google Scholar] [CrossRef]
- Schimmel, P. Development of tRNA synthetases and connection to genetic code and disease. Protein Sci. 2008, 17, 1643–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trifonov, E.N. Consensus temporal order of amino acids and evolution of the triplet code. Gene 2000, 261, 139–151. [Google Scholar] [CrossRef]
- Trifonov, E.N. The triplet code from first principles. J. Biomol. Struct. Dyn. 2004, 22, 1. [Google Scholar] [CrossRef]
- Trifonov, E.N.; Kirzhner, A.; Kirzhner, V.M.; Berezovsky, I.N. Distinc stage of protein evolution as suggested by protein sequence analysis. J. Mol. Evol. 2001, 53, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Widmann, J.; Di Giulio, M.; Yarus, M.; Knight, R. tRNA creation by hairpin duplication. J. Mol. Evol. 2005, 61, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Rodin, S.N.; SOhno, S. Two types of aminoacyl-trna synthetases could be originally encoded by complementary strands of the same nucleic acid. Orig. Life Evol. Biosph. 1995, 25, 565–589. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Rodriguez, L.; Erdogan, O.; Jimenez-Rodriguez, M.; Gonzalez-Rivera, K.; Williams, T.; Li, L.; Weinreb, V.; Collier, M.; Chandrasekaran, S.N.; Ambroggio, X.; et al. Functional class I and II amino acid-activating enzymes can be coded by opposite strands of the same gene. J. Biol. Chem. 2015, 290, 19710–19725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gestel, R.F. (Ed.) The RNA World, 3rd ed.; Cold Spring Harbor Laboratory: New York, NY, USA, 2006. [Google Scholar]



| Stability | CG*N | GC*N | TA*N | AT*N |
|---|---|---|---|---|
| (−) | GC*A | AT*C AT*A | ||
| (+) | CG*G | GC*C GC*G | TA*C TA*G TA*A | AT*T |
| (++) | CG*A CG*T | GC*T | ||
| (3+) | AT*G | |||
| (4+) | CG*C | TA*T | ||
| (+)CG*G → (++)CG*A increase in stability | (+)GC*C → (−)GC*A unstable | (+)TA*A → (+)TA*G no increase in stability | (+)AT*T → (3+)AT*G | |
| (+)CG*G → (4+)CG*C increase in stability | (+)GC*C → (+)GC*G no increase in stability | (+)TA*A → (4+)TA*T | (+)AT*T → (−)AT*A unstable | |
| (+)GC*C → (++)GC*T increase in stability | (+)CG*G → (++)CG*T | (+)AT*T → (+)AT*C no increase in stability | (+)TA*A → (+)TA*C no increase in stability | |
| POSSIBLE (Roadmap) | Impossible | Impossible | Impossible | |
| (+)CG*G → (++)CG*T | (+)GC*C → (++)GC*T | (+)TA*A → (+)TA*C no increase in stability | (+)AT*T → (−)AT*C unstable | |
| (+)CG*G → (4+)CG*C | (+)GC*C → (+)GC*G no increase in stability | (+)TA*A → (4+)TA*T | (+)AT*T → (−)AT*A unstable | |
| (+)GC*C → (−)GC*A unstable | (+)CG*G → (++)CG*A | (+)AT*T → (3+)AT*G | (+)TA*A → (+)TA*G no increase in stability | |
| Impossible | Impossible | Impossible | Impossible |
| Hierarchy 1 to Hierarchy 2 | Hierarchy 2 to Hierarchy 3 | Hierarchy 3 to Hierarchy 4 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Route 0 | 1Gly | 6Pro | 3Glu | 7Ser | 8Leu | 10Arg | 17Phe | 20Lys | ||||||||
| Route 1 | 1Gly | 2Ala | 4Asp | 5Val | 9Thr | 7Ser | 15Ile | 19Asn | ||||||||
| Route 2 | 2Ala | 10Arg | 5Val | 9Thr | 11Cys | 13His | 16Met | 18Tyr | ||||||||
| Route 3 | 6Pro | 10Arg | 7Ser | 8Leu | 12Trp | 14Gln | 8Leu | stop | ||||||||
| Codon box | GGN | GCN | CCN | CGN | GAN | GUN | UCN | CUN | ACN | AGN | UGN | CAN | AUN | UUN | UAN | AAN |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, D.J. Formation of the Codon Degeneracy during Interdependent Development between Metabolism and Replication. Genes 2021, 12, 2023. https://doi.org/10.3390/genes12122023
Li DJ. Formation of the Codon Degeneracy during Interdependent Development between Metabolism and Replication. Genes. 2021; 12(12):2023. https://doi.org/10.3390/genes12122023
Chicago/Turabian StyleLi, Dirson Jian. 2021. "Formation of the Codon Degeneracy during Interdependent Development between Metabolism and Replication" Genes 12, no. 12: 2023. https://doi.org/10.3390/genes12122023
APA StyleLi, D. J. (2021). Formation of the Codon Degeneracy during Interdependent Development between Metabolism and Replication. Genes, 12(12), 2023. https://doi.org/10.3390/genes12122023