Advances in Genetics and Epigenetic Alterations in Alzheimer’s Disease: A Notion for Therapeutic Treatment
Abstract
1. Background
2. The Genetics of AD
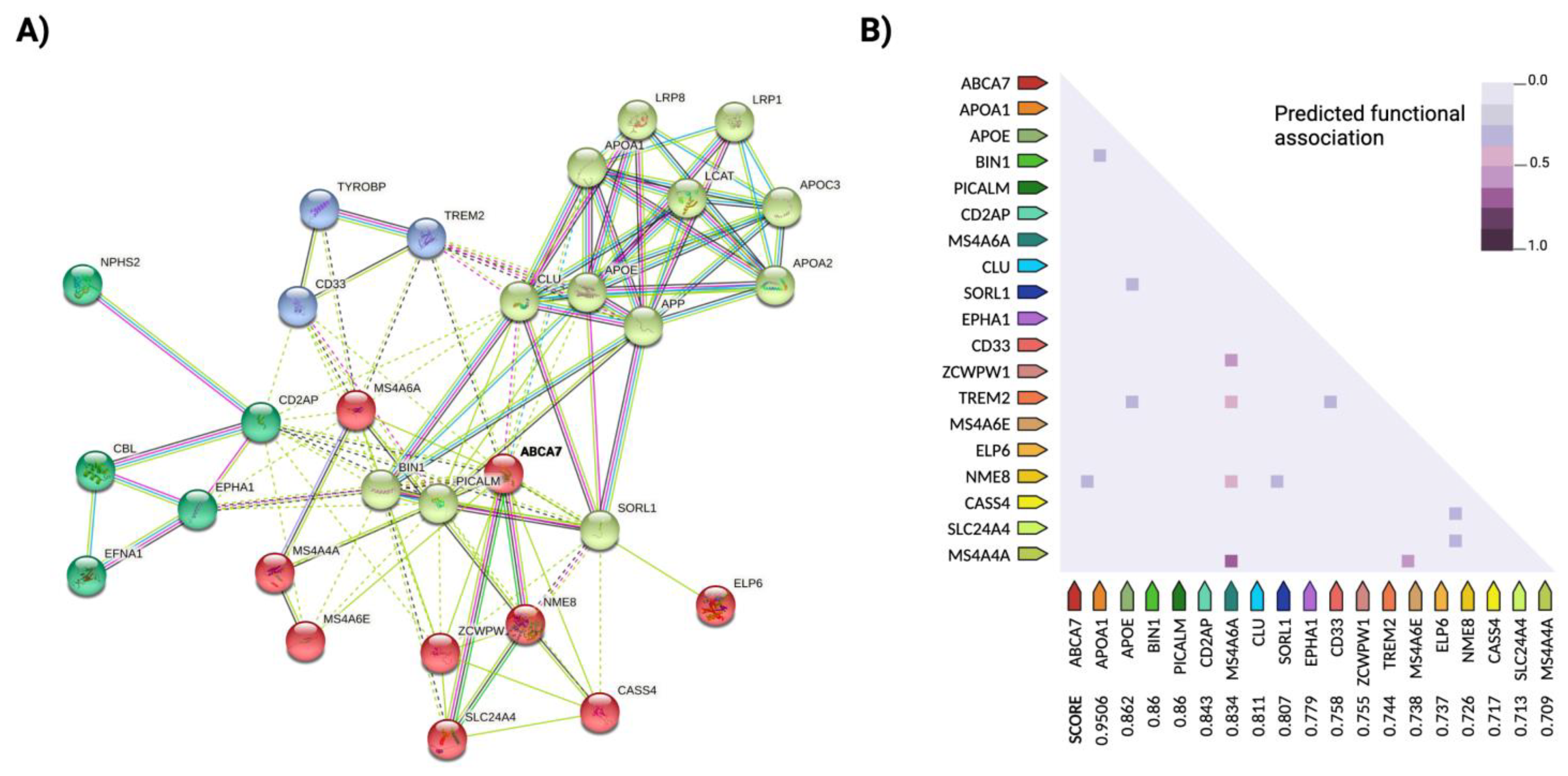
2.1. Important Susceptibility Genes
2.2. Missing Heritability: Epistasis Explains Undetected Associations in Risk Loci
3. Alzheimer’s and the Epigenome: Filling the Gap?
3.1. Epigenetics Alterations of AD
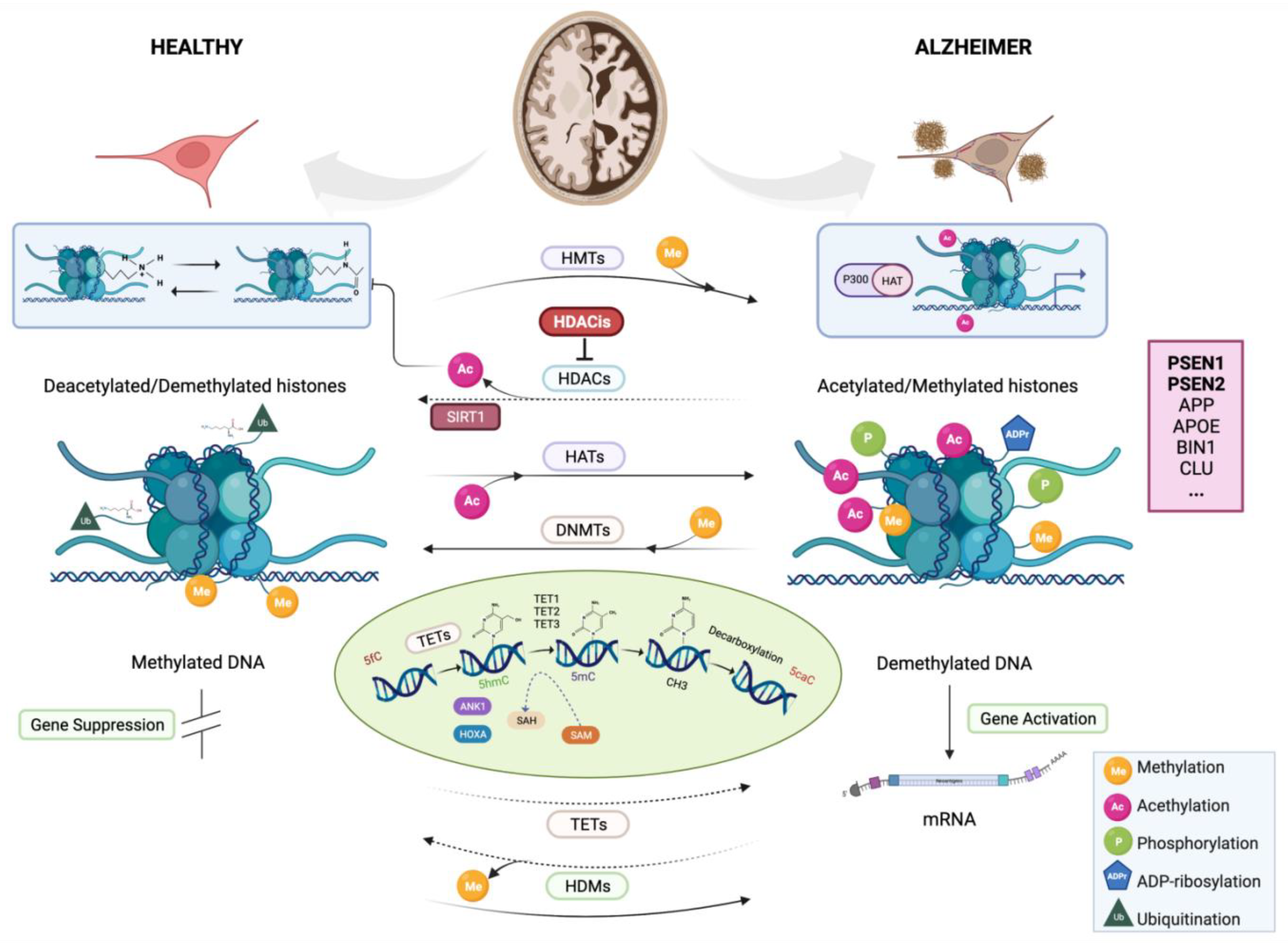
3.1.1. DNA Methylation
3.1.2. Histone Modifications
3.1.3. Noncoding RNAs
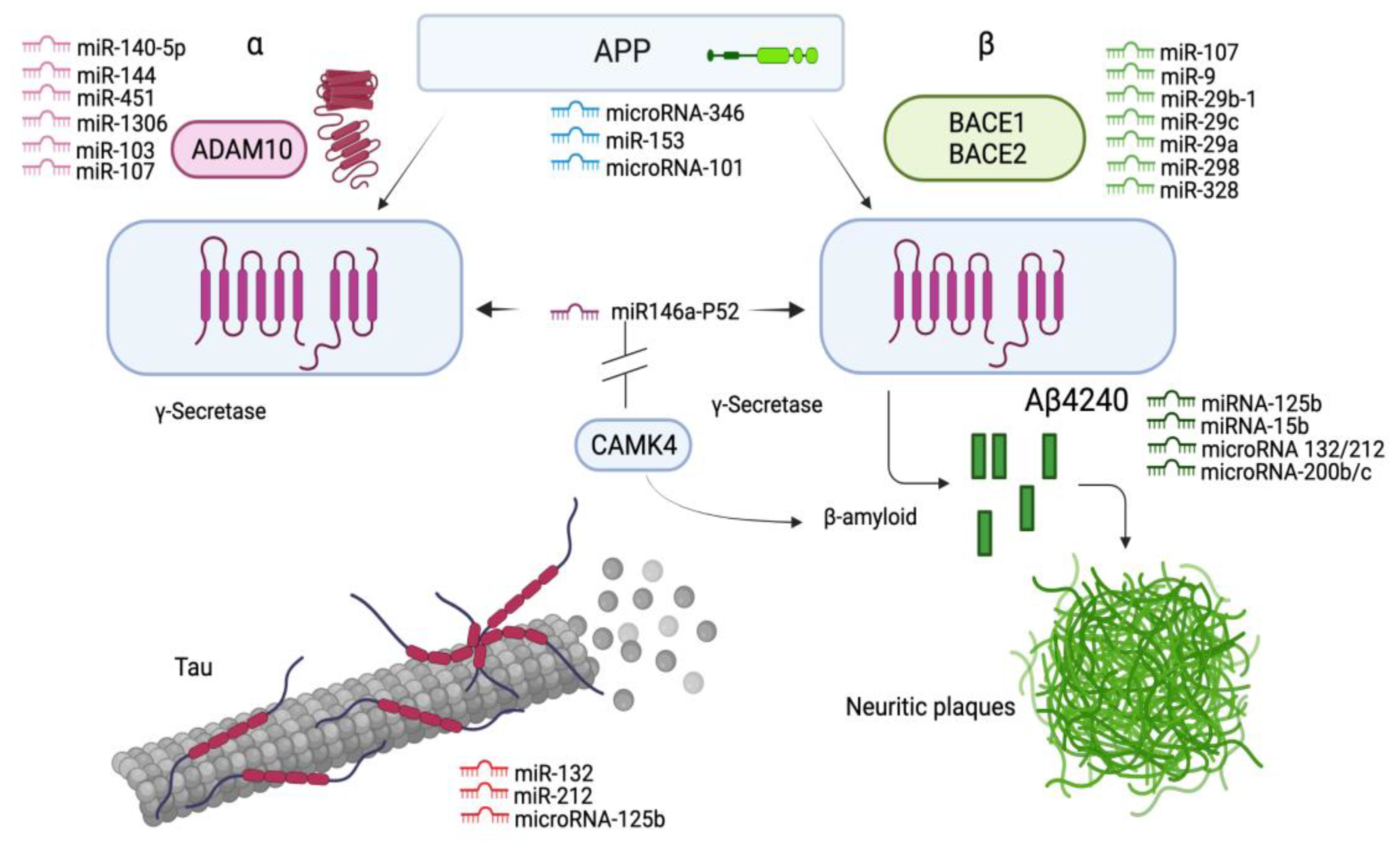
3.1.4. MiRNAs
3.1.5. Piwi-Interacting RNAs
4. Targets for Therapeutic Treatment of AD
4.1. Targeting Key Genes for AD
4.2. Epigenomic Biomarkers for the Treatment of AD
4.3. Aβ Immunotherapy
4.4. HDAC Inhibitors as Therapeutics for AD
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Glossary
| AD | Alzheimer disease |
| LOAD | Late-Onset Alzheimer Disease |
| EOAD | Early-Onset Alzheimer Disease |
| MCI | Mild cognitive impairment |
| CNS | Central nervous system |
| CSF | Cerebrospinal fluid |
| PFC | prefrontal cortex |
| EWAS | Epigenome-wide association study |
| GWAS | Genome-wide association study |
| NGS | Next-generation sequencing |
| WES | Whole-exome sequencing |
| WGS | Whole-genome sequencing |
| MGAS | Multivariate gene association analysis |
| PPI | Protein–protein interaction |
| p-Tau | Phosphorylated protein tau |
| CGIs | CpG islands |
| Aβ | Amyloid-β peptide |
| Aβ42 | Amyloid-β 42 |
| NFTs | Neurofibrillary tangles |
| APP | Amyloid-β precursor protein |
| APP-Aβ | β-amyloid domain containing APP |
| APP-CTF | APP C-terminal fragment |
| BACE1 | β-site amyloid precursor protein cleaving enzyme 1 |
| BACE2 | β-site amyloid precursor protein cleaving enzyme 2 |
| ApoE | Apolipoprotein E |
| ApoE2 | Apolipoprotein ε2 |
| ApoE3 | Apolipoprotein ε3 |
| ApoE4 | Apolipoprotein ε4 |
| APOC1 | Apolipoprotein C1 |
| BIN1 | Bridging Integrator 1 |
| CLU | Clusterin |
| PICALM | Phosphatidylinositol-binding clathrin assembly protein |
| TOMM40 | Translocase of outer mitochondrial membrane 40 |
| CR1 | Complement receptor type 1 |
| ANK1 | Ankyrin 1 |
| CDH23 | Cadherin-related 23 |
| CaMKIV | Calcium/calmodulin-dependent protein kinase type IV |
| DIP2A | Disco-interacting protein 2 homolog A |
| RHBDF2 | Inactive rhomboid protein 2 |
| RPL13 | 60S ribosomal protein L13 |
| SERPINA3 | Serpin family A member 3 |
| SERPINF | Serpin family F member |
| AICD | APP intracellular domain |
| CBP | CREB-binding protein |
| MAPT | Microtubule-associated protein Tau |
| IGF-I | Insulin-like growth factor 1 |
| GLUT4 | Glucose transporter Type 4 |
| PSEN1 | Presenilin 1 |
| PSEN2 | Presenilin 2 |
| SORL1 | Sortilin-related receptor 1 |
| ADAM10 | A disintegrin and metalloprotease 10 |
| PTM-Hs | Post-translational modification of histones |
| DMRs | Differentially methylated regions |
| SAM | Methyl donor S-adenosylmethionine |
| SIRT1 | Sirtuin 1 |
| RNase P | Ribonuclease P |
| TER | Telomerase RNA |
| TET | Ten–eleven translocation |
| SUMO | Small ubiquitin-like modifier |
| DGCR8 | Microprocessor complex subunit DGCR8 |
| DNMT | DNA methyltransferase |
| HAT | Histone acetyltransferase |
| HDAC | Histone deacetylase |
| HDACi | HDAC inhibitors |
| ICBs | Immune checkpoint blockers |
| siRC | silencing RNA-induced complex |
| RBL2 | Retinoblastoma-like 2 protein |
| HDL | High-density lipoproteins |
| VLDL | Very low-density lipoprotein |
| NF-κB | Nuclear transcription factor kappa B |
| ncRNA | Noncoding RNA |
| miRNA | MicroRNA |
| piRNA | Piwi-interacting RNA |
| rRNA | Ribosomal RNA |
| snRNA | Small nuclear RNA |
| tRNA | Transfer RNA |
| snoRNA | Small nucleolar RNA |
References
- Oddo, S.; Caccamo, A.; Shepherd, J.D.; Murphy, M.P.; Golde, T.E.; Kayed, R.; Metherate, R.; Mattson, M.P.; Akbari, Y.; LaFerla, F.M. Triple-Transgenic Model of Alzheimer’s Disease with Plaques and Tangles: Intracellular Aβ and Synaptic Dysfunction. Neuron 2003, 39, 409–421. [Google Scholar] [CrossRef]
- Fuhrmann, M.; Bittner, T.; Jung, C.K.E.; Burgold, S.; Page, R.M.; Mitteregger, G.; Haass, C.; LaFerla, F.M.; Kretzschmar, H.; Herms, J. Microglial Cx3cr1 Knockout Prevents Neuron Loss in a Mouse Model of Alzheimer’s Disease. Nat. Neurosci. 2010, 13, 411–413. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Pozo, A.; Qian, J.; Monsell, S.E.; Blacker, D.; Gómez-Isla, T.; Betensky, R.A.; Growdon, J.H.; Johnson, K.A.; Frosch, M.P.; Sperling, R.A.; et al. Mild to Moderate Alzheimer Dementia with Insufficient Neuropathological Changes. Ann. Neurol. 2014, 75, 597–601. [Google Scholar] [CrossRef]
- Monsell, S.E.; Kukull, W.A.; Roher, A.E.; Maarouf, C.L.; Serrano, G.; Beach, T.G.; Caselli, R.J.; Montine, T.J.; Reiman, E.M. Characterizing Apolipoprotein E Ε4 Carriers and Noncarriers With the Clinical Diagnosis of Mild to Moderate Alzheimer Dementia and Minimal β-Amyloid Peptide Plaques. JAMA Neurol. 2015, 72, 1124–1131. [Google Scholar] [CrossRef] [PubMed]
- Jack, C.; Bennett, D.; Blennow, K.; Carrillo, M.; Dunn, B.; Haeberlein, S.; Holtzman, D.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a Biological Definition of Alzheimer’s Disease. Alzheimer’s Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef] [PubMed]
- Sabbagh, M.N.; Lue, L.-F.; Fayard, D.; Shi, J. Increasing Precision of Clinical Diagnosis of Alzheimer’s Disease Using a Combined Algorithm Incorporating Clinical and Novel Biomarker Data. Neurol. Ther. 2017, 6, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Meakin, P.; Coull, B.; Tuharska, Z.; McCaffery, C.; Akoumianakis, I.; Antoniades, C.; Brown, J.; Griffin, K.; Platt, F.; Ozber, C.; et al. Elevated Circulating Amyloid Concentrations in Obesity and Diabetes Promote Vascular Dysfunction. J. Clin. Investig. 2020, 130, 4104–4117. [Google Scholar] [CrossRef] [PubMed]
- Heneka, M.T.; Carson, M.J.; El Khoury, J.; Landreth, G.E.; Brosseron, F.; Feinstein, D.L.; Jacobs, A.H.; Wyss-Coray, T.; Vitorica, J.; Ransohoff, R.M.; et al. Neuroinflammation in Alzheimer’s Disease. Lancet. Neurol. 2015, 14, 388–405. [Google Scholar] [CrossRef]
- Jäkel, L.; Boche, D.; Nicoll, J.; Verbeek, M. Aβ43 in Human Alzheimer’s Disease: Effects of Active Aβ42 Immunization. Acta Neuropathol. Commun. 2019, 7, 141. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Cejudo, J.; Wisniewski, T.; Marmar, C.; Zetterberg, H.; Blennow, K.; de Leon, M.J.; Fossati, S. Traumatic Brain Injury and Alzheimer’s Disease: The Cerebrovascular Link. EBioMedicine 2018, 28, 21–30. [Google Scholar] [CrossRef]
- Del Prete, D.; Suski, J.; Oulès, B.; Debayle, D.; Gay, A.-S.; Lacas-Gervais, S.; Bussiere, R.; Bauer, C.; Pinton, P.; Paterlini, P.; et al. Localization and Processing of the Amyloid-β Protein Precursor in Mitochondria-Associated Membranes. J. Alzheimer’s Dis. 2016, 55, 1549–1570. [Google Scholar] [CrossRef]
- Pulina, M.V.; Hopkins, M.; Haroutunian, V.; Greengard, P.; Bustos, V. C99 Selectively Accumulates in Vulnerable Neurons in Alzheimer’s Disease. Alzheimer’s Dement. 2019. [Google Scholar] [CrossRef]
- Vaillant-Beuchot, L.; Mary, A.; Pardossi-Piquard, R.; Bourgeois, A.; Lauritzen, I.; Eysert, F.; Kinoshita, P.F.; Cazareth, J.; Badot, C.; Fragaki, K.; et al. Accumulation of Amyloid Precursor Protein C-terminal Fragments Triggers Mitochondrial Structure, Function, and Mitophagy Defects in Alzheimer’s Disease Models and Human Brains. Acta Neuropathol. 2021, 141, 39–65. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, P.-H.; Wang, H.; Dislich, B.; Colombo, A.; Zeitschel, U.; Ellwart, J.W.; Kremmer, E.; Rossner, S.; Lichtenthaler, S.F. ADAM10 Is the Physiologically Relevant, Constitutive α-Secretase of the Amyloid Precursor Protein in Primary Neurons. EMBO J. 2010, 29, 3020–3032. [Google Scholar] [CrossRef]
- Kojro, E.; Gimpl, G.; Lammich, S.; Marz, W.; Fahrenholz, F. Low Cholesterol Stimulates the Nonamyloidogenic Pathway by Its Effect on the α -Secretase ADAM 10. Proc. Natl. Acad. Sci. USA 2001, 98, 5815–5820. [Google Scholar] [CrossRef]
- Simons, M.; Keller, P.; De Strooper, B.; Beyreuther, K.; Dotti, C.G.; Simons, K. Cholesterol Depletion Inhibits the Generation of β-Amyloid in Hippocampal Neurons. Proc. Natl. Acad. Sci. USA 1998, 95, 6460–6464. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, Q.; Cai, F.; Liu, X.; Wu, Y.; Song, W. BACE2, a Conditional β-Secretase, Contributes to Alzheimer’s Disease Pathogenesis. JCI Insight 2019, 4, e123431. [Google Scholar] [CrossRef]
- Miller, J.; Woltjer, R.; Goodenbour, J.; Horvath, S.; Geschwind, D. Genes and Pathways Underlying Regional and Cell Type Changes in Alzheimer’s Disease. Genome Med. 2013, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Greenwood, A.; Binder, L.; Bigio, E.; Denial, S.; Nicholson, L.; Zhou, X.; Lu, K.P. Proline Isomer-Specific Antibodies Reveal the Early Pathogenic Tau Conformation in Alzheimer’s Disease. Cell 2012, 149, 232–244. [Google Scholar] [CrossRef]
- Neu, S.C.; Pa, J.; Kukull, W.; Beekly, D.; Kuzma, A.; Gangadharan, P.; Wang, L.-S.; Romero, K.; Arneric, S.P.; Redolfi, A.; et al. Apolipoprotein E Genotype and Sex Risk Factors for Alzheimer Disease: A Meta-Analysis. JAMA Neurol. 2017, 74, 1178–1189. [Google Scholar] [CrossRef]
- Bomfim, T.; Forny-Germano, L.; Sathler, L.; Brito-Moreira, J.; Houzel, J.-C.; Decker, H.; Silverman, M.; Kazi, H.; de Melo, H.; Mcclean, P.; et al. An Anti-Diabetes Agent Protects the Mouse Brain from Defective Insulin Signaling Caused by Alzheimer?S Disease-Associated A? Oligomers. J. Clin. Invest. 2012, 122, 1339–1353. [Google Scholar] [CrossRef]
- Ayton, S.; Bush, A. β-Amyloid: The Known Unknowns. Ageing Res. Rev. 2021, 65, 101212. [Google Scholar] [CrossRef] [PubMed]
- Gyenesei, A.; Moody, J.; Semple, C.; Haley, C.; Wei, W. HHigh-Throughput Analysis of Epistasis in Genome-Wide Association Studies with BiForce. Bioinformatics 2012, 28, 1957–1964. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van den Hove, D.; Riemens, R.; Koulousakis, P.; Pishva, E. Epigenome-wide Association Studies in Alzheimer’s Disease; Achievements and Challenges. Brain Pathol. 2020, 30, 978–983. [Google Scholar] [CrossRef]
- Area-Gomez, E.; Schon, E.A. On the Pathogenesis of Alzheimer’s Disease: The MAM Hypothesis. FASEB J. 2017, 31, 864–867. [Google Scholar] [CrossRef] [PubMed]
- Panegyres, P.K.; Chen, H.-Y. Differences between Early and Late Onset Alzheimer’s Disease. Am. J. Neurodegener. Dis. 2013, 2, 300–306. [Google Scholar]
- Mendez, M.F. Early-Onset Alzheimer Disease. Neurol. Clin. 2017, 35, 263–281. [Google Scholar] [CrossRef]
- Braak, H.; Del Tredici, K.; Braak, H.; Del Tredici, K. The Pathological Process Underlying Alzheimer’s Disease in Individuals under Thirty. Acta Neuropathol 121: 171-181. Acta Neuropathol. 2011, 121, 171–181. [Google Scholar] [CrossRef]
- Gatz, M.; Pedersen, N.L. Study of Dementia in Swedish Twins. Twin Res. Hum. Genet. 2013, 16, 313–316. [Google Scholar] [CrossRef]
- Yokoyama, A.; Rutledge, J.; Medici, V. DNA Methylation Alterations in Alzheimer’s Disease. Environ. Epigenetics 2017, 3, dvx008. [Google Scholar] [CrossRef] [PubMed]
- D’addario, C.; Bastías-Candia, S.; Arosio, B.; Di Bartolomeo, M.; Abbate, C.; Casè, A.; Candeletti, S.; Romualdi, P.; Damanti, S.; Maccarrone, M.; et al. Transcriptional and Epigenetic Phenomena in Peripheral Blood Cells of Monozygotic Twins Discordant for Alzheimer’s Disease, a Case Report. J. Neurol. Sci. 2016, 372, 211–216. [Google Scholar] [CrossRef]
- Levy, E.; Carman, M.; Fernandez-Madrid, I.J.; Power, M.; Lieberburg, I.; Duinen, S.G.; Bots, G.; Luyendijk, W.; Frangione, B. Mutation of the Alzheimer’s Disease Amyloid Gene in Hereditary Cerebral Hemorrhage, Dutch Type. Science 1990, 248, 1124–1126. [Google Scholar] [CrossRef]
- Rogaev, E.; Sherrington, R.; Rogaeva, E.A.; Levesque, G.; Ikeda, M.; Liang, Y.; Chi, H.; Lin, C.; Holman, K.; Tsuda, T.; et al. Familial Alzheimer’s Disease in Kindreds with Missense Mutations in a Gene on Chromosome 1 Related to the Alzheimer’s Disease Type 3 Gene. Nature 1995, 376, 775–778. [Google Scholar] [CrossRef]
- Genin, E.; Hannequin, D.; Wallon, D.; Sleegers, K.; Hiltunen, M.; Combarros, O.; Bullido, M.; Engelborghs, S.; Paul, D.; Berr, C.; et al. APOE and Alzheimer Disease: A Major Gene with Semi-Dominant Inheritance. Mol. Psychiatry 2011, 16, 903–907. [Google Scholar] [CrossRef]
- McKay, G.J.; Silvestri, G.; Chakravarthy, U.; Dasari, S.; Fritsche, L.G.; Weber, B.H.; Keilhauer, C.N.; Klein, M.L.; Francis, P.J.; Klaver, C.C.; et al. Variations in Apolipoprotein E Frequency with Age in a Pooled Analysis of a Large Group of Older People. Am. J. Epidemiol. 2011, 173, 1357–1364. [Google Scholar] [CrossRef]
- Martínez-Magaña, J.; Genis-Mendoza, A.; Tovilla-Zarate, C.A.; González-Castro, T.; Juárez-Rojop, I.; Hernández-Díaz, Y.; Martínez-H, A.; García-Ortiz, H.; Orozco, L.; Narváez, L.; et al. Association between APOE Polymorphisms and Lipid Profile in Mexican Amerindian Population. Mol. Genet. Genom. Med. 2019, 7, e958. [Google Scholar] [CrossRef]
- Keeney, J.; Ibrahimi, S.; Zhao, L. Human ApoE Isoforms Differentially Modulate Glucose and Amyloid Metabolic Pathways in Female Brain: Evidence of the Mechanism of Neuroprotection by ApoE2 and Implications for Alzheimer’s Disease Prevention and Early Intervention. J. Alzheimers. Dis. 2015, 48, 411–424. [Google Scholar] [CrossRef]
- Escott-Price, V.; Shoai, M.; Pither, R.; Williams, J.; Hardy, J. Polygenic Score Prediction Captures Nearly All Common Genetic Risk for Alzheimer’s Disease. Neurobiol. Aging 2016, 49, 214.e7–214.e11. [Google Scholar] [CrossRef] [PubMed]
- Gatz, M.; Reynolds, C.; Fratiglioni, L.; Johansson, B.; Mortimer, J.; Berg, S.; Fiske, A.; Pedersen, N. Role of Genes and Environments for Explaining Alzheimer Disease. Arch. Gen. Psychiatry 2006, 63, 168–174. [Google Scholar] [CrossRef]
- Zhang, S.; Cai, F.; Wu, Y.; Bozorgmehr, T.; Wang, Z.; Zhang, S.; Huang, D. A Presenilin-1 Mutation Causes Alzheimer Disease without Affecting Notch Signaling. Mol. Psychiatry 2020, 25, 603–613. [Google Scholar] [CrossRef]
- Barber, I.; Braae, A.; Clement, N.; Patel, T.; Guetta-Baranes, T.; Brookes, K.-J.; Medway, C.; Chappell, S.; Guerreiro, R.; Bras, J.; et al. Mutation Analysis of Sporadic Early-Onset Alzheimer’s Disease Using the NeuroX Array. Neurobiol. Aging 2016, 49, 215.e1–215.e8. [Google Scholar] [CrossRef]
- Lanoiselee, H.; Nicolas, G.; Wallon, D.; Rovelet-Lecrux, A.; Lacour, M.; Rousseau, S.; Richard, A.-C.; Pasquier, F.; Rollin-Sillaire, A.; Martinaud, O.; et al. APP, PSEN1, and PSEN2 Mutations in Early-Onset Alzheimer Disease: A Genetic Screening Study of Familial and Sporadic Cases. PLoS Med. 2017, 14, e1002270. [Google Scholar] [CrossRef]
- Giaccone, G.; Morbin, M.; Moda, F.; Botta, M.; Mazzoleni, G.; Uggetti, A.; Catania, M.; Redaelli, V.; Spagnoli, A.; Rossi, R.; et al. Neuropathology of the Recessive A673V APP Mutation: Alzheimer Disease with Distinctive Features. Acta Neuropathol. 2010, 120, 803–812. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Z.; Cai, F.; Zhang, M.; Wu, Y.; Zhang, J.; Song, W. BACE1 Cleavage Site Selection Critical for Amyloidogenesis and Alzheimer’s Pathogenesis. J. Neurosci. 2017, 37, 317–340. [Google Scholar] [CrossRef]
- Holler, C.J.; Davis, P.R.; Beckett, T.L.; Platt, T.L.; Webb, R.L.; Head, E.; Murphy, M.P. Bridging Integrator 1 (BIN1) Protein Expression Increases in the Alzheimer’s Disease Brain and Correlates with Neurofibrillary Tangle Pathology. J. Alzheimers. Dis. 2014, 42, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Franzmeier, N.; Rubinski, A.; Neitzel, J.; Ewers, M.; Weiner, M.W.; Aisen, P.; Petersen, R.; Jack, C.R.; Jagust, W.; Trojanowski, J.Q.; et al. The BIN1 Rs744373 SNP Is Associated with Increased Tau-PET Levels and Impaired Memory. Nat. Commun. 2019, 10, 1766. [Google Scholar] [CrossRef] [PubMed]
- Chapuis, J.; Hansmannel, F.; Gistelinck, M.; Mounier, A.; Van Cauwenberghe, C.; Kolen, K.; Geller, F.; Sottejeau, Y.; Harold, D.; Dourlen, P.; et al. Increased Expression of BIN1 Mediates Alzheimer Genetic Risk by Modulating Tau Pathology. Mol. Psychiatry 2013, 18, 1225–1234. [Google Scholar] [CrossRef]
- Engelhart, M.; Geerlings, M.; Meijer, J.; Kiliaan, A.; Ruitenberg, A.; Swieten, J.; Stijnen, T.; Hofman, A.; Witteman, J.; Breteler, M. Inflammatory Proteins in Plasma and the Risk of Dementia: The Rotterdam Study. Arch. Neurol. 2004, 61, 668–672. [Google Scholar] [CrossRef]
- Blalock, E.; Buechel, H.; Popovic, J.; Geddes, J.; Landfield, P. Microarray Analyses of Laser-Captured Hippocampus Reveal Distinct Gray and White Matter Signatures Associated with Incipient Alzheimer’s Disease. J. Chem. Neuroanat. 2011, 42, 118–126. [Google Scholar] [CrossRef]
- Padmanabhan, J.; Judge, M.; Dickson, D.; Potter, H. α 1-Antichymotrypsin, an Inflammatory Protein Overexpressed in Alzheimer’s Disease Brain, Induces Tau Phosphorylation in Neurons. Brain 2006, 129, 3020–3034. [Google Scholar] [CrossRef]
- Kamboh, M.I.; Minster, R.L.; Kenney, M.; Ozturk, A.; Desai, P.P.; Kammerer, C.M.; DeKosky, S.T. α-1-Antichymotrypsin (ACT or SERPINA3) Polymorphism May Affect Age-at-Onset and Disease Duration of Alzheimer’s Disease. Neurobiol. Aging 2006, 27, 1435–1439. [Google Scholar] [CrossRef][Green Version]
- Nilsson, L.N.; Bales, K.R.; DiCarlo, G.; Gordon, M.N.; Morgan, D.; Paul, S.M.; Potter, H. α-1-Antichymotrypsin Promotes β-Sheet Amyloid Plaque Deposition in a Transgenic Mouse Model of Alzheimer’s Disease. J. Neurosci. 2001, 21, 1444–1451. [Google Scholar] [CrossRef]
- Xiao, Q.; Gil, S.-C.; Yan, P.; Wang, Y.; Han, S.; Gonzales, E.; Perez, R.; Cirrito, J.; Lee, J.-M. Role of Phosphatidylinositol Clathrin Assembly Lymphoid-Myeloid Leukemia (PICALM) in Intracellular Amyloid Precursor Protein (APP) Processing and Amyloid Plaque Pathogenesis. J. Biol. Chem. 2012, 287, 21279–21289. [Google Scholar] [CrossRef]
- Zhou, Q.; Peng, D.; Yuan, X.; Lv, Z.; Pang, S.; Jiang, W.; Yang, C.; Shi, X.; Pang, G.; Yang, Y.; et al. APOE and APOC1 Gene Polymorphisms Are Associated with Cognitive Impairment Progression in Chinese Patients with Late-Onset Alzheimer’s Disease. Neural Regen. Res. 2014, 9, 653–660. [Google Scholar] [CrossRef]
- Cuccaro, M.; Carney, R.; Zhang, Y.; Bohm, C.; Kunkle, B.; Vardarajan, B.; Whitehead, P.; Cukier, H.; Mayeux, R.; George-Hyslop, P.; et al. SORL1 Mutations in Early- and Late-Onset Alzheimer Disease. Neurol. Genet. 2016, 2, e116. [Google Scholar] [CrossRef] [PubMed]
- Vardarajan, B.; Zhang, Y.; Lee, J.; Cheng, R.; Bohm, C.; Ghani, M.; Reitz, C.; Reyes-Dumeyer, D.; Shen, Y.; Rogaeva, E.; et al. Coding Mutations in SORL1 and Alzheimer Disease. Ann. Neurol. 2014, 77, 215–227. [Google Scholar] [CrossRef]
- Li, X.; Zhu, X.; Zhang, W.; Yang, F.; Hui, J.; Tan, J.; Xie, H.; Peng, D.; Ma, L.; Cui, L.; et al. The Etiological Effect of a New Low-Frequency ESR1 Variant on Mild Cognitive Impairment and Alzheimer’s Disease: A Population-Based Study. Aging 2018, 10, 2316–2337. [Google Scholar] [CrossRef]
- Naj, A.C.; Jun, G.; Beecham, G.W.; Wang, L.-S.; Vardarajan, B.N.; Buros, J.; Gallins, P.J.; Buxbaum, J.D.; Jarvik, G.P.; Crane, P.K.; et al. Common Variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 Are Associated with Late-Onset Alzheimer’s Disease. Nat. Genet. 2011, 43, 436–441. [Google Scholar] [CrossRef]
- Lacour, A.; Espinosa, A.; Louwersheimer, E.; Heilmann, S.; Hernández, I.; Wolfsgruber, S.; Fernández, V.; Wagner, H.; Rosende-Roca, M.; Mauleón, A.; et al. Genome-Wide Significant Risk Factors for Alzheimer’s Disease: Role in Progression to Dementia Due to Alzheimer’s Disease among Subjects with Mild Cognitive Impairment. Mol. Psychiatry 2017, 22, 153–160. [Google Scholar] [CrossRef]
- Kang, S.; Kurti, A.; Wojtas, A.; Baker, K.; Liu, C.-C.; Kanekiyo, T.; Deming, Y.; Cruchaga, C.; Estus, S.; Bu, G.; et al. Identification of Plexin A4 as a Novel Clusterin Receptor Links Two Alzheimer’s Disease Risk Genes. Hum. Mol. Genet. 2016, 25, ddw188. [Google Scholar] [CrossRef]
- Tan, L.; Wang, H.-F.; Tan, M.-S.; Tan, C.-C.; Zhu, X.-C.; Miao, D.; Yu, W.-J.; Jiang, T.; Tan, L.; Yu, J.-T.; et al. Effect of CLU Genetic Variants on Cerebrospinal Fluid and Neuroimaging Markers in Healthy, Mild Cognitive Impairment and Alzheimer’s Disease Cohorts. Sci. Rep. 2016, 6, 26027. [Google Scholar] [CrossRef] [PubMed]
- Bailey, C.; DeVaux, L.; Farzan, M. The Triggering Receptor Expressed on Myeloid Cells 2 Binds Apolipoprotein E. J. Biol. Chem. 2015, 290, 26033–26042. [Google Scholar] [CrossRef] [PubMed]
- Hoeijmakers, L.; Ruigrok, S.R.; Amelianchik, A.; Ivan, D.; van Dam, A.-M.; Lucassen, P.J.; Korosi, A. Early-Life Stress Lastingly Alters the Neuroinflammatory Response to Amyloid Pathology in an Alzheimer’s Disease Mouse Model. Brain. Behav. Immun. 2017, 63, 160–175. [Google Scholar] [CrossRef]
- Foster, E.M.; Dangla-Valls, A.; Lovestone, S.; Ribe, E.M.; Buckley, N.J. Clusterin in Alzheimer’s Disease: Mechanisms, Genetics, and Lessons From Other Pathologies. Front. Neurosci. 2019, 13, 164. [Google Scholar] [CrossRef] [PubMed]
- Kumita, J.; Poon, S.; Caddy, G.; Hagan, C.; Dumoulin, M.; Yerbury, J.; Stewart, E.; Robinson, C.; Wilson, M.; Dobson, C. The Extracellular Chaperone Clusterin Potently Inhibits Human Lysozyme Amyloid Formation by Interacting with Prefibrillar Species. J. Mol. Biol. 2007, 369, 157–167. [Google Scholar] [CrossRef][Green Version]
- Wojtas, A.M.; Kang, S.S.; Olley, B.M.; Gatherer, M.; Shinohara, M.; Lozano, P.A.; Liu, C.-C.; Kurti, A.; Baker, K.E.; Dickson, D.W.; et al. Loss of Clusterin Shifts Amyloid Deposition to the Cerebrovasculature via Disruption of Perivascular Drainage Pathways. Proc. Natl. Acad. Sci. USA 2017, 114, E6962–E6971. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, T.; Stefansson, H.; Steinberg, S.; Jonsdottir, I.; Jonsson, P.; Snaedal, J.; Bjornsson, S.; Huttenlocher, J.; Levey, A.; Lah, J.; et al. Variant of TREM2 Associated with the Risk of Alzheimer’s Disease. N. Engl. J. Med. 2013, 368, 107–116. [Google Scholar] [CrossRef]
- Slattery, C.F.; Beck, J.A.; Harper, L.; Adamson, G.; Abdi, Z.; Uphill, J.; Campbell, T.; Druyeh, R.; Mahoney, C.J.; Rohrer, J.D.; et al. R47H TREM2 Variant Increases Risk of Typical Early-Onset Alzheimer’s Disease but Not of Prion or Frontotemporal Dementia. Alzheimer’s Dement. 2014, 10, 602–608.e4. [Google Scholar] [CrossRef]
- Ulrich, J.; Finn, M.; Wang, Y.; Shen, A.; Mahan, T.; Jiang, H.; Stewart, F.; Piccio, L.; Colonna, M.; Holtzman, D. Altered Microglial Response to Aβ Plaques in APPPS1-21 Mice Heterozygous for TREM2. Mol. Neurodegener. 2014, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- McQuade, A.; Kang, Y.; Hasselmann, J.; Jairaman, A.; Sotelo, A.; Coburn, M.; Kiani Shabestari, S.; Chadarevian, J.; Fote, G.; Tu, C.; et al. Gene Expression and Functional Deficits Underlie TREM2-Knockout Microglia Responses in Human Models of Alzheimer’s Disease. Nat. Commun. 2020, 11, 5370. [Google Scholar] [CrossRef]
- Lill, C.; Rengmark, A.; Pihlstrøm, L.; Fogh, I.; Shatunov, A.; Sleiman, P.; Wang, L.-S.; Liu, T.; Funch Lassen, C.; Meissner, E.; et al. The Role of TREM2 R47H as a Risk Factor for Alzheimer’s Disease, Frontotemporal Lobar Degeneration, Amyotrophic Lateral Sclerosis, and Parkinson’s Disease. Alzheimers. Dement. 2015, 11, 1407–1416. [Google Scholar] [CrossRef]
- Atagi, Y.; Liu, C.-C.; Painter, M.M.; Chen, X.-F.; Verbeeck, C.; Zheng, H.; Li, X.; Rademakers, R.; Kang, S.S.; Xu, H.; et al. Apolipoprotein E Is a Ligand for Triggering Receptor Expressed on Myeloid Cells 2 (TREM2). J. Biol. Chem. 2015, 290, 26043–26050. [Google Scholar] [CrossRef]
- Vo, G.; Bagyinszky, E.; Yang, Y.; Youn, Y.C.; An, S.; Kim, S. Genetic Analyses of Early-Onset Alzheimer’s Disease Using next-Generation Sequencing. Sci. Rep. 2019, 9, 8368. [Google Scholar] [CrossRef]
- Cochran, J.; Rush, T.; Buckingham, S.; Roberson, E. The Alzheimer’s Disease Risk Factor CD2AP Maintains Blood-Brain Barrier Integrity. Hum. Mol. Genet. 2015, 24, 6667–6674. [Google Scholar] [CrossRef]
- Tao, Q.-Q.; Zhijun, L.; Sun, Y.-M.; Li, H.-L.; Yang, P.; Jiang, B.; Li, X.-Y.; Xu, J.-F.; Wu, Z.-Y. Decreased Gene Expression of CD2AP in Chinese Patients with Sporadic Alzheimer’s Disease. Neurobiol. Aging 2017, 56, 212.e5–212.e10. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zhao, A.; Qui, Y.; Li, Y.; Yan, R.; Wang, Y.; Xu, W.; Deng, Y. Genetic Association of FERMT2, HLA-DRB1, CD2AP, and PTK2B Polymorphisms With Alzheimer’s Disease Risk in the Southern Chinese Population. Front. Aging Neurosci. 2020, 12, 16. [Google Scholar] [CrossRef]
- Myers, A.; Pittman, A.; Zhao, A.; Rohrer, K.; Kaleem, M.; Marlowe, L.; Lees, A.; Leung, D.; Mckeith, I.; Perry, R.; et al. The MAPT H1c Risk Haplotype Is Associated with Incresed Expression of Tau and Especially of 4 Repeat Containing Transcripts. Neurobiol. Dis. 2007, 25, 561–570. [Google Scholar] [CrossRef]
- Abraham, R.; Sims, R.; Carroll, L.; Hollingworth, P.; O’Donovan, M.; Williams, J.; Owen, M. An Association Study of Common Variation at the MAPT Locus With Late-Onset Alzheimer’s Disease. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009, 150B, 1152–1155. [Google Scholar] [CrossRef]
- Allen, M.; Kachadoorian, M.; Quicksall, Z.; Zou, F.; Chai, H.; Younkin, C.; Crook, J.; Pankratz, V.; Carrasquillo, M.; Krishnan, S.; et al. Association of MAPT Haplotypes with Alzheimer’s Disease Risk and MAPT Brain Gene Expression Levels. Alzheimers. Res. Ther. 2014, 6, 39. [Google Scholar] [CrossRef]
- Lee, E.-G.; Chen, S.; Leong, L.; Tulloch, J.; Yu, C.-E. TOMM40 RNA Transcription in Alzheimer’s Disease Brain and Its Implication in Mitochondrial Dysfunction. Genes 2021, 12, 871. [Google Scholar] [CrossRef]
- Chiba-Falek, O.; Gottschalk, W.K.; Lutz, M.W. The Effects of the TOMM40 Poly-T Alleles on Alzheimer’s Disease Phenotypes. Alzheimers. Dement. 2018, 14, 692–698. [Google Scholar] [CrossRef]
- Soyal, S.M.; Kwik, M.; Kalev, O.; Lenz, S.; Zara, G.; Strasser, P.; Patsch, W.; Weis, S. A TOMM40/APOE Allele Encoding APOE-E3 Predicts High Likelihood of Late-Onset Alzheimer’s Disease in Autopsy Cases. Mol. Genet. Genom. Med. 2020, 8, e1317. [Google Scholar] [CrossRef]
- Nelson, P.; Alafuzoff, I.; Bigio, E.; Bouras, C.; Braak, H.; Cairns, N.; Castellani, R.; Crain, B.; Davies, P.; Del Tredici, K.; et al. Correlation of Alzheimer Disease Neuropathologic Changes With Cognitive Status. J. Neuropathol. Exp. Neurol. 2012, 71, 362–381. [Google Scholar] [CrossRef]
- Vassar, R. ADAM10 Prodomain Mutations Cause Late-Onset Alzheimer’s Disease: Not Just the Latest FAD. Neuron 2013, 80, 250–253. [Google Scholar] [CrossRef]
- Lambert, J.-C.; Ibrahim-Verbaas, C.; Harold, D.; Naj, A.; Sims, R.; Bellenguez, C.; Jun, G.; DeStefano, A.; Bis, J.; Beecham, G.; et al. Meta-Analysis of 74,046 Individuals Identifies 11 New Susceptibility Loci for Alzheimer’s Disease. Nat. Genet. 2013, 45, 1452–1458. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shen, N.; Zhang, S.; Liu, J.; Jiang, Q.; Liao, M.; Feng, R.; Zhang, L.; Wang, G.; Ma, G.; et al. CD33 Rs3865444 Polymorphism Contributes to Alzheimer’s Disease Susceptibility in Chinese, European, and North American Populations. Mol. Neurobiol. 2015, 52, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.; Whetzel, A.; Serrano, G.; Sue, L.; Beach, T.; Lue, L.-F. Association of CD33 Polymorphism Rs3865444 with Alzheimer’s Disease Pathology and CD33 Expression in Human Cerebral Cortex. Neurobiol. Aging 2014, 36, 571–582. [Google Scholar] [CrossRef]
- Bertram, L.; Lange, C.; Mullin, K.; Parkinson, M.; Hsiao, M.; Hogan, M.; Schjeide, B.; Hooli, B.; Divito, J.; Ionita, I.; et al. Genome-Wide Association Analysis Reveals Putative Alzheimer’s Disease Susceptibility Loci in Addition to APOE. Am. J. Hum. Genet. 2008, 83, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Wijsman, E.; Pankratz, N.; Choi, Y.; Rothstein, J.; Faber, K.; Cheng, R.; Lee, J.; Bird, T.; Bennett, D.; Diaz-Arrastia, R.; et al. Genome-Wide Asociation of Familial Late-Onset Alzheimer’s Disease Replicates BIN1 and CLU and Nominates CUGBP2 in Interaction with APOE. PLoS Genet. 2011, 7, e1001308. [Google Scholar] [CrossRef]
- Jun, G.; Asai, H.; Zeldich, E.; Drapeau, E.; Chen, C.; Chung, J.; Park, J.; Kim, S.; Haroutunian, V.; Foroud, T.; et al. PLXNA4 Is Associated with Alzheimer Disease and Modulates Tau Phosphorylation. Ann. Neurol. 2014, 76, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Ebbert, M.T.W.; Boehme, K.L.; Wadsworth, M.E.; Staley, L.A.; Initiative, A.D.N.; Consortium, A.D.G.; Mukherjee, S.; Crane, P.K.; Ridge, P.G.; Kauwe, J.S.K. Interaction between Variants in CLU and MS4A4E Modulates Alzheimer’s Disease Risk. Alzheimers. Dement. 2016, 12, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Chan, G.; White, C.; Winn, P.; Cimpean, M.; Replogle, J.; Glick, L.; Cuerdon, N.; Ryan, K.; Johnson, K.; Schneider, J.; et al. CD33 Modulates TREM2: Convergence of Alzheimer Loci. Nat. Neurosci. 2015, 18, 1556–1558. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.-H.; Bi, Y.-L.; Tan, M.-S.; Xu, W.; Li, J.-Q.; Shen, X.-N.; Dou, K.-X.; Tan, C.-C.; Tan, L.; Yu, J.-T. Genome-Wide Association Study Identifies Alzheimer’s Risk Variant in MS4A6A Influencing Cerebrospinal Fluid STREM2 Levels. Neurobiol. Aging 2019, 84, 241.e13–241.e20. [Google Scholar] [CrossRef] [PubMed]
- Griciuc, A.; Patel, S.; Federico, A.; Choi, S.H.; Innes, B.; Oram, M.; Cereghetti, G.; McGinty, D.; Anselmo, A.; Sadreyev, R.; et al. TREM2 Acts Downstream of CD33 in Modulating Microglial Pathology in Alzheimer’s Disease. Neuron 2019, 103, 820–835.e7. [Google Scholar] [CrossRef]
- Wißfeld, J.; Mathews, M.; Mossad, O.; Picardi, P.; Cinti, A.; Redaelli, L.; Pradier, L.; Brüstle, O.; Neumann, H. Reporter Cell Assay for Human CD33 Validated by Specific Antibodies and Human IPSC-Derived Microglia. Sci. Rep. 2021, 11, 13462. [Google Scholar] [CrossRef] [PubMed]
- Goñi, J.; Esteban, F.; Velez de Mendizabal, N.; Sepulcre, J.; Ardanza-Trevijano, S.; Agirrezabal, I.; Villoslada, P. A Computational Analysis of Protein-Protein Interaction Networks in Neurodegenerative Diseases. BMC Syst. Biol. 2008, 2, 52. [Google Scholar] [CrossRef] [PubMed]
- Marín, M.; Esteban, F.; Ramirez, H.; Ros, E.; Saez-Lara, M. An Integrative Methodology Based on Protein-Protein Interaction Networks for Identification and Functional Annotation of Disease-Relevant Genes Applied to Channelopathies. BMC Bioinform. 2019, 20, 565. [Google Scholar] [CrossRef]
- Meng, X.; Li, J.; Zhang, Q.; Chen, F.; Bian, C.; Yao, X.; Yan, J.; Xu, Z.; Risacher, S.; Saykin, A.; et al. Multivariate Genome Wide Association and Network Analysis of Subcortical Imaging Phenotypes in Alzheimer’s Disease. BMC Genom. 2020, 21, 896. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Fang, K.; Wang, W.; Lin, W.; Guo, L.; Wang, J. Identification of Key Genes and Pathways for Alzheimer’s Disease via Combined Analysis of Genome-Wide Expression Profiling in the Hippocampus. Biophys. Rep. 2019, 5, 98–109. [Google Scholar] [CrossRef]
- Chung, J.; Jun, G.; Dupuis, J.; Farrer, L. Comparison of Methods for Multivariate Gene-Based Association Tests for Complex Diseases Using Common Variants. Eur. J. Hum. Genet. 2019, 27, 811–823. [Google Scholar] [CrossRef]
- Ziller, M.J.; Gu, H.; Müller, F.; Donaghey, J.; Tsai, L.T.-Y.; Kohlbacher, O.; De Jager, P.L.; Rosen, E.D.; Bennett, D.A.; Bernstein, B.E.; et al. Charting a Dynamic DNA Methylation Landscape of the Human Genome. Nature 2013, 500, 477–481. [Google Scholar] [CrossRef]
- John, R.; Rougeulle, C. Developmental Epigenetics: Phenotype and the Flexible Epigenome. Front. Cell Dev. Biol. 2018, 6, 130. [Google Scholar] [CrossRef]
- Choy, M.-K.; Movassagh, M.; Goh, H.G.; Bennett, M.; Down, T.; Foo, R. Genome-Wide Conserved Consensus Transcription Factor Binding Motifs Are Hyper-Methylated. BMC Genom. 2010, 11, 519. [Google Scholar] [CrossRef]
- Choy, M.-K.; Movassagh, M.; Foo, R. The Human Variome: Genomic and Epigenomic Diversity. EMBO Mol. Med. 2011, 3, 573–574. [Google Scholar] [CrossRef] [PubMed]
- Koch, L. Altered Splicing in Alzheimer Transcriptomes. Nat. Rev. Genet. 2018, 19, 738–739. [Google Scholar] [CrossRef]
- Wang, F.; Yang, Y.-Z.; Shi, C.-Z.; Zhang, P.; Moyer, M.P.; Zhang, H.-Z.; Zou, Y.; Qin, H.-L. UHRF1 Promotes Cell Growth and Metastasis Through Repression of P16ink4a in Colorectal Cancer. Ann. Surg. Oncol. 2012, 19, 2753–2762. [Google Scholar] [CrossRef]
- Gasparoni, G.; Bultmann, S.; Lutsik, P.; Kraus, T.; Sordon, S.; Vlcek, J.; Dietinger, V.; Steinmaurer, M.; Haider, M.; Mulholland, C.; et al. DNA Methylation Analysis on Purified Neurons and Glia Dissects Age and Alzheimer’s Disease-Specific Changes in the Human Cortex. Epigenetics Chromatin 2018, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Aristizabal, M.; Anreiter, I.; Halldorsdottir, T.; Odgers, C.; Mcdade, T.; Goldenberg, A.; Mostafavi, S.; Kobor, M.; Binder, E.; Sokolowski, M.; et al. Biological Embedding of Experience: A Primer on Epigenetics. Proc. Natl. Acad. Sci. USA 2019, 117, 23261–23269. [Google Scholar] [CrossRef]
- Li, S.; Nguyen, T.; Wong, E.; Dugué, P.-A.; Dite, G.; Armstrong, N.; Craig, J.; Mather, K.; Sachdev, P.; Saffery, R.; et al. Genetic and Environmental Causes of Variation in Epigenetic Aging across the Lifespan. Clin. Epigenetics 2020, 12, 158. [Google Scholar] [CrossRef] [PubMed]
- Mcdaniell, R.; Lee, B.-K.; Song, L.; Liu, Z.; Boyle, A.; Erdos, M.; Scott, L.; Morken, M.; Kucera, K.; Battenhouse, A.; et al. Heritable Individual-Specific and Allele-Specific Chromatin Signatures in Humans. Science 2010, 328, 235–239. [Google Scholar] [CrossRef]
- Griñán-Ferré, C.; Izquierdo, V.; Otero, E.; Puigoriol-Illamola, D.; Corpas, R.; Sanfeliu, C.; Ortuño-Sahagún, D.; Pallàs, M. Environmental Enrichment Improves Cognitive Deficits, AD Hallmarks and Epigenetic Alterations Presented in 5xFAD Mouse Model. Front. Cell. Neurosci. 2018, 12, 224. [Google Scholar] [CrossRef] [PubMed]
- Watson, C.; Roussos, P.; Garg, P.; Ho, D.; Azam, N.; Katsel, P.; Haroutunian, V.; Sharp, A. Genome-Wide DNA Methylation Profiling in the Superior Temporal Gyrus Reveals Epigenetic Signatures Associated with Alzheimer’s Disease. Genome Med. 2016, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Tsang, V.; Fry, R.; Niculescu, M.; Rager, J.; Saunders, J.; Paul, D.; Zeisel, S.; Waalkes, M.; Stýblo, M.; Drobná, Z. The Epigenetic Effects of a High Prenatal Folate Intake in Male Mouse Fetuses Exposed In Utero to Arsenic. Toxicol. Appl. Pharmacol. 2012, 264, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Zeilinger, S.; Kühnel, B.; Klopp, N.; Baurecht, H.; Kleinschmidt, A.; Gieger, C.; Weidinger, S.; Lattka, E.; Adamski, J.; Peters, A.; et al. Tobacco Smoking Leads to Extensive Genome-Wide Changes in DNA Methylation. PLoS ONE 2013, 8, e63812. [Google Scholar] [CrossRef] [PubMed]
- Ambatipudi, S.; Cuenin, C.; Hernández-Vargas, H.; Ghantous, A.; Calvez-Kelm, F.; Kaaks, R.; Barrdahl, M.; Boeing, H.; Aleksandrova, K.; Trichopoulou, A.; et al. Tobacco Smoking-Associated Genome-Wide DNA Methylation Changes in the EPIC Study. Epigenomics 2016, 8, 599–618. [Google Scholar] [CrossRef]
- Yoon, H.-G.; Chan, D.; Reynolds, A.; Qin, J.; Wong, J. N-CoR Mediates DNA Methylation-Dependent Repression through a Methyl CpG Binding Protein Kaiso. Mol. Cell 2003, 12, 723–734. [Google Scholar] [CrossRef]
- Rottach, A.; Frauer, C.; Pichler, G.; Bonapace, I.; Spada, F.; Leonhardt, H. The Multi-Domain Protein Np95 Connects DNA Methylation and Histone Modification. Nucleic Acids Res. 2009, 38, 1796–1804. [Google Scholar] [CrossRef] [PubMed]
- Berdasco, M.; Esteller, M. Genetic Syndromes Caused by Mutations in Epigenetic Genes. Hum. Genet. 2013, 132, 359–383. [Google Scholar] [CrossRef]
- Schröder, C.; Leitão, E.; Wallner, S.; Schmitz, G.; Klein-Hitpass, L.; Sinha, A.; Jöckel, K.-H.; Heilmann, S.; Hoffmann, P.; Nöthen, M.; et al. Regions of Common Inter-Individual DNA Methylation Differences in Human Monocytes: Genetic Basis and Potential Function. Epigenetics Chromatin 2017, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.; Christensen, J.; Pedersen, M.T.; Johansen, J.V.; Cloos, P.A.C.; Rappsilber, J.; Helin, K. TET1 and Hydroxymethylcytosine in Transcription and DNA Methylation Fidelity. Nature 2011, 473, 343–348. [Google Scholar] [CrossRef]
- Ficz, G.; Branco, M.R.; Seisenberger, S.; Santos, F.; Krueger, F.; Hore, T.A.; Marques, C.J.; Andrews, S.; Reik, W. Dynamic Regulation of 5-Hydroxymethylcytosine in Mouse ES Cells and during Differentiation. Nature 2011, 473, 398–402. [Google Scholar] [CrossRef]
- Kuehner, J.N.; Chen, J.; Bruggeman, E.C.; Wang, F.; Li, Y.; Xu, C.; McEachin, Z.T.; Li, Z.; Chen, L.; Hales, C.M.; et al. 5-Hydroxymethylcytosine Is Dynamically Regulated during Forebrain Organoid Development and Aberrantly Altered in Alzheimer’s Disease. Cell Rep. 2021, 35, 109042. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z.; Li, L.; Xu, K.; Ma, Z.; Chow, H.-M.; Herrup, K.; Li, J. Selective Loss of 5hmC Promotes Neurodegeneration in the Mouse Model of Alzheimer’s Disease. FASEB J. 2020, 34, 16364–16382. [Google Scholar] [CrossRef]
- Davies, M.; Volta, M.; Pidsley, R.; Lunnon, K.; Dixit, A.; Lovestone, S.; Coarfa, C.; Harris, R.; Milosavljevic, A.; Troakes, C.; et al. Functional Annotation of the Human Brain Methylome Identifies Tissue-Specific Epigenetic Variation across Brain and Blood. Genome Biol. 2012, 13, R43. [Google Scholar] [CrossRef] [PubMed]
- Chouliaras, L.; Mastroeni, D.; Delvaux, E.; Grover, A.; Kenis, G.; Hof, P.; Steinbusch, H.; Coleman, P.; Rutten, B.; Van den Hove, D. Consistent Decrease in Global DNA Methylation and Hydroxymethylation in the Hippocampus of Alzheimer’s Disease Patients. Neurobiol. Aging 2013, 34, 2091–2099. [Google Scholar] [CrossRef]
- Coppieters, N.; Dieriks, B.; Lill, C.; Faull, R.; Curtis, M.; Dragunow, M. Global Changes in DNA Methylation and Hydroxymethylation in Alzheimer’s Disease Human Brain. Neurobiol. Aging 2013, 35, 1334–1344. [Google Scholar] [CrossRef]
- Lord, J.; Cruchaga, C. The Epigenetic Landscape of Alzheimer’s Disease. Nat. Neurosci. 2014, 17, 1138–1140. [Google Scholar] [CrossRef]
- De Jager, P.L.; Srivastava, G.; Lunnon, K.; Burgess, J.; Schalkwyk, L.C.; Yu, L.; Eaton, M.L.; Keenan, B.T.; Ernst, J.; McCabe, C.; et al. Alzheimer’s Disease: Early Alterations in Brain DNA Methylation at ANK1, BIN1, RHBDF2 and Other Loci. Nat. Neurosci. 2014, 17, 1156–1163. [Google Scholar] [CrossRef] [PubMed]
- Lunnon, K.; Smith, R.; Hannon, E.; Lee, P.; Srivastava, G.; Volta, M.; Troakes, C.; Al-Sarraj, S.; Burrage, J.; Macdonald, R.; et al. Methylomic Profiling Implicates Cortical Deregulation of ANK1 in Alzheimer’s Disease. Nat. Neurosci. 2014, 17, 1164–1170. [Google Scholar] [CrossRef] [PubMed]
- Bakulski, K.M.; Dolinoy, D.C.; Sartor, M.A.; Paulson, H.L.; Konen, J.R.; Lieberman, A.P.; Albin, R.L.; Hu, H.; Rozek, L.S. Genome-Wide DNA Methylation Differences between Late-Onset Alzheimer’s Disease and Cognitively Normal Controls in Human Frontal Cortex. J. Alzheimers. Dis. 2012, 29, 571–588. [Google Scholar] [CrossRef]
- Mulder, C.; Schoonenboom, N.; Jansen, E.; Verhoeven, N.; Kamp, G.; Jakobs, C.; Scheltens, P. The Transmethylation Cycle in the Brain of Alzheimer Patients. Neurosci. Lett. 2005, 386, 69–71. [Google Scholar] [CrossRef] [PubMed]
- Lashley, T.; Gami-Patel, P.; Valizadeh, N.; Li, A.; Revesz, T.; Balazs, R. Alterations in Global DNA Methylation and Hydroxymethylation Are Not Detected in Alzheimer’s Disease: Global DNA Methylation in AD. Neuropathol. Appl. Neurobiol. 2014, 41, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Iwata, A.; Nagata, K.; Hatsuta, H.; Takuma, H.; Bundo, M.; Iwamoto, K.; Tamaoka, A.; Murayama, S.; Saido, T.; Tsuji, S. Altered CpG Methylation in Sporadic Alzheimer’s Disease Is Associated with APP and MAPT Dysregulation. Hum. Mol. Genet. 2014, 23, 648–656. [Google Scholar] [CrossRef]
- Foraker, J.; Millard, S.P.; Leong, L.; Thomson, Z.; Chen, S.; Keene, C.D.; Bekris, L.M.; Yu, C.-E. The APOE Gene Is Differentially Methylated in Alzheimer’s Disease. J. Alzheimers. Dis. 2015, 48, 745–755. [Google Scholar] [CrossRef]
- Yu, L.; Chibnik, L.; Srivastava, G.; Pochet, N.; Yang, J.; Xu, J.; Kozubek, J.; Obholzer, N.; Leurgans, S.; Schneider, J.; et al. Association of Brain DNA Methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with Pathological Diagnosis of Alzheimer Disease. JAMA Neurol. 2014, 72, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Tan, L.; Bi, Y.-L.; Xu, W.; Tan, L.; Shen, X.-N.; Hou, X.-H.; Ma, Y.-H.; Dong, Q.; Yu, J.-T. Association between Methylation of BIN1 Promoter in Peripheral Blood and Preclinical Alzheimer’s Disease. Transl. Psychiatry 2021, 11, 89. [Google Scholar] [CrossRef] [PubMed]
- Satoh, K.; Abe-Dohmae, S.; Yokoyama, S.; George-Hyslop, P.; Fraser, P. ATP-Binding Cassette Transporter A7 (ABCA7) Effects on Amyloid Processing and Relevance to Alzheimer’s Disease. Alzheimer’s Dement. 2012, 8, P473. [Google Scholar] [CrossRef]
- Smith, R.; Hannon, E.; Lee, P.; Chibnik, L.; Lott, S.; Condliffe, D.; Smith, A.; Haroutunian, V.; Troakes, C.; Al-Sarraj, S.; et al. Elevated DNA Methylation across a 48-Kb Region Spanning the HOXA Gene Cluster Is Associated with Alzheimer’s Disease Neuropathology. Alzheimer’s Dement. 2018, 14, 1580–1588. [Google Scholar] [CrossRef] [PubMed]
- Roubroeks, J.; Smith, A.; Smith, R.; Pishva, E.; Ibrahim, Z.; Sattlecker, M.; Hannon, E.; Kloszewska, I.; Mecocci, P.; Soininen, H.; et al. An Epigenome-Wide Association Study of Alzheimer’s Disease Blood Highlights Robust DNA Hypermethylation in the HOXB6 Gene. Neurobiol. Aging 2020, 95, 26–45. [Google Scholar] [CrossRef]
- Narayan, P.; Lill, C.; Faull, R.; Curtis, M.; Dragunow, M. Increased Acetyl and Total Histone Levels in Post-Mortem Alzheimer’s Disease Brain. Neurobiol. Dis. 2014, 74, 281–294. [Google Scholar] [CrossRef]
- Wang, S.-C.; Oelze, B.; Schumacher, A. Age-Specific Epigenetic Drift in Late-Onset Alzheimer’s Disease. PLoS ONE 2008, 3, e2698. [Google Scholar] [CrossRef]
- lu, X.; Wang, L.; Yu, C.; Yu, D.; Yu, G. Histone Acetylation Modifiers in the Pathogenesis of Alzheimer’s Disease. Front. Cell. Neurosci. 2015, 9, 226. [Google Scholar] [CrossRef]
- Plagg, B.; Ehrlich, D.; Kniewallner, K.; Marksteiner, J.; Humpel, C. Increased Acetylation of Histone H4 at Lysine 12 (H4K12) in Monocytes of Transgenic Alzheimer’s Mice and in Human Patients. Curr. Alzheimer Res. 2015, 12, 752–760. [Google Scholar] [CrossRef]
- Klein, H.-U.; McCabe, C.; Gjoneska, E.; Sullivan, S.E.; Kaskow, B.J.; Tang, A.; Smith, R.V.; Xu, J.; Pfenning, A.R.; Bernstein, B.E.; et al. Epigenome-Wide Study Uncovers Large-Scale Changes in Histone Acetylation Driven by Tau Pathology in Aging and Alzheimer’s Human Brains. Nat. Neurosci. 2019, 22, 37–46. [Google Scholar] [CrossRef]
- Wang, W.-X.; Rajeev, B.; Stromberg, A.; Ren, N.; Tang, G.; Huang, Q.; Rigoutsos, I.; Nelson, P. The Expression of MicroRNA MiR-107 Decreases Early in Alzheimer’s Disease and May Accelerate Disease Progression through Regulation of β-Site Amyloid Precursor Protein-Cleaving Enzyme 1. J. Neurosci. 2008, 28, 1213–1223. [Google Scholar] [CrossRef]
- Schueller, E.; Paiva, I.; Blanc, F.; Wang, X.-L.; Cassel, J.-C.; Boutillier, A.-L.; Bousiges, O. Dysregulation of Histone Acetylation Pathways in Hippocampus and Frontal Cortex of Alzheimer’s Disease Patients. Eur. Neuropsychopharmacol. 2020, 33, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Nativio, R.; Donahue, G.; Berson, A.; Lan, Y.; Amlie-Wolf, A.; Tuzer, F.; Toledo, J.; Gosai, S.; Gregory, B.; Torres, C.; et al. Dysregulation of the Epigenetic Landscape of Normal Aging in Alzheimer’s Disease. Nat. Neurosci. 2018, 21, 497–505. [Google Scholar] [CrossRef]
- Gräff, J.; Rei, D.; Guan, J.-S.; Wang, W.-Y.; Seo, J.; Hennig, K.; Nieland, T.; Fass, D.; Kao, P.; Kahn, M.; et al. An Epigenetic Blockade of Cognitive Functions in the Neurodegenerating Brain. Nature 2012, 483, 222–226. [Google Scholar] [CrossRef]
- Zhang, K.; Schrag, M.; Crofton, A.; Trivedi, R.; Vinters, H.; Kirsch, W. Targeted Proteomics for Quantification of Histone Acetylation in Alzheimer’s Disease. Proteomics 2012, 12, 1261–1268. [Google Scholar] [CrossRef]
- Park, H.J.; Kim, S.S.; Seong, Y.M.; Kim, K.H.; Hui, G.G.; Eun, J.Y.; Do, S.M.; Kang, S.; Rhim, H. β-Amyloid Precursor Protein Is a Direct Cleavage Target of HtrA2 Serine Protease: Implications for the Physiological Function of HtrA2 in the Mitochondria. J. Biol. Chem. 2006, 281, 34277–34287. [Google Scholar] [CrossRef]
- Chaeyoung, K.; Choi, H.; Jung, E.; Lee, W.; Oh, S.; Jeon, N.; Mook-Jung, I. HDAC6 Inhibitor Blocks Amyloid β-Induced Impairment of Mitochondrial Transport in Hippocampal Neurons. PLoS ONE 2012, 7, e42983. [Google Scholar] [CrossRef]
- Onishi, T.; Maeda, R.; Terada, M.; Sato, S.; Fujii, T.; Ito, M.; Hashikami, K.; Kawamoto, T.; Tanaka, M. A Novel Orally Active HDAC6 Inhibitor T-518 Shows a Therapeutic Potential for Alzheimer’s Disease and Tauopathy in Mice. Sci. Rep. 2021, 11, 15423. [Google Scholar] [CrossRef] [PubMed]
- Cohen, T.; Guo, J.; Hurtado, D.; Kwong, L.; Mills, I.; Trojanowski, J.; Lee, V. The Acetylation of Tau Inhibits Its Function and Promotes Pathological Tau Aggregation. Nat. Commun. 2011, 2, 252. [Google Scholar] [CrossRef]
- Cook, C.; Carlomagno, Y.; Gendron, T.; Dunmore, J.; Scheffel, K.; Stetler, C.; Davis, M.; Dickson, D.; Jarpe, M.; Deture, M.; et al. Acetylation of the KXGS Motifs in Tau Is a Critical Determinant in Modulation of Tau Aggregation and Clearance. Hum. Mol. Genet. 2013, 23, 104–116. [Google Scholar] [CrossRef]
- Marambaud, P.; Wen, P.; Dutt, A.; Shioi, J.; Takashima, A.; Siman, R.; Robakis, N. A CBP Binding Transcriptional Repressor Produced by the PS1/ϵ-Cleavage of N-Cadherin Is Inhibited by PS1 FAD Mutations. Cell 2003, 114, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Irwin, D.; Cohen, T.; Grossman, M.; Arnold, S.; Xie, S.; Lee, V.; Trojanowski, J. Acetylated Tau, a Novel Pathological Signature in Alzheimer’s Disease and Other Tauopathies. Brain 2012, 135, 807–818. [Google Scholar] [CrossRef]
- Park, S.-Y.; Kim, M.J.; Kim, Y.; Lee, Y.-H.; Bae, D.; Kim, S.; Na, Y.; Yoon, H.-G. Selective PCAF Inhibitor Ameliorates Cognitive and Behavioral Deficits by Suppressing NF-ΚB-Mediated Neuroinflammation Induced by Aβ in a Model of Alzheimer’s Disease. Int. J. Mol. Med. 2015, 35, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Min, S.-W.; Sohn, P.; Li, Y.; Devidze, N.; Johnson, J.; Krogan, N.; Masliah, E.; Mok, S.-A.; Gestwicki, J.; Gan, L. SIRT1 Deacetylates Tau and Reduces Pathogenic Tau Spread in a Mouse Model of Tauopathy. J. Neurosci. 2018, 38, 2317–2369. [Google Scholar] [CrossRef]
- Min, S.-W.; Cho, S.H.; Zhou, Y.; Schröder, S.; Haroutunian, V.; Seeley, W.; Huang, E.; Shen, Y.; Masliah, E.; Mukherjee, C.; et al. Acetylation of Tau Inhibits Its Degradation and Contributes to Tauopathy. Neuron 2010, 67, 953–966. [Google Scholar] [CrossRef]
- Kim, D.; Nguyen, M.; Dobbin, M.; Fischer, A.; Sananbenesi, F.; Rodgers, J.; Delalle, I.; Baur, J.; Sui, G.; Armour, S.; et al. SIRT1 Deacetylase Protects against Neurodegeneration in Models for Alzheimer’s Disease and Amyotrophic Lateral Sclerosis. EMBO J. 2007, 26, 3169–3179. [Google Scholar] [CrossRef] [PubMed]
- Aubry, S.; Shin, W.; Crary, J.F.; Lefort, R.; Qureshi, Y.H.; Lefebvre, C.; Califano, A.; Shelanski, M.L. Assembly and Interrogation of Alzheimer’s Disease Genetic Networks Reveal Novel Regulators of Progression. PLoS ONE 2015, 10, e0120352. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Kim, S.-Y.; Hong, Y.-M.; Jo, D.-G.; Lee, J.-Y.; Shim, S.M.; Chung, C.-W.; Seo, S.J.; Yoo, Y.J.; Koh, J.-Y.; et al. Essential Role of E2-25K/Hip-2 in Mediating Amyloid-β Neurotoxicity. Mol. Cell 2003, 12, 553–563. [Google Scholar] [CrossRef]
- Oddo, S.; Billings, L.; Kesslak, J.P.; Cribbs, D.H.; LaFerla, F.M. Aβ Immunotherapy Leads to Clearance of Early, but Not Late, Hyperphosphorylated Tau Aggregates via the Proteasome. Neuron 2004, 43, 321–332. [Google Scholar] [CrossRef]
- Prasanth, K.; Spector, D. Prasanth, K.V.; Spector, D.L. Eukaryotic Regulatory RNAs: An Answer to the “genome Complexity” Conundrum. Genes Dev. 2007, 21, 11–42. [Google Scholar] [CrossRef] [PubMed]
- Ciarlo, E.; Massone, S.; Penna, I.; Nizzari, M.; Gigoni, A.; Dieci, G.; Russo, C.; Florio, T.; Cancedda, R.; Pagano, A. An Intronic NcRNA-Dependent Regulation of SORL1 Expression Affecting Aβ Formation Is Upregulated in Post-Mortem Alzheimer’s Disease Brain Samples. Dis. Model. Mech. 2013, 6, 424–433. [Google Scholar] [CrossRef]
- Kim, J.; Inoue, K.; Ishii, J.; Vanti, W.; Voronov, S.; Murchison, E.; Hannon, G.; Abeliovich, A. A MicroRNA Feedback Circuit in Midbrain Dopamine Neurons. Science 2007, 317, 1220–1224. [Google Scholar] [CrossRef] [PubMed]
- Zovoilis, A.; Agbemenyah, H.; Agís-Balboa, R.; Stilling, R.; Edbauer, D.; Rao, P.; Farinelli, L.; Delalle, I.; Schmitt, A.; Falkai, P.; et al. MicroRNA-34c Is a Novel Target to Treat Dementias. EMBO J. 2011, 30, 4299–4308. [Google Scholar] [CrossRef] [PubMed]
- Schonrock, N.; Ke, Y.D.; Humphreys, D.; Staufenbiel, M.; Ittner, L.M.; Preiss, T.; Götz, J. Neuronal MicroRNA Deregulation in Response to Alzheimer’s Disease Amyloid-β. PLoS ONE 2010, 5, e11070. [Google Scholar] [CrossRef]
- Jain, G.; Stuendl, A.; Rao, P.; Berulava, T.; Pena Centeno, T.; Kaurani, L.; Burkhardt, S.; Delalle, I.; Kornhuber, J.; Hüll, M.; et al. A Combined MiRNA–PiRNA Signature to Detect Alzheimer’s Disease. Transl. Psychiatry 2019, 9, 250. [Google Scholar] [CrossRef]
- Kumar, S.; Reddy, P.H. MicroRNA-455-3p as a Potential Biomarker for Alzheimer’s Disease: An Update. Front. Aging Neurosci. 2018, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Manzine, P.; Pelucchi, S.; Horst, M.; Vale, F.; Pavarini, S.; Audano, M.; Mitro, N.; Di Luca, M.; Marcello, E. MicroRNA 221 Targets ADAM10 MRNA and Is Downregulated in Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 61, 113–123. [Google Scholar] [CrossRef]
- Lau, P.; Bossers, K.; Janky, R.; Salta, E.; Sala Frigerio, C.; Barbash, S.; Rothman, R.; Sierksma, A.; Thathiah, A.; Greenberg, D.; et al. Alteration of the MicroRNA Network during the Progression of Alzheimer’s Disease. EMBO Mol. Med. 2013, 5, 1613–1634. [Google Scholar] [CrossRef]
- Geekiyanage, H.; Jicha, G.; Nelson, P.; Chan, C. Blood Serum MiRNA: Non-Invasive Biomarkers for Alzheimer’s Disease. Exp. Exp. Neurol. 2011, 235, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Hosseinian, S.; Arefian, E.; Rakhsh-Khorshid, H.; Eivani, M.; Rezayof, A.; Pezeshk, H.; Marashi, S.-A. A Meta-Analysis of Gene Expression Data Highlights Synaptic Dysfunction in the Hippocampus of Brains with Alzheimer’s Disease. Sci. Rep. 2020, 10, 8384. [Google Scholar] [CrossRef]
- Mairet-Coello, G.; Courchet, J.; Pieraut, S.; Courchet, V.; Maximov, A.; Polleux, F. The CAMKK2-AMPK Kinase Pathway Mediates the Synaptotoxic Effects of Aβ Oligomers through Tau Phosphorylation. Neuron 2013, 78, 94–108. [Google Scholar] [CrossRef]
- Yin, Y.; Gao, D.; Wang, Y.; Wang, Z.-H.; Xin, W.; Jinwang, Y.; Wu, D.; Fang, L.; Pi, G.; Yang, Y.; et al. Tau Accumulation Induces Synaptic Impairment and Memory Deficit by Calcineurin-Mediated Inactivation of Nuclear CaMKIV/CREB Signaling. Proc. Natl. Acad. Sci. USA 2016, 113, E3773–E3781. [Google Scholar] [CrossRef] [PubMed]
- Benetti, R.; Gonzalo, S.; Jaco, I.; Muñoz, P.; Gonzalez, S.; Schoeftner, S.; Murchison, E.; Andl, T.; Chen, T.; Klatt, P.; et al. A Mammalian MicroRNA Cluster Controls DNA Methylation and Telomere Recombination via Rbl2-Dependent Regulation of DNA Methyltransferases. Nat. Struct. Mol. Biol. 2008, 15, 998. [Google Scholar] [CrossRef]
- Sinkkonen, L.; Hugenschmidt, T.; Berninger, P.; Gaidatzis, D.; Mohn, F.; Artus-Revel, C.; Zavolan, M.; Svoboda, P.; Filipowicz, W. MicroRNAs Control de Novo DNA Methylation through Regulation of Transcriptional Repressors in Mouse Embryonic Stem Cells. Nat. Struct. Mol. Biol. 2008, 15, 259–267. [Google Scholar] [CrossRef]
- Zhang, S.; Pointer, B.; Kelleher, E. Rapid Evolution of PiRNA-Mediated Silencing of an Invading Transposable Element Was Driven by Abundant de Novo Mutations. Genome Res. 2020, 30, 566–575. [Google Scholar] [CrossRef]
- Homolka, D.; Pandey, R.R.; Goriaux, C.; Brasset, E.; Vaury, C.; Sachidanandam, R.; Fauvarque, M.-O.; Pillai, R.S. PIWI Slicing and RNA Elements in Precursors Instruct Directional Primary PiRNA Biogenesis. Cell Rep. 2015, 12, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Siomi, M. Piwi-Interacting RNAs: Biological Functions and Biogenesis. Essays Biochem. 2013, 54, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Guo, X.; Lin, X.; Yang, Q.; Zhang, W.; Zhang, Y.; Zuo, L.; Zhu, Y.; Li, C.-S.R.; Ma, C.; et al. Transcriptome-Wide PiRNA Profiling in Human Brains of Alzheimer’s Disease. Neurobiol. Aging 2017, 57, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Roy, J.; Sarkar, A.; Parida, S.; Ghosh, Z.; Mallick, B. Small RNA Sequencing Revealed Dysregulated PiRNAs in Alzheimer’s Disease and Their Probable Role in Pathogenesis. Mol. Biosyst. 2017, 13, 565–576. [Google Scholar] [CrossRef]
- Mendiola-Precoma, J.; Berumen, L.; Padilla, K.; Garcia-Alcocer, G. Therapies for Prevention and Treatment of Alzheimer’s Disease. BioMed Res. Int. 2016, 2016, 2589276. [Google Scholar] [CrossRef]
- Varidaki, A.; Hong, Y.; Coffey, E.T. Repositioning Microtubule Stabilizing Drugs for Brain Disorders. Front. Cell. Neurosci. 2018, 12, 226. [Google Scholar] [CrossRef] [PubMed]
- Perez-Gonzalez, R.; Kim, Y.; Miller, C.; Pacheco-Quinto, J.; Eckman, E.; Levy, E. Extracellular Vesicles: Where the Amyloid Precursor Protein Carboxyl-Terminal Fragments Accumulate and Amyloid-β Oligomerizes. FASEB J. 2020, 34, 12922–12931. [Google Scholar] [CrossRef]
- Gratuze, M.; Leyns, C.; Holtzman, D. New Insights into the Role of TREM2 in Alzheimer’s Disease. Mol. Neurodegener. 2018, 13, 1–6. [Google Scholar] [CrossRef]
- Chávez-Gutiérrez, L.; Bammens, L.; Benilova, I.; Vandersteen, A.; Benurwar, M.; Borgers, M.; Lismont, S.; Zhou, L.; Cleynenbreugel, S.; Esselmann, H.; et al. The Mechanism of γ-Secretase Dysfunction in Familial Alzheimer Disease. EMBO J. 2012, 31, 2261–2274. [Google Scholar] [CrossRef] [PubMed]
- Bai, B. U1 SnRNP Alteration and Neuronal Cell Cycle Reentry in Alzheimer Disease. Front. Aging Neurosci. 2018, 10, 75. [Google Scholar] [CrossRef]
- Schwartz, M.; Arad, M.; Ben-Yehuda, H. Potential Immunotherapy for Alzheimer Disease and Age-Related Dementia. Dialogues Clin. Neurosci. 2019, 21, 21–25. [Google Scholar] [CrossRef]
- Baruch, K.; Rosenzweig, N.; Kertser, A.; Deczkowska, A.; Sharif, A.; Spinrad, A.; Tsitsou-Kampeli, A.; Sarel, A.; Cahalon, L.; Schwartz, M. Breaking Immune Tolerance by Targeting Foxp3+ Regulatory T Cells Mitigates Alzheimer’s Disease Pathology. Nat. Commun. 2015, 6, 7967. [Google Scholar] [CrossRef]
- Rogers, N.K.; Romero, C.; SanMartín, C.D.; Ponce, D.P.; Salech, F.; López, M.N.; Gleisner, A.; Tempio, F.; Behrens, M.I. Inverse Relationship Between Alzheimer’s Disease and Cancer: How Immune Checkpoints Might Explain the Mechanisms Underlying Age-Related Diseases. J. Alzheimer’s Dis. 2020, 73, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Obst, J.; Mancuso, R.; Simon, E.; Gomez-Nicola, D. PD-1 Deficiency Is Not Sufficient to Induce Myeloid Mobilization to the Brain or Alter the Inflammatory Profile during Chronic Neurodegeneration. Brain. Behav. Immun. 2018, 73, 708–716. [Google Scholar] [CrossRef]
- Rosenzweig, N.; Dvir-Szternfeld, R.; Tsitsou-Kampeli, A.; Keren-Shaul, H.; Ben Yehuda, H.; Weill-Raynal, P.; Cahalon, L.; Kertser, A.; Baruch, K.; Amit, I.; et al. PD-1/PD-L1 Checkpoint Blockade Harnesses Monocyte-Derived Macrophages to Combat Cognitive Impairment in a Tauopathy Mouse Model. Nat. Commun. 2019, 10, 465. [Google Scholar] [CrossRef]
- Guan, J.-S.; Haggarty, S.; Giacometti, E.; Dannenberg, J.-H.; Joseph, N.; Gao, J.; Nieland, T.; Zhou, Y.; Wang, X.; Mazitschek, R.; et al. HDAC2 Negatively Regulates Memory Formation and Synaptic Plasticity. Nature 2009, 459, 55–60. [Google Scholar] [CrossRef]
- Mahgoub, M.; Monteggia, L. A Role for Histone Deacetylases in the Cellular and Behavioral Mechanisms Underlying Learning and Memory. Learn. Mem. 2014, 21, 564–568. [Google Scholar] [CrossRef]
- Lim, J.; Song, Y.; Jang, J.-H.; Jeong, C.-H.; Lee, S.; Park, B.; Seo, Y. Aspirin-Inspired Acetyl-Donating HDACs Inhibitors. Arch. Pharm. Res. 2018, 41, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Bang, S.; Ambavade, S.; Jagdale, P.; Adkar, D.P.; Waghmare, A.; Ambavade, P. Lacosamide Reduces HDAC Levels in the Brain and Improves Memory: Potential for Treatment of Alzheimer’s Disease. Pharmacol. Biochem. Behav. 2015, 134, 65–69. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, C.; Wu, J.; Tao, J.-J.; Sui, X.-L.; Yao, Z.-G.; Xu, Y.-F.; Huang, L.; Zhu, H.; Sheng, S.-L.; et al. Tubastatin A/ACY-1215 Improves Cognition in Alzheimer’s Disease Transgenic Mice. J. Alzheimers. Dis. 2014, 41, 1193–1205. [Google Scholar] [CrossRef]
- Arts, J.; King, P.; Mariën, A.; Floren, W.; Beliën, A.; Janssen, L.; Pilatte, I.; Roux, B.; Decrane, L.; Gilissen, R.; et al. JNJ-26481585, a Novel “Second-Generation” Oral Histone Deacetylase Inhibitor, Shows Broad-Spectrum Preclinical Antitumoral Activity. Clin. Cancer Res. 2009, 15, 6841–6851. [Google Scholar] [CrossRef]
- Francis, Y.; Fa, M.; Ashraf, H.; Zhang, H.; Staniszewski, A.; Latchman, D.; Arancio, O. Dysregulation of Histone Acetylation in the APP/PS1 Mouse Model of Alzheimer’s Disease. J. Alzheimers. Dis. 2009, 18, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Volmar, C.-H.; Salah-Uddin, H.; Janczura, K.; Halley, P.; Lambert, G.; Wodrich, A.; Manoah, S.; Patel, N.; Sartor, G.; Mehta, N.; et al. M344 Promotes Nonamyloidogenic Amyloid Precursor Protein Processing While Normalizing Alzheimer’s Disease Genes and Improving Memory. Proc. Natl. Acad. Sci. USA 2017, 114, E9135–E9144. [Google Scholar] [CrossRef]
- Qing, H.; He, G.; Ly, P.T.T.; Fox, C.J.; Staufenbiel, M.; Cai, F.; Zhang, Z.; Wei, S.; Sun, X.; Chen, C.-H.; et al. Valproic Acid Inhibits Abeta Production, Neuritic Plaque Formation, and Behavioral Deficits in Alzheimer’s Disease Mouse Models. J. Exp. Med. 2008, 205, 2781–2789. [Google Scholar] [CrossRef]
- Ricobaraza, A.; Cuadrado-Tejedor, M.; Marco, S.; Pérez-Otaño, I.; García-Osta, A. Phenylbutyrate Rescues Dendritic Spine Loss Associated with Memory Deficits in a Mouse Model of Alzheimer Disease. Hippocampus 2012, 22, 1040–1050. [Google Scholar] [CrossRef]
- Fan, S.-J.; Huang, F.-I.; Liou, J.-P.; Yang, C.-R. The Novel Histone de Acetylase 6 Inhibitor, MPT0G211, Ameliorates Tau Phosphorylation and Cognitive Deficits in an Alzheimer’s Disease Model. Cell Death Dis. 2018, 9, 655. [Google Scholar] [CrossRef]
- Green, K.; Steffan, J.; Martinez-Coria, H.; Sun, X.; Schreiber, S.; Thompson, L.; Laferla, F. Nicotinamide Restores Cognition in Alzheimer’s Disease Transgenic Mice via a Mechanism Involving Sirtuin Inhibition and Selective Reduction of Thr231-Phosphotau. J. Neurosci. 2008, 28, 11500–11510. [Google Scholar] [CrossRef]
- Hu, J.; An, B.; Pan, T.; Zhengcunxiao, L.; Huang, L.; Li, X. Design, Synthesis, and Biological Evaluation of Histone Deacetylase Inhibitors Possessing Glutathione Peroxidase-like and Antioxidant Activities against Alzheimer’s Disease. Bioorg. Med. Chem. 2018, 26, 5718–5729. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado-Tejedor, M.; Garcia-Barroso, C.; Sanchez Arias, J.A.; Rabal, O.; Pérez-González, M.; Mederos, S.; Ugarte, A.; Franco, R.; Segura, V.; Perea, G.; et al. A First-in-Class Small-Molecule That Acts as a Dual Inhibitor of HDAC and PDE5, and That Rescues Hippocampal Synaptic Impairment in Alzheimer’s Disease Mice. Neuropsychopharmacology 2016, 42, 524–539. [Google Scholar] [CrossRef]
- Cuadrado-Tejedor, M.; Pérez-González, M.; García-Muñoz, C.; Muruzabal, D.; García-Barroso, C.; Rabal, O.; Segura, V.; Sanchez Arias, J.A.; Oyarzabal, J.; García-Osta, A. Taking Advantage of the Selectivity of Histone Deacetylases and Phosphodiesterase Inhibitors to Design Better Therapeutic Strategies to Treat Alzheimer’s DiseaseData_Sheet_1.Docx. Front. Aging Neurosci. 2019, 11, 149. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rabaneda-Bueno, R.; Mena-Montes, B.; Torres-Castro, S.; Torres-Carrillo, N.; Torres-Carrillo, N.M. Advances in Genetics and Epigenetic Alterations in Alzheimer’s Disease: A Notion for Therapeutic Treatment. Genes 2021, 12, 1959. https://doi.org/10.3390/genes12121959
Rabaneda-Bueno R, Mena-Montes B, Torres-Castro S, Torres-Carrillo N, Torres-Carrillo NM. Advances in Genetics and Epigenetic Alterations in Alzheimer’s Disease: A Notion for Therapeutic Treatment. Genes. 2021; 12(12):1959. https://doi.org/10.3390/genes12121959
Chicago/Turabian StyleRabaneda-Bueno, Rubén, Beatriz Mena-Montes, Sara Torres-Castro, Norma Torres-Carrillo, and Nora Magdalena Torres-Carrillo. 2021. "Advances in Genetics and Epigenetic Alterations in Alzheimer’s Disease: A Notion for Therapeutic Treatment" Genes 12, no. 12: 1959. https://doi.org/10.3390/genes12121959
APA StyleRabaneda-Bueno, R., Mena-Montes, B., Torres-Castro, S., Torres-Carrillo, N., & Torres-Carrillo, N. M. (2021). Advances in Genetics and Epigenetic Alterations in Alzheimer’s Disease: A Notion for Therapeutic Treatment. Genes, 12(12), 1959. https://doi.org/10.3390/genes12121959





