Age-Related Changes of Gene Expression Profiles in Drosophila
Abstract
:1. Introduction
2. Results
2.1. Comparison of the Expression Profile between Young and Old Age Groups
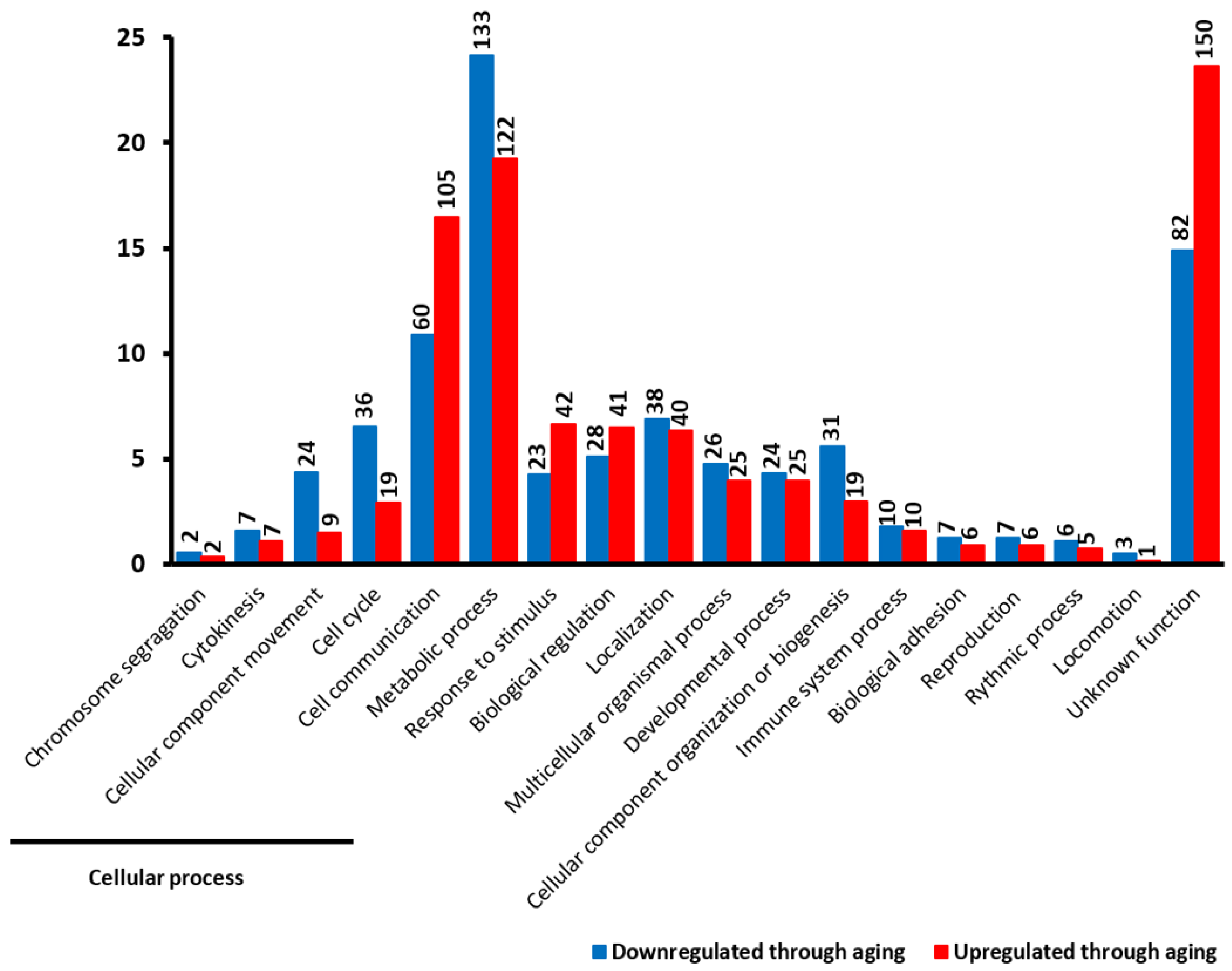
2.2. Cellular Functions of DEGs
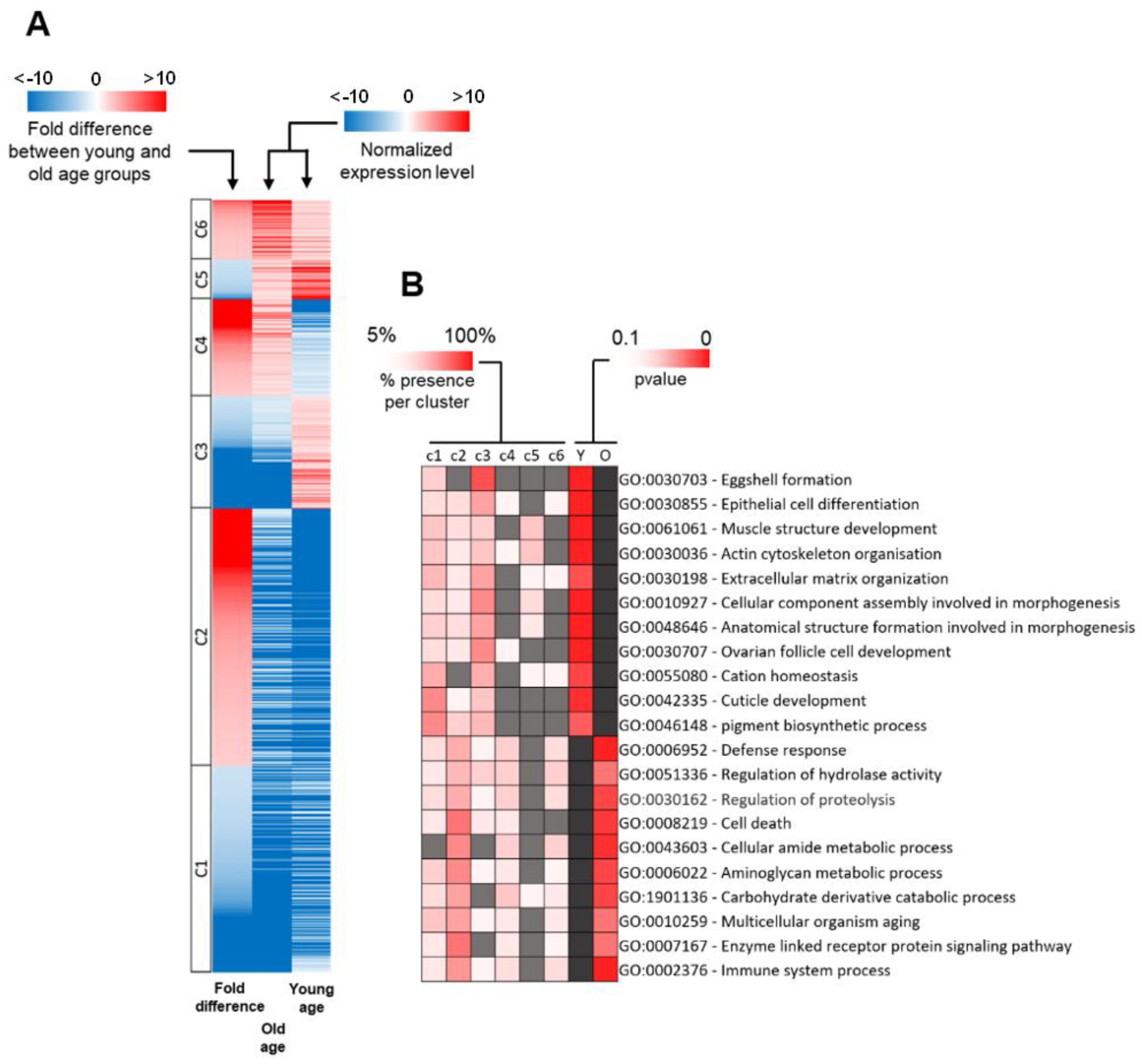
2.3. Distribution of DEG Types between Young and Old Age Groups
2.4. Functional Classification of DEGs
2.5. Overrepresentation of Genes Important for Flight Muscles and Sarcomere
2.6. Genes Regulating Programmed Cell Death Are Overrepresented among DEGs
2.7. Members of Cytochrome Family Are Overrepresented among DEGs
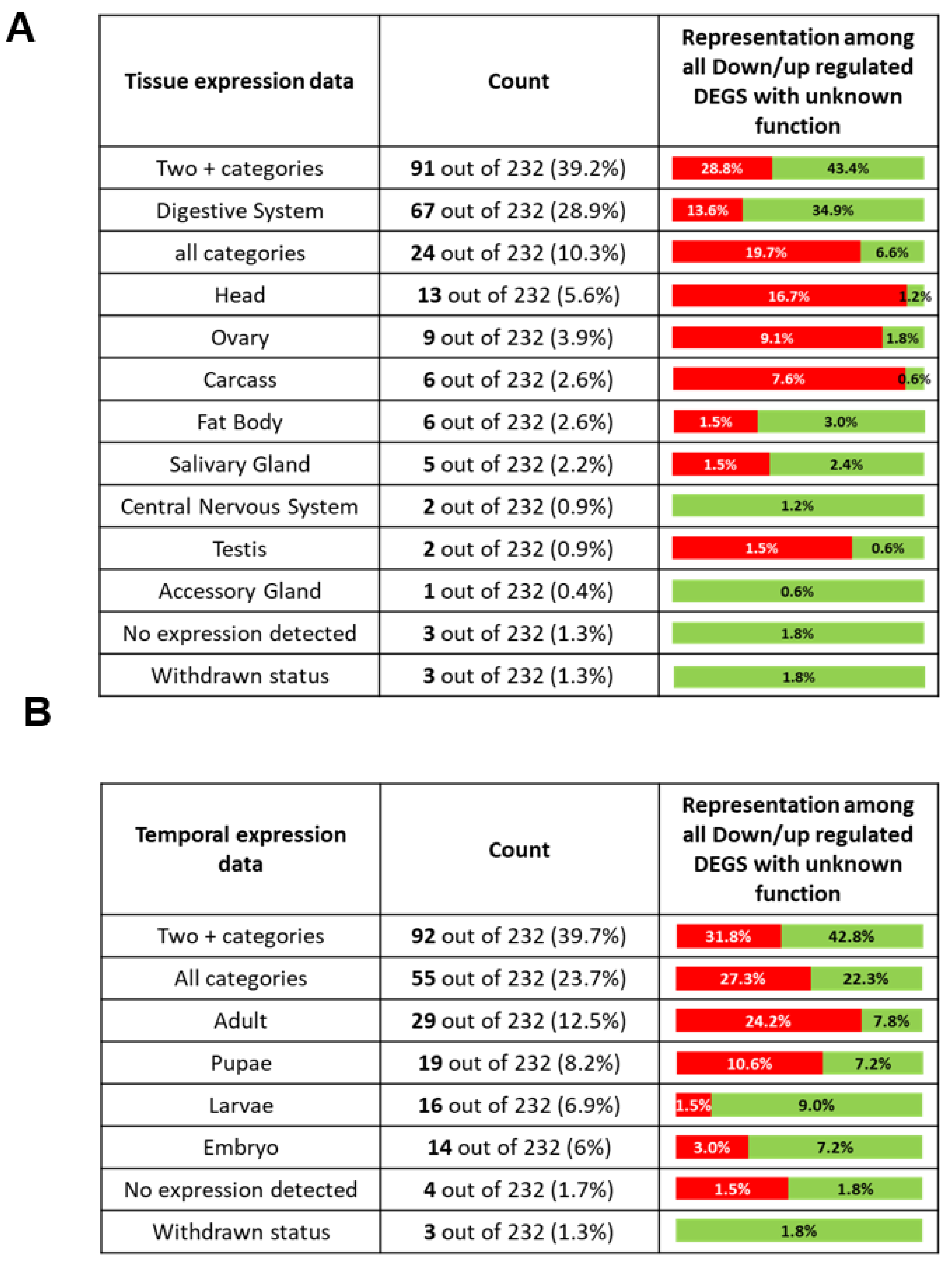
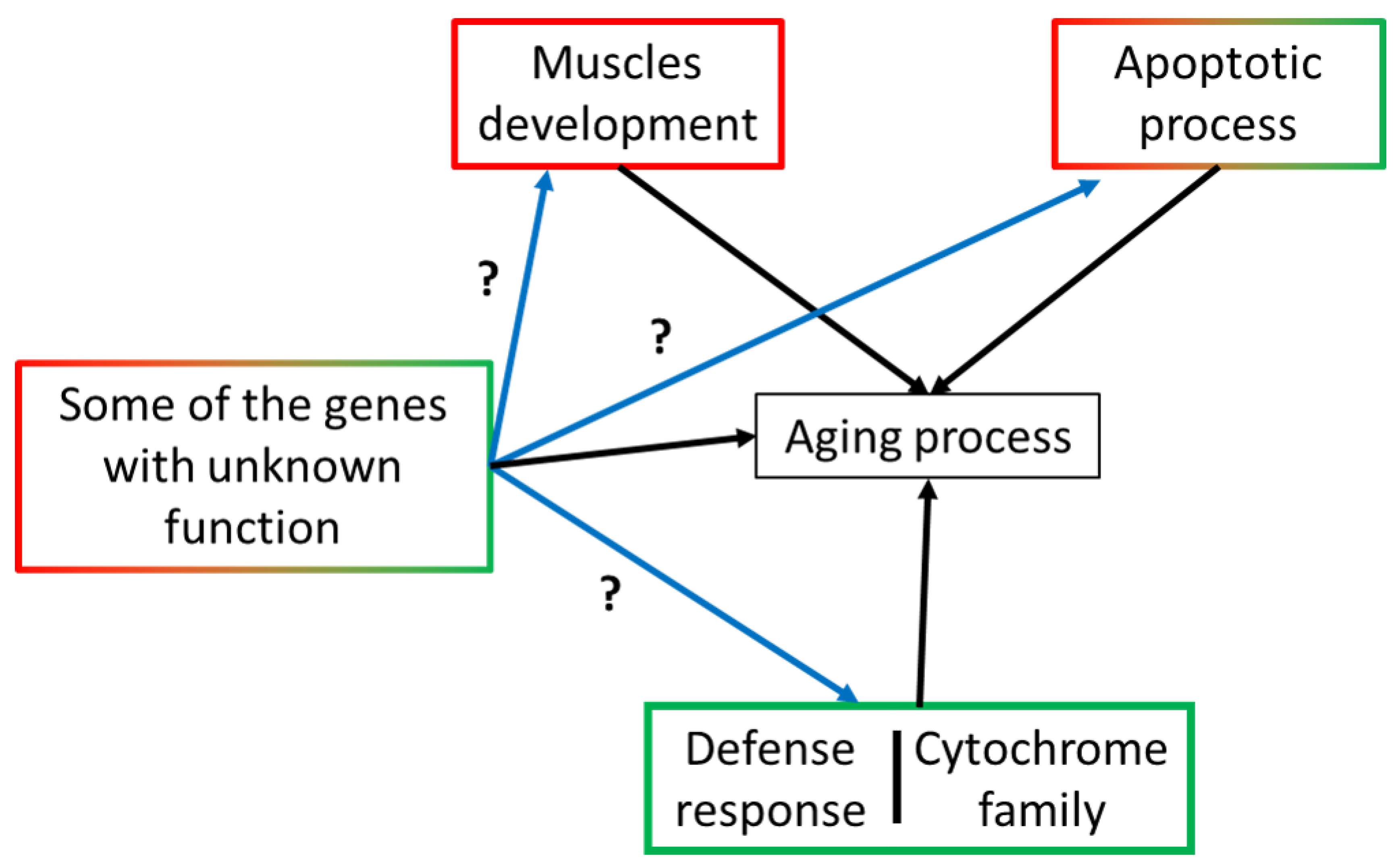
2.8. DEGs with Unknown Function
3. Discussion
4. Materials and Methods
4.1. Drosophila Strains and Genetics and Construction of Transgenic Drosophila
4.2. RNA Isolation and Microarray
4.3. Microarray Data Analysis
4.4. DEG Cellular Functions and GO-Terms Overrepresentation Analysis
4.5. Lifespan Experiments
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- DePinho, R.A. The age of cancer. Nature 2000, 408, 248–254. [Google Scholar] [CrossRef]
- Campisi, J. Cancer and Ageing: Rival Demons? Nature 2003, 3, 339–349. [Google Scholar] [CrossRef]
- Lakatta, E.G.; Levy, D. Arterial and Cardian Aging: Major Shareholders in Cardiovascular Disease Enterprises, Part II The Aging Heart in Health: Links to Heart Disease. Circulation 2003, 107, 346–354. [Google Scholar] [CrossRef] [Green Version]
- Allikmets, R.; Shroyer, N.F.; Singh, N.; Seddon, J.M.; Lewis, R.A.; Bernstein, P.; Peiffer, A.; Zabriskie, N.; Li, Y.; Hutchinson, A.; et al. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science 1997, 277, 1805–1807. [Google Scholar] [CrossRef] [Green Version]
- Sato, N.; Hori, O.; Yamaguchi, A.; Lambert, J.C.; Chartier-Harlin, M.C.; Robinson, P.A.; Delacourte, A.; Schmidt, A.M.; Furuyama, T.; Imaizumi, K.; et al. A Novel Presenilin-2 Splice Variant in Human Alzheimer’s Disease Brain Tissue. J. Neurochem. 1999, 72, 2498–2505. [Google Scholar] [CrossRef]
- Pietschmann, P.; Rauner, M.; Sipos, W.; Kerschan-Schindl, K. Osteoporosis: An Age Related and Gender Specific Disease—A Mini-Review. Gerontology 2008, 55, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, R.P.; Porter Abate, J.; Dilks, K.; Landis, J.; Ashraf, J.; Murphy, C.T.; Blackwell, T.K. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 2009, 8, 524–541. [Google Scholar] [CrossRef] [Green Version]
- Hazane-Puch, F.; Bonnet, M.; Valenti, K.; Schnebert, S.; Kurfurst, R.; Favier, A.; Sauvaigo, S. Study of fibroblast gene expression in response to oxidative stress induced by hydrogen peroxide or UVA with skin aging. Eur. J. Derm. 2010, 20, 308–320. [Google Scholar] [CrossRef]
- Kim, K.; Lin, Y.R.; Park, Y. Enhancement of stress resistances and downregulation of Imd pathway by lower developmental temperature in Drosophila melanogaster. Exp. Gerontol. 2010, 45, 984–987. [Google Scholar] [CrossRef]
- Linford, N.; Bilgir, C.; Ro, J.; Pletcher, S.D. Measurement of lifespan in Drosophila melanogaster. J. Vis. Exp. 2013, 71, e50068. [Google Scholar]
- Mi, H.; Dong, Q.; Muruganujan, A.; Gaudet, P.; Lewis, S.; Thomas, P.D. PANTHER version 7: Improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010, 38, D204–D210. [Google Scholar] [CrossRef] [Green Version]
- Oyelade, J.; Isewon, I.; Oladipupo, F.; Aromolaran, O.; Uwoghiren, E.; Ameh, F.; Achas, M.; Adebiyi, E. Clustering Algorithms: Their Application to Gene Expression Data. Bioinform. Biol. Insights 2016, 10, 237–253. [Google Scholar] [CrossRef]
- Carlson, K.A.; Gardner, K.; Pashaj, A.; Carlson, D.J.; Yu, F.; Eudy, J.D.; Zhang, C.; Harshman, L.G. Genome-Wide Gene Expression in relation to Age in Large Laboratory Cohorts of Drosophila melanogaster. Genet. Res. Int. 2015, 2015, 835624. [Google Scholar] [CrossRef] [Green Version]
- Pletcher, S.D.; Macdonald, S.J.; Marguerie, R.; Certa, U.; Stearns, S.C.; Goldstein, D.B.; Partridge, L. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr. Biol. 2002, 12, 712–723. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; Mackay, T.; Anholt, R.R. Transcriptional and epigenetic responses to mating and aging in Drosophila melanogaster. BMC Genom. 2014, 15, 927. [Google Scholar] [CrossRef] [Green Version]
- Jasper, H. Exploring the physiology and pathology of aging in the intestine of Drosophila melanogaster. Invertebr. Reprod. Dev. 2015, 59, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Zhan, M.; Yamaza, H.; Sun, Y.; Sinclair, J.; Li, H.; Zou, S. Temporal and spatial transcriptional profiles of aging in Drosophila melanogaster. Genome. Res. 2007, 17, 1236–1243. [Google Scholar] [CrossRef] [Green Version]
- Demontis, F.; Piccirillo, R.; Goldberg, A.L.; Perrimon, N. Mechanisms of skeletal muscle aging: Insights from Drosophila and mammalian models. Dis. Model. Mech. 2013, 6, 1339–1352. [Google Scholar] [CrossRef] [Green Version]
- Highfill, C.A.; Reeves, G.A.; Macdonald, S.J. Genetic analysis of variation in lifespan using a multiparental advanced intercross Drosophila mapping population. BMC Genet. 2016, 17, 113. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Edelman, S.W.; Tharmarajah, G.; Walker, D.W.; Pletcher, S.D.; Seroude, L. Differential patterns of apoptosis in response to aging in Drosophila. Proc. Natl. Acad. Sci. USA 2005, 102, 1083–1088. [Google Scholar] [CrossRef] [Green Version]
- Schonbauer, C.; Distler, J.; Jahrling, N.; Radolf, M.; Dodt, H.U.; Frasch, M.; Schnorrer, F. Spalt mediates an evolutionarily conserved switch to fibrillar muscle fate in insects. Nature 2011, 479, 406–409. [Google Scholar] [CrossRef]
- Liu, Y.H.; Jakobsen, J.S.; Valentin, G.; Amarantos, I.; Gilmour, D.T.; Furlong, E.E. A systematic analysis of Tinman function reveals Eya and JAK-STAT signaling as essential regulators of muscle development. Dev. Cell 2009, 16, 280–291. [Google Scholar] [CrossRef] [Green Version]
- Courchesne-Smith, C.L.; Tobin, S.L. Tissue-specific expression of the 79B actin gene during Drosophila development. Dev. Biol. 1989, 133, 313–321. [Google Scholar] [CrossRef]
- Fyrberg, E.A.; Mahaffey, J.W.; Bond, B.J.; Davidson, N. Transcripts of the six Drosophila actin genes accumulate in a stage- and tissue-specific manner. Cell 1983, 33, 115–123. [Google Scholar] [CrossRef]
- Clark, K.A.; Lesage-Horton, H.; Zhao, C.; Beckerle, M.C.; Swank, D.M. Deletion of Drosophila muscle LIM protein decreases flight muscle stiffness and power generation. Am. J. Physiol. Cell Physiol. 2011, 301, C373–C382. [Google Scholar] [CrossRef] [Green Version]
- Herranz, R.; Diaz-Castillo, C.; Nguyen, T.P.; Lovato, T.L.; Cripps, R.M.; Marco, R. Expression patterns of the whole troponin C gene repertoire during Drosophila development. Gene Expr. Patterns 2004, 4, 183–190. [Google Scholar] [CrossRef]
- Chakravorty, S.; Tanner, B.C.W.; Foelber, V.L.; Vu, H.; Rosenthal, M.; Ruiz, T.; Vigoreaux, J.O. Flightin maintains myofilament lattice organization required for optimal flight power and courtship song quality in Drosophila. Proc. Biol. Sci. 2017, 284, 20170431. [Google Scholar] [CrossRef] [Green Version]
- Spletter, M.L.; Barz, C.; Yeroslaviz, A.; Schonbauer, C.; Ferreira, I.R.; Sarov, M.; Gerlach, D.; Stark, A.; Habermann, B.H.; Schnorrer, F. The RNA-binding protein Arrest (Bruno) regulates alternative splicing to enable myofibril maturation in Drosophila flight muscle. EMBO Rep. 2015, 16, 178–191. [Google Scholar] [CrossRef]
- Katzemich, A.; West, R.J.; Fukuzawa, A.; Sweeney, S.T.; Gautel, M.; Sparrow, J.; Bullard, B. Binding partners of the kinase domains in Drosophila obscurin and their effect on the structure of the flight muscle. J. Cell Sci. 2015, 128, 3386–3397. [Google Scholar]
- Lee, G.; Wang, Z.; Sehgal, R.; Chen, C.H.; Kikuno, K.; Hay, B.; Park, J.H. Drosophila caspases involved in developmentally regulated programmed cell death of peptidergic neurons during early metamorphosis. J. Comp. Neurol. 2011, 519, 34–48. [Google Scholar] [CrossRef]
- Brown, J.B.; Boley, N.; Eisman, R.; May, G.E.; Stoiber, M.H.; Duff, M.O.; Booth, B.W.; Wen, J.; Park, S.; Suzuki, A.M.; et al. Diversity and dynamics of the Drosophila transcriptome. Nature 2014, 512, 393–399. [Google Scholar] [CrossRef] [Green Version]
- Merino, M.M.; Rhiner, C.; Portela, M.; Moreno, E. “Fitness fingerprints” mediate physiological culling of unwanted neurons in Drosophila. Curr. Biol. 2013, 23, 1300–1309. [Google Scholar] [CrossRef] [Green Version]
- Bass, B.P.; Cullen, K.; McCall, K. The axon guidance gene lola is required for programmed cell death in the Drosophila ovary. Dev. Biol. 2007, 304, 771–785. [Google Scholar] [CrossRef] [Green Version]
- Huang, Q.; Tang, X.; Wang, G.; Fan, Y.; Ray, L.; Bergmann, A.; Belenkaya, T.Y.; Ling, X.; Yan, D.; Lin, Y.; et al. Ubr3 E3 ligase regulates apoptosis by controlling the activity of DIAP1 in Drosophila. Cell Death Differ. 2014, 21, 1961–1970. [Google Scholar] [CrossRef] [Green Version]
- Balan, V.; Miller, G.S.; Kaplun, L.; Balan, K.; Chong, Z.Z.; Li, F.; Kaplun, A.; VanBerkum, M.F.; Arking, R.; Freeman, D.C.; et al. Life span extension and neuronal cell protection by Drosophila nicotinamidase. J. Biol. Chem. 2008, 283, 27810–27819. [Google Scholar] [CrossRef] [Green Version]
- Han, Z.; Yi, P.; Li, X.; Olson, E.N. Hand, an evolutionarily conserved bHLH transcription factor required for Drosophila cardiogenesis and hematopoiesis. Development 2006, 133, 1175–1182. [Google Scholar] [CrossRef] [Green Version]
- Cao, C.; Liu, Y.; Lehmann, M. Fork head controls the timing and tissue selectivity of steroid-induced developmental cell death. J. Cell Biol. 2007, 176, 843–845. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, F.J.; Gelboin, H.V. Human cytochromes P450: Evolution and cDNA-directed expression. Environ. Health Perspect. 1992, 98, 81–85. [Google Scholar] [CrossRef]
- Thurmond, J.; Goodman, J.L.; Strelets, V.B.; Attrill, H.; Gramates, L.S.; Marygold, S.J.; Matthews, B.B.; Millburn, G.; Antonazzo, G.; Trovisco, V.; et al. FlyBase 2.0: The next generation. Nucleic Acids Res. 2019, 47, D759–D765. [Google Scholar] [CrossRef] [Green Version]
- UniProt Consortium, T. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef] [Green Version]
- Seong, K.M.; Coates, B.S.; Pittendrigh, B.R. Cytochrome P450s Cyp4p1 and Cyp4p2 associated with the DDT tolerance in the Drosophila melanogaster strain 91-R. Pestic. Biochem. Physiol. 2019, 159, 136–143. [Google Scholar] [CrossRef]
- Schmidt, J.M.; Battlay, P.; Gledhill-Smith, R.S.; Good, R.T.; Lumb, C.; Fournier-Level, A.; Robin, C. Insights into DDT Resistance from the Drosophila melanogaster Genetic Reference Panel. Genetics 2017, 207, 1181–1193. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Valero, M.C.; Seong, K.M.; Steele, L.D.; Huang, I.T.; Lee, C.H.; Clark, J.M.; Qiu, X.; Pittendrigh, B.R. A glycine insertion in the estrogen-related receptor (ERR) is associated with enhanced expression of three cytochrome P450 genes in transgenic Drosophila melanogaster. PLoS ONE 2015, 10, e0118779. [Google Scholar] [CrossRef]
- Terhzaz, S.; Cabrero, P.; Brinzer, R.A.; Halberg, K.A.; Dow, J.A.; Davies, S.A. A novel role of Drosophila cytochrome P450-4e3 in permethrin insecticide tolerance. Insect. Biochem. Mol. Biol. 2015, 67, 38–46. [Google Scholar] [CrossRef] [Green Version]
- Thomas, A.M.; Hui, C.; South, A.; McVey, M. Common variants of Drosophila melanogaster Cyp6d2 cause camptothecin sensitivity and synergize with loss of Brca2. G3 (Bethesda) 2013, 3, 91–99. [Google Scholar] [CrossRef] [Green Version]
- Guittard, E.; Blais, C.; Maria, A.; Parvy, J.P.; Pasricha, S.; Lumb, C.; Lafont, R.; Daborn, P.J.; Dauphin-Villemant, C. CYP18A1, a key enzyme of Drosophila steroid hormone inactivation, is essential for metamorphosis. Dev. Biol. 2011, 349, 35–45. [Google Scholar] [CrossRef]
- Lederer, F. The cytochrome b5-fold: An adaptable module. Biochimie 1994, 76, 674–692. [Google Scholar] [CrossRef]
- Malatesta, F.; Antonini, G.; Sarti, P.; Brunori, M. Structure and function of a molecular machine: Cytochrome c oxidase. Biophys. Chem. 1995, 54, 1–33. [Google Scholar] [CrossRef]
- Asard, H.; Barbaro, R.; Trost, P.; Berczi, A. Cytochromes b561: Ascorbate-mediated trans-membrane electron transport. Antioxid. Redox. Signal. 2013, 19, 1026–1035. [Google Scholar] [CrossRef] [Green Version]
- Graveley, B.R.; Brooks, A.N.; Carlson, J.W.; Duff, M.O.; Landolin, J.M.; Yang, L.; Artieri, C.G.; van Baren, M.J.; Boley, N.; Booth, B.W.; et al. The developmental transcriptome of Drosophila melanogaster. Nature 2011, 471, 472–479. [Google Scholar] [CrossRef] [Green Version]
- DeVeale, B.; Brummel, T.; Seroude, L. Immunity and aging: The enemy within? Aging Cell 2004, 3, 195–208. [Google Scholar] [CrossRef]
- Fagiolo, U.; Cossarizza, A.; Scala, E.; Fanales-Belasio, E.; Ortolani, C.; Cozzi, E.; Monti, D.; Franceschi, C.; Paganelli, R. Increased cytokine production in mononuclear cells of healthy elderly people. Eur. J. Immunol. 1993, 23, 2375–2378. [Google Scholar] [CrossRef]
- Yu, G.; Hyun, S. Proteostasis-associated aging: Lessons from a Drosophila model. Genes Genom. 2021, 43, 289–294. [Google Scholar] [CrossRef]
- Mauruzs, T.; Simon-Vecsei, Z.; Kiss, V.; Csizmadia, T.; Juhasz, G. On the Fly: Recent Progress on Autophagy and Aging in Drosophila. Front. Cell Dev. Biol. 2019, 7, 140. [Google Scholar] [CrossRef]
- Sanson, B.; White, P.; Vincent, J.P. Uncoupling cadherin-based adhesion from wingless signalling in Drosophila. Nature 1996, 383, 627–630. [Google Scholar] [CrossRef]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 188, 401–415. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 16, 9440–9445. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
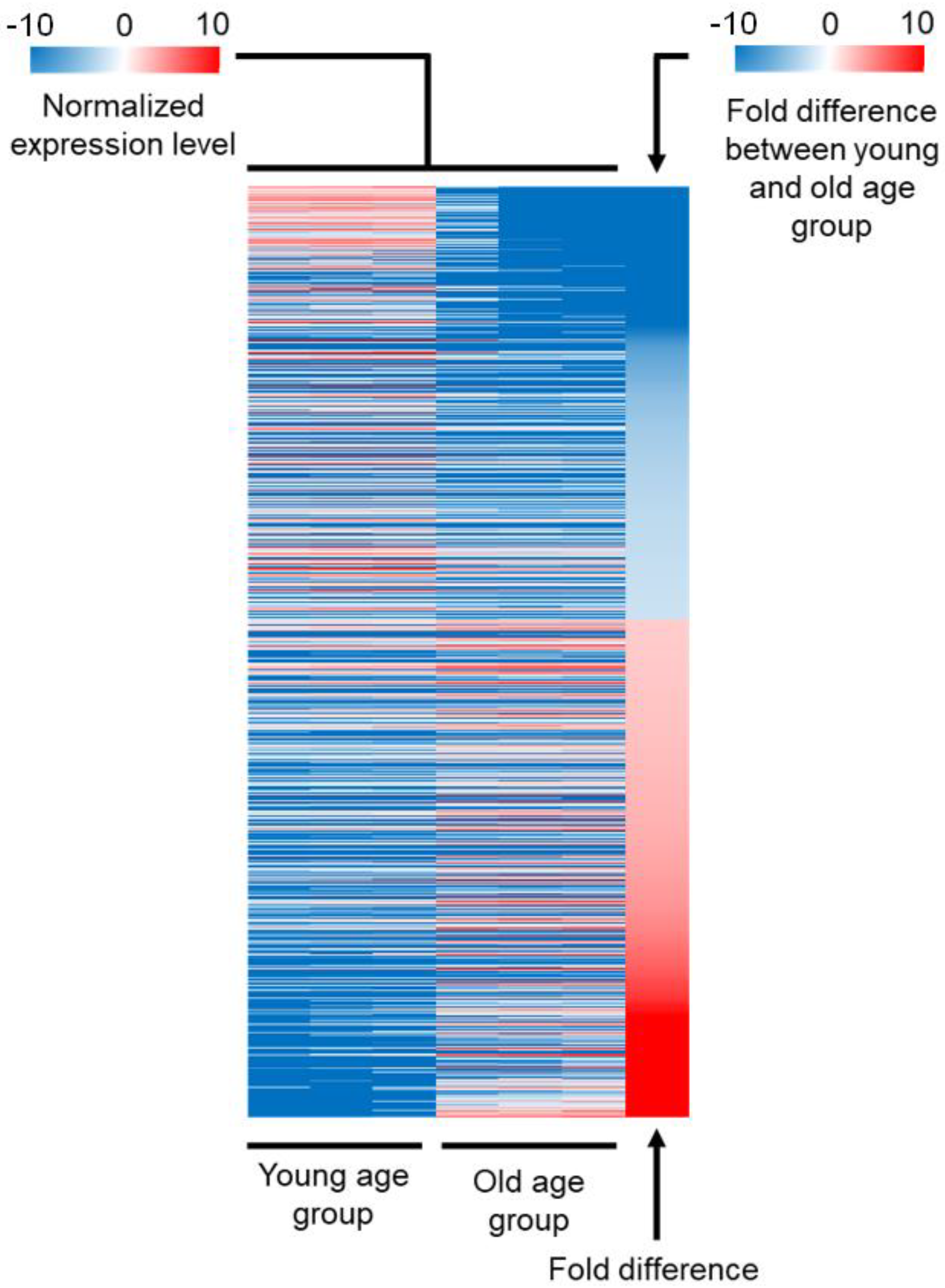
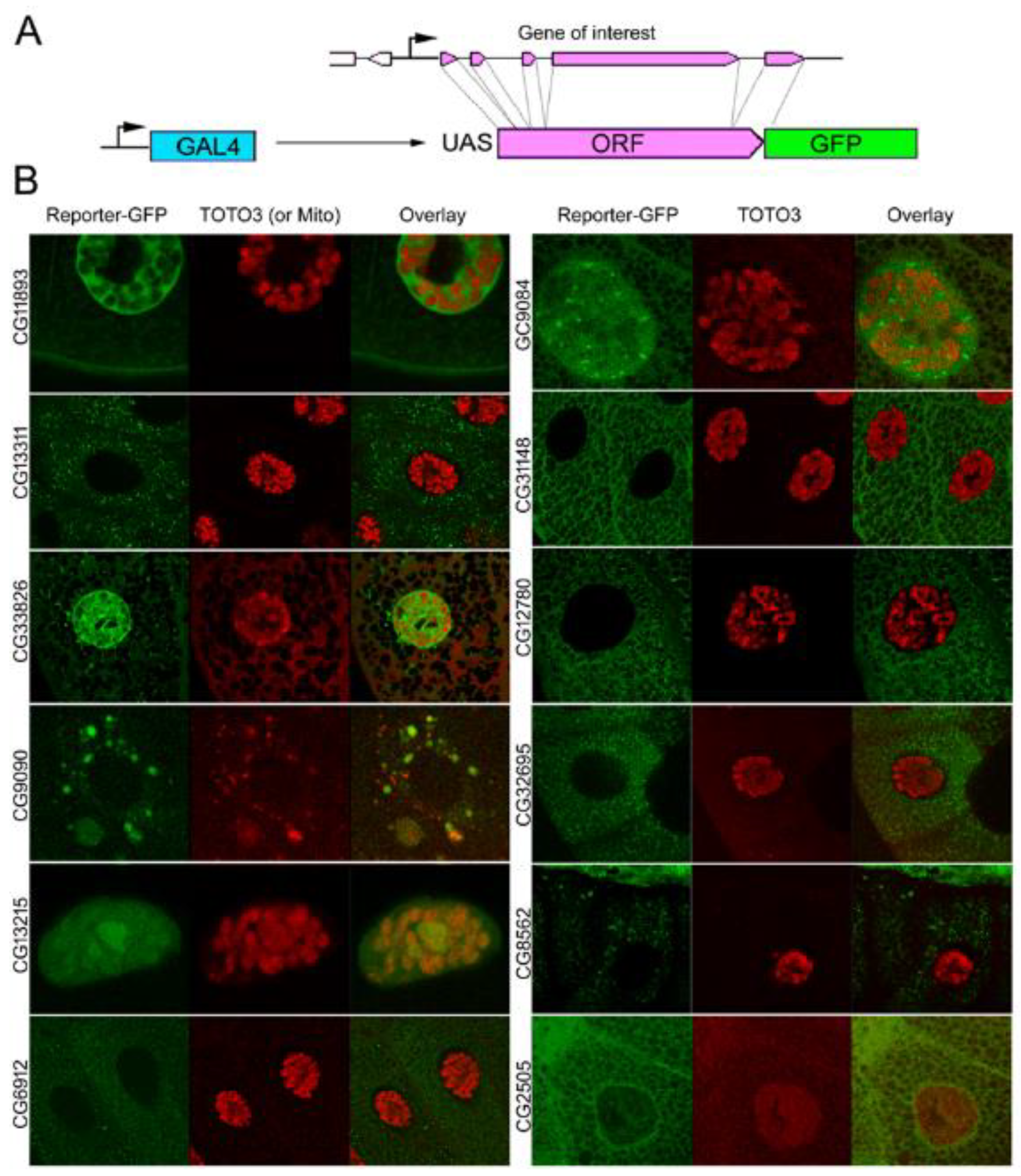
| Gene Class | Number of Genes Included | Expression in the Young Age Group Compared to Average | Expression in the Old Age Group Compared to Average | Down- or Upregulated during Aging |
|---|---|---|---|---|
| 1 | 328 | Lower | Lower | Downregulated |
| 2 | 408 | Lower | Lower | Upregulated |
| 3 | 178 | Higher | Lower | Downregulated |
| 4 | 154 | Lower | Higher | Upregulated |
| 5 | 63 | Higher | Higher | Downregulated |
| 6 | 94 | Higher | Higher | Upregulated |
| Gene | Differential Expression Level between ‘Old Age’ and ‘Young Age’ Flies (Fold Difference) | Gene Class | Protein Group | Human Ortholog |
|---|---|---|---|---|
| Sls | −29.01 | 1 | Titin | HMCN1 |
| MLP60A | −20.92 | 1 | Z-line-associated protein | CSRP3 |
| TPnC25D | −9.97 | 1 | Troponin C | CALML6 |
| Salm | −4.9 | 1 | Zinc finger transcription factor | SALL1 |
| Eya | −3.44 | 1 | Tyrosine phosphatase | EYA2 |
| TPnC47D | −118 | 3 | Troponin C | CALML6 |
| Fln | −85.68 | 3 | A-band-located protein | / |
| Strn-Mlck | −75.2 | 3 | Calcuim/calmodulin-dependent protein kinase | MYLK |
| Unc-89 | −35.71 | 3 | obscurin | SPEG |
| Act88F | −20.58 | 3 | Actin | ACTB |
| TPnC41C | −19.2 | 3 | Troponin C | CALML6 |
| Act79B | −13.2 | 3 | Actin | ACTB |
| Prm | −3.48 | 3 | Paramyosin | / |
| Mlp84B | −2.79 | 3 | Z-line-associated protein | CSRP3 |
| Tn | −2.06 | 3 | Z-line-associated protein | TRIM2 |
| TPnC4 | −6.56 | 5 | Troponin C | CALM1 |
| Tm2 | −3.65 | 5 | Tropomyosin | TPM3 |
| Up | −3.24 | 5 | Troponin T | TNNT2 |
| mlc2 | −3.2 | 5 | Myosin light chain | MYL |
| Tm1 | −3.16 | 5 | Tropomyosin | TPM1 |
| Fhos | −3.04 | 5 | Formin -like | FHOD3 |
| mlc1 | −2.78 | 5 | Myosin light chain | MYL |
| Act87E | −2.75 | 5 | Actin | ACTB |
| mhc | −2.47 | 5 | Myosin heavy chain | MYH |
| Mf | −2.44 | 5 | A-band-located protein | / |
| Bt | −2.27 | 5 | Projectin | TTN |
| wupA | −2.24 | 5 | Troponin I | TNNI2 |
| Gene | Differential Expression Level between ‘Old Age’ and ‘Young Age’ Flies (Fold Difference) | Gene Class | Protein Group | Human Ortholog |
|---|---|---|---|---|
| Fkh | 3.86 | 2 | Fork head box transcription factor | FOXA2 |
| Strica | 3.84 | 2 | Caspase | CASP3 |
| Ubr3 | 2.37 | 2 | RING finger protein | UBR3 |
| Hand | 11.25 | 2 | bHLH transcription factor | HAND2 |
| Lola | 3.33 | 2 | C2H2 zinc finger transcription factor | ZBTB20 |
| Naam | 3.11 | 2 | Nicotinamide amidase | / |
| Fwe | 3.11 | 2 | Other calcium channel-forming subunit | CACFD1 |
| Decay | 6.72 | 4 | Caspase | CASP3 |
| Damm | 13.09 | 4 | Caspase | CASP3 |
| Diap1 | 2.7 | 6 | RING finger protein | BIRC2 |
| Gene | Differential Expression Level between ‘Old Age’ and ‘Young Age’ Flies (Fold Difference) | Gene Class | Protein Group | Function | Human Closest Ortholog |
|---|---|---|---|---|---|
| Cyp313a1 | −16.6 | 1 | Other cytochrome P450 | Unknown | CYP26B1 |
| Cyp4ac2 | −7.3 | 1 | Other cytochrome P450 | Toxic chemicals breakdown (predicted) | CYP4V2 |
| Cyp4e1 | −5.0 | 1 | Other cytochrome P450 | Permethrin resistance | CYP4V2 |
| Cyp6a21 | −6.2 | 3 | Other cytochrome P450 | Toxic chemicals breakdown (predicted) | CYP3A4 |
| Cyp305a1 | −2.7 | 3 | Other cytochrome P450 | Unknown | CYP2J2 |
| Cyp18a1 | −2.2 | 1 | Other cytochrome P450 | Steroid hormone inhibition | CYP2J2 |
| CG34172 | −2.9 | 5 | Cytochrome-C oxidase | Unknown | COX7A1 |
| CG6870 | −2.1 | 5 | Cytochrome b5 | Unknown | CYB5A |
| Cyp4g15 | 2.1 | 2 | Other cytochrome P450 | Toxic chemicals breakdown (predicted) | CYP4V2 |
| Cyp4ac3 | 8.4 | 2 | Other cytochrome P450 | Toxic chemicals breakdown (predicted) | CYP4V2 |
| Cyp4p1 | 7.9 | 4 | Other cytochrome P450 | DDT resistance | CYP4V2 |
| Cyp4p3 | 13.2 | 4 | Other cytochrome P450 | DDT resistance | CYP4V2 |
| Cyp6w1 | 2.5 | 4 | Other cytochrome P450 | DDT resistance | CYP3A4 |
| Cyp9c1 | 6.1 | 2 | Other cytochrome P450 | DDT resistance | CYP3A4 |
| Cyp6d2 | 31.0 | 2 | Other cytochrome P450 | Camptothecin resistance | CYP3A4 |
| Cyp28d1 | 2.6 | 4 | Other cytochrome P450 | Toxic chemicals breakdown (predicted) | CYP3A4 |
| Cyp6d4 | 3.2 | 2 | Other cytochrome P450 | Toxic chemicals breakdown (predicted) | CYP3A4 |
| Cyp309a1 | 10.2 | 2 | Other cytochrome P450 | Unknown | CYP3A4 |
| Cyp9b2 | 2.2 | 6 | Other cytochrome P450 | Toxic chemicals breakdown (predicted) | CYP3A5 |
| Cyp9b1 | 2.3 | 2 | Other cytochrome P450 | Toxic chemicals breakdown (predicted) | CYP3A5 |
| Cyp9h1 | 14.1 | 2 | Other cytochrome P450 | Toxic chemicals breakdown (predicted) | CYP3A5 |
| Cyp6a13 | 3.7 | 4 | Other cytochrome P450 | Toxic chemicals breakdown (predicted) | CYP3A7 |
| Cyp304a1 | 53.7 | 2 | Other cytochrome P450 | Toxic chemicals breakdown (predicted) | CYP2F1 |
| Cyp12a5 | 2.2 | 2 | Other cytochrome P450 | Unknown | CYP24A1 |
| Cyp12e1 | 4.3 | 2 | Other cytochrome P450 | Unknown | CYP24A1 |
| CG10337 | 2.2 | 6 | Cytochrome b561 | Unknown | CYB561D1 |
| CG13077 | 11.8 | 2 | Cytochrome b561 | Unknown | CYB561D1 |
| Gene | Differential Expression Level between ‘Old Age’ and ‘Young Age’ Flies (Fold Difference) | GFP-Reporter Localization | Secreted | Ectopic Expression Effect on Longevity | siRNA Transgene (Vienna) | Phenotype of the Knockdown |
|---|---|---|---|---|---|---|
| CG11893 | 4.6 | Soluble nucleoplasm; Secretory granules | Yes | No | v16356 v105267 | No |
| CG13311 | 44.3 | Cytoplasmic particles | No | No | v101655 v51574 | −17% |
| CG33926 | 33.8 | Soluble nucleoplasm; Secretory granules | Yes | −11% | v107542 | −46% |
| CG9090 | −179.7 | Mitochondria | No | lethal | v101848 v44297 | lethal |
| CG13215 | 59.9 | Nuclear chromatin and nucleolus | No | +5% | v37166 v37167 | −22% |
| CG6912 | 3.7 | Secretory granules | Yes | −7% | v102806 v31093 | −9% |
| CG9084 | 6.0 | Soluble cytoplasm; nucleoplasmic granules; Soluble nucleoplasm | No | −28% | v26908 v106414 | +6% |
| CG31148 | 254.5 | Soluble cytoplasm | No | No | v14698 v14697 | +6% |
| CG12780 | 2.6 | Secretory granules | Yes | +5% | v101969 v24613 | +5% |
| CG32695 | 4.3 | Cytoplasmic granules | No | −9% | v44751 v102678 | −12% |
| CG8562 | −121.6 | Secretory granules | Yes | −6% | v44577 v44578 | −24% |
| CG2505 | −5.9 | Perinuclear and peri-plasma membrane space | No | -−31% | v105578 v20879 | +11% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bordet, G.; Lodhi, N.; Kossenkov, A.; Tulin, A. Age-Related Changes of Gene Expression Profiles in Drosophila. Genes 2021, 12, 1982. https://doi.org/10.3390/genes12121982
Bordet G, Lodhi N, Kossenkov A, Tulin A. Age-Related Changes of Gene Expression Profiles in Drosophila. Genes. 2021; 12(12):1982. https://doi.org/10.3390/genes12121982
Chicago/Turabian StyleBordet, Guillaume, Niraj Lodhi, Andrew Kossenkov, and Alexei Tulin. 2021. "Age-Related Changes of Gene Expression Profiles in Drosophila" Genes 12, no. 12: 1982. https://doi.org/10.3390/genes12121982
APA StyleBordet, G., Lodhi, N., Kossenkov, A., & Tulin, A. (2021). Age-Related Changes of Gene Expression Profiles in Drosophila. Genes, 12(12), 1982. https://doi.org/10.3390/genes12121982







