MiR-138-5p Suppresses Cell Growth and Migration in Melanoma by Targeting Telomerase Reverse Transcriptase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Detection of pTERTm
2.3. Cell Transfection and Molecule Extractions
2.4. Luciferase Assay
2.5. Cell Proliferation Assays
2.6. Cell Cycle Assays
2.7. Telomerase Activity
2.8. Cell Migration Assay
2.9. Western Blot Assay
2.10. Real Time qPCR (RT-qPCR)
2.11. Statistics
3. Results
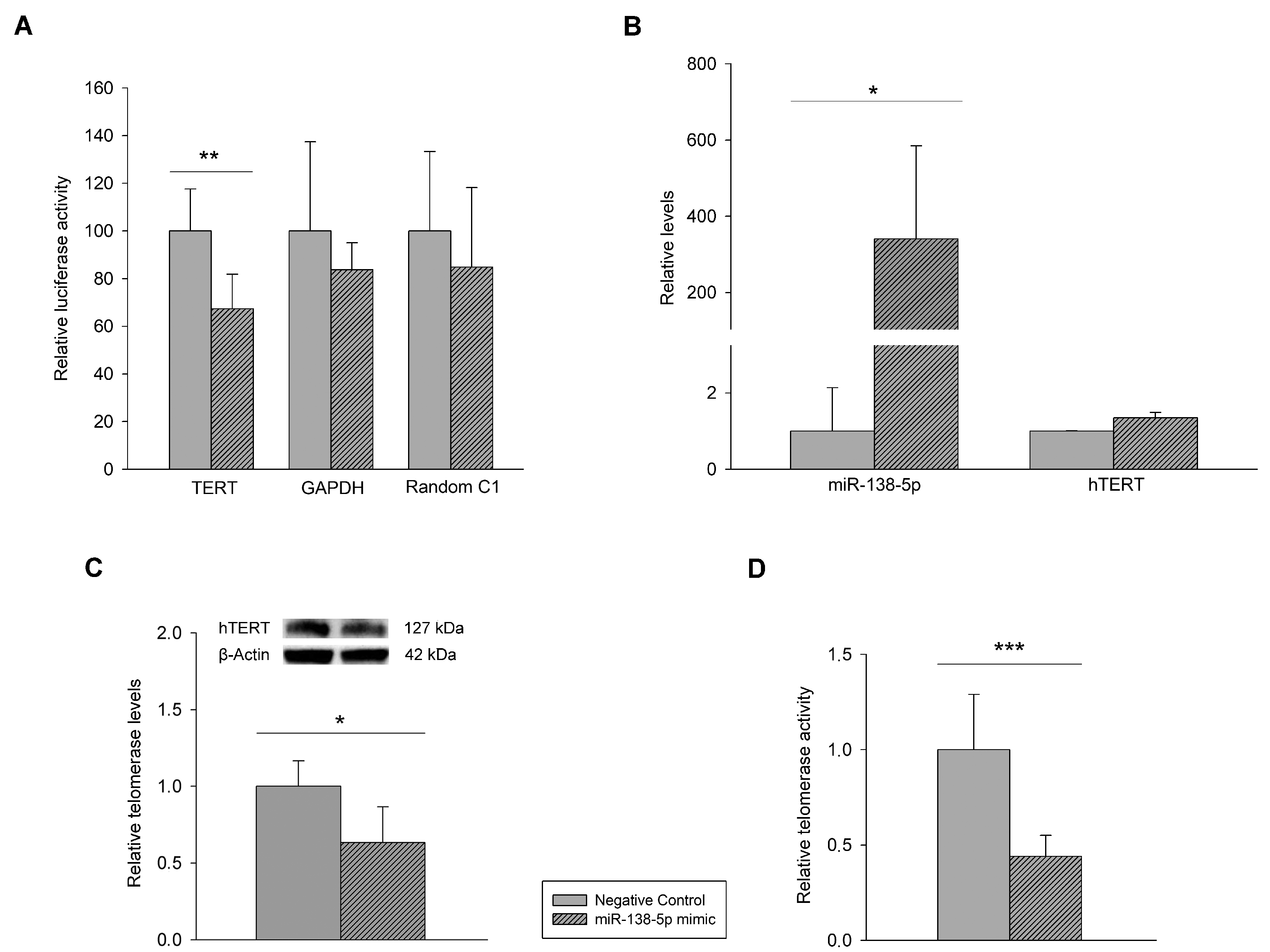
3.1. Mutational Characterization of hTERT Promoter and hTERT and miR-138-5p Expression in Melanoma Cell Lines and Human Epidermal Melanocytes
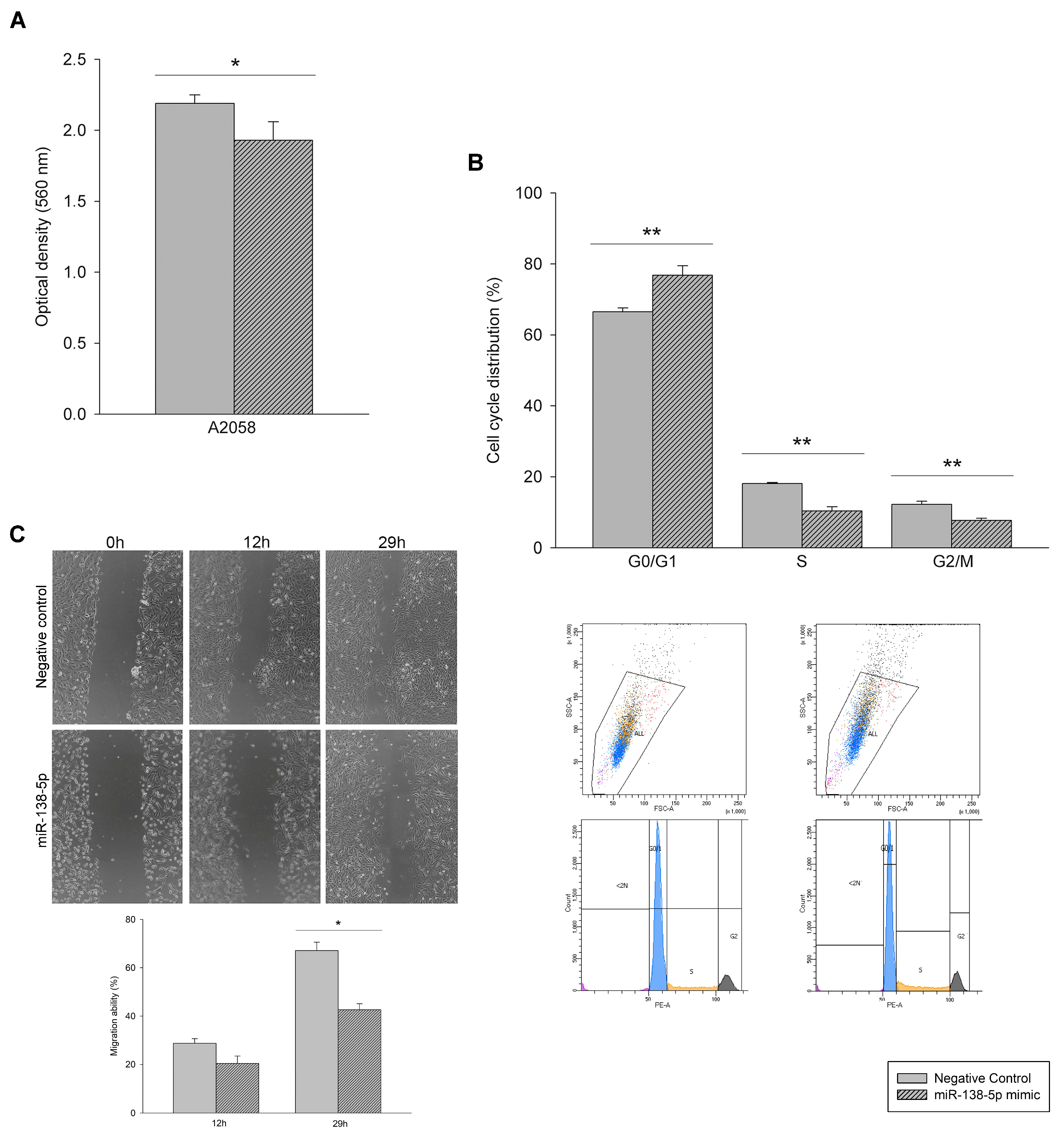
3.2. Effect of miR-138-5p Overexpression on Melanoma Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Bandarchi, B.; Ma, L.; Navab, R.; Seth, A.; Rasty, G. From melanocyte to metastatic malignant melanoma. Dermatol. Res. Pract. 2010, 2010, 583748. [Google Scholar] [CrossRef]
- Little, E.G.; Eide, M.J. Update on the current state of melanoma incidence. Dermatol. Clin. 2012, 30, 355–361. [Google Scholar] [CrossRef]
- Lombardo, N.; Della Corte, M.; Pelaia, C.; Piazzetta, G.; Lobello, N.; Del Duca, E.; Bennardo, L.; Nistico, S.P. Primary Mucosal Melanoma Presenting with a Unilateral Nasal Obstruction of the Left Inferior Turbinate. Medicina 2021, 57, 359. [Google Scholar] [CrossRef] [PubMed]
- Horn, S.; Figl, A.; Rachakonda, P.S.; Fischer, C.; Sucker, A.; Gast, A.; Kadel, S.; Moll, I.; Nagore, E.; Hemminki, K.; et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013, 339, 959–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly recurrent TERT promoter mutations in human melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Unamuno Bustos, B.; Murria Estal, R.; Perez Simo, G.; Oliver Martinez, V.; Llavador Ros, M.; Palanca Suela, S.; Botella Estrada, R. Lack of TERT promoter mutations in melanomas with extensive regression. J. Am. Acad. Dermatol. 2016, 74, 570–572. [Google Scholar] [CrossRef] [Green Version]
- Griewank, K.G.; Murali, R.; Puig-Butille, J.A.; Schilling, B.; Livingstone, E.; Potrony, M.; Carrera, C.; Schimming, T.; Moller, I.; Schwamborn, M.; et al. TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. J. Natl. Cancer Inst. 2014, 106, dju246. [Google Scholar] [CrossRef]
- Heidenreich, B.; Nagore, E.; Rachakonda, P.S.; Garcia-Casado, Z.; Requena, C.; Traves, V.; Becker, J.; Soufir, N.; Hemminki, K.; Kumar, R. Telomerase reverse transcriptase promoter mutations in primary cutaneous melanoma. Nat. Commun. 2014, 5, 3401. [Google Scholar] [CrossRef]
- Populo, H.; Boaventura, P.; Vinagre, J.; Batista, R.; Mendes, A.; Caldas, R.; Pardal, J.; Azevedo, F.; Honavar, M.; Guimaraes, I.; et al. TERT promoter mutations in skin cancer: The effects of sun exposure and X-irradiation. J. Investig. Dermatol. 2014, 134, 2251–2257. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Opresko, P.; Pappo, A.; Kirkwood, J.M.; Bahrami, A. Association of TERT promoter mutations with telomerase expression in melanoma. Pigment Cell Melanoma Res. 2016, 29, 391–393. [Google Scholar] [CrossRef] [Green Version]
- Vallarelli, A.F.; Rachakonda, P.S.; Andre, J.; Heidenreich, B.; Riffaud, L.; Bensussan, A.; Kumar, R.; Dumaz, N. TERT promoter mutations in melanoma render TERT expression dependent on MAPK pathway activation. Oncotarget 2016, 7, 53127–53136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Unamuno Bustos, B.; Sahuquillo Torralba, A.; Moles Poveda, P.; Perez Simo, G.; Simarro Farinos, J.; Llavador Ros, M.; Palanca Suela, S.; Botella Estrada, R. Telomerase Expression in a Series of Melanocytic Neoplasms. Actas Dermosifiliogr. 2019, 110, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Hugdahl, E.; Kalvenes, M.B.; Mannelqvist, M.; Ladstein, R.G.; Akslen, L.A. Prognostic impact and concordance of TERT promoter mutation and protein expression in matched primary and metastatic cutaneous melanoma. Br. J. Cancer 2018, 118, 98–105. [Google Scholar] [CrossRef] [Green Version]
- Kohli, J.S.; Mir, H.; Wasif, A.; Chong, H.; Akhras, V.; Kumar, R.; Nagore, E.; Bennett, D.C. ETS1, nucleolar and non-nucleolar TERT expression in nevus to melanoma progression. Oncotarget 2017, 8, 104408–104417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masui, K.; Komori, T.; Kato, Y.; Masutomi, K.; Ichimura, K.; Ogasawara, S.; Kaneko, M.K.; Oki, H.; Suzuki, H.; Nitta, M.; et al. Elevated TERT Expression in TERT-Wildtype Adult Diffuse Gliomas: Histological Evaluation with a Novel TERT-Specific Antibody. Biom. Res. Int. 2018, 2018, 7945845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz, A.; Puig-Butille, J.A.; Munoz, C.; Costa, D.; Diez, A.; Garcia-Herrera, A.; Carrera, C.; Badenas, C.; Sole, F.; Malvehy, J.; et al. TERT gene amplification is associated with poor outcome in acral lentiginous melanoma. J. Am. Acad. Dermatol. 2014, 71, 839–841. [Google Scholar] [CrossRef]
- Zhang, C.; Song, C.; Liu, T.; Tang, R.; Chen, M.; Gao, F.; Xiao, B.; Qin, G.; Shi, F.; Li, W.; et al. KMT2A promotes melanoma cell growth by targeting hTERT signaling pathway. Cell Death Dis. 2017, 8, e2940. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Lee, S.; Wu, G.; Easton, J.; Yergeau, D.; Dummer, R.; Vogel, P.; Kirkwood, J.M.; Barnhill, R.L.; Pappo, A.; et al. Telomerase Expression by Aberrant Methylation of the TERT Promoter in Melanoma Arising in Giant Congenital Nevi. J. Investig. Dermatol. 2016, 136, 339–342. [Google Scholar] [CrossRef] [Green Version]
- Guilleret, I.; Yan, P.; Grange, F.; Braunschweig, R.; Bosman, F.T.; Benhattar, J. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int. J. Cancer 2002, 101, 335–341. [Google Scholar] [CrossRef]
- Qin, Y.Z.; Xie, X.C.; Liu, H.Z.; Lai, H.; Qiu, H.; Ge, L.Y. Screening and preliminary validation of miRNAs with the regulation of hTERT in colorectal cancer. Oncol. Rep. 2015, 33, 2728–2736. [Google Scholar] [CrossRef] [Green Version]
- Hrdlickova, R.; Nehyba, J.; Bargmann, W.; Bose, H.R., Jr. Multiple tumor suppressor microRNAs regulate telomerase and TCF7, an important transcriptional regulator of the Wnt pathway. PLoS ONE 2014, 9, e86990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- de Unamuno, B.; Palanca, S.; Botella, R. Update on melanoma epigenetics. Curr. Opin. Oncol. 2015, 27, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Bartels, C.L.; Tsongalis, G.J. MicroRNAs: Novel biomarkers for human cancer. Clin. Chem. 2009, 55, 623–631. [Google Scholar] [CrossRef] [Green Version]
- Nair, V.S.; Maeda, L.S.; Ioannidis, J.P. Clinical outcome prediction by microRNAs in human cancer: A systematic review. J. Natl. Cancer Inst. 2012, 104, 528–540. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Lee, C.G. MicroRNA and cancer—Focus on apoptosis. J. Cell Mol. Med. 2009, 13, 12–23. [Google Scholar] [CrossRef]
- Chen, J.; Feilotter, H.E.; Pare, G.C.; Zhang, X.; Pemberton, J.G.; Garady, C.; Lai, D.; Yang, X.; Tron, V.A. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am. J. Pathol. 2010, 176, 2520–2529. [Google Scholar] [CrossRef]
- Xu, Y.; Brenn, T.; Brown, E.R.; Doherty, V.; Melton, D.W. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br. J. Cancer 2012, 106, 553–561. [Google Scholar] [CrossRef] [Green Version]
- Chai, L.; Kang, X.J.; Sun, Z.Z.; Zeng, M.F.; Yu, S.R.; Ding, Y.; Liang, J.Q.; Li, T.T.; Zhao, J. MiR-497-5p, miR-195-5p and miR-455-3p function as tumor suppressors by targeting hTERT in melanoma A375 cells. Cancer Manag. Res. 2018, 10, 989–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohira, T.; Naohiro, S.; Nakayama, Y.; Osaki, M.; Okada, F.; Oshimura, M.; Kugoh, H. miR-19b regulates hTERT mRNA expression through targeting PITX1 mRNA in melanoma cells. Sci. Rep. 2015, 5, 8201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, G.; Wang, R.; Guo, J.; Liu, X.; Wang, F.; Qi, Y.; Wan, H.; Liu, M.; Li, X.; Tang, H. miR-346 and miR-138 competitively regulate hTERT in GRSF1- and AGO2-dependent manners, respectively. Sci. Rep. 2015, 5, 15793. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhao, Y.; Cao, W.; Wang, C.; Sun, B.; Chen, J.; Li, S.; Chen, J.; Cui, M.; Zhang, B.; et al. miR-138-5p acts as a tumor suppressor by targeting hTERT in human colorectal cancer. Int. J. Clin. Exp. Pathol. 2017, 10, 11516–11525. [Google Scholar]
- Zhang, X.L.; Xu, L.L.; Wang, F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol. Int. 2017, 41, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Mitomo, S.; Maesawa, C.; Ogasawara, S.; Iwaya, T.; Shibazaki, M.; Yashima-Abo, A.; Kotani, K.; Oikawa, H.; Sakurai, E.; Izutsu, N.; et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008, 99, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Cho, W.C. OncomiRs: The discovery and progress of microRNAs in cancers. Mol. Cancer 2007, 6, 60. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Wang, C.; Chen, Z.; Jin, Y.; Wang, Y.; Kolokythas, A.; Dai, Y.; Zhou, X. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem. J. 2011, 440, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Jiang, L.; Wang, A.; Yu, J.; Shi, F.; Zhou, X. MicroRNA-138 suppresses invasion and promotes apoptosis in head and neck squamous cell carcinoma cell lines. Cancer Lett. 2009, 286, 217–222. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Lu, M.H.; Zhang, D.; Hao, N.B.; Fan, Y.H.; Wu, Y.Y.; Wang, S.M.; Xie, R.; Fang, D.C.; Zhang, H.; et al. miR-1207-5p and miR-1266 suppress gastric cancer growth and invasion by targeting telomerase reverse transcriptase. Cell Death Dis. 2014, 5, e1034. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Cao, K.E.; Wang, S.; Chen, J.; He, B.; He, G.U.; Chen, Y.; Peng, B.; Zhou, J. MicroRNA-138 suppresses proliferation, invasion and glycolysis in malignant melanoma cells by targeting HIF-1alpha. Exp. Ther. Med. 2016, 11, 2513–2518. [Google Scholar] [CrossRef] [Green Version]
- Meng, F.; Zhang, Y.; Li, X.; Wang, J.; Wang, Z. Clinical significance of miR-138 in patients with malignant melanoma through targeting of PDK1 in the PI3K/AKT autophagy signaling pathway. Oncol. Rep. 2017, 38, 1655–1662. [Google Scholar] [CrossRef] [PubMed]
- Salgado, C.; Roelse, C.; Nell, R.; Gruis, N.; van Doorn, R.; van der Velden, P. Interplay between TERT promoter mutations and methylation culminates in chromatin accessibility and TERT expression. PLoS ONE 2020, 15, e0231418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandal, T. Molecular aspects of the mammalian cell cycle and cancer. Oncologist 2002, 7, 73–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rapanotti, M.C.; Cugini, E.; Nuccetelli, M.; Terrinoni, A.; Di Raimondo, C.; Lombardo, P.; Costanza, G.; Cosio, T.; Rossi, P.; Orlandi, A.; et al. MCAM/MUC18/CD146 as a Multifaceted Warning Marker of Melanoma Progression in Liquid Biopsy. Int. J. Mol. Sci. 2021, 22, 12416. [Google Scholar] [CrossRef]
- Salloum, R.; Hummel, T.R.; Kumar, S.S.; Dorris, K.; Li, S.; Lin, T.; Daryani, V.M.; Stewart, C.F.; Miles, L.; Poussaint, T.Y.; et al. A molecular biology and phase II study of imetelstat (GRN163L) in children with recurrent or refractory central nervous system malignancies: A pediatric brain tumor consortium study. J. Neurooncol. 2016, 129, 443–451. [Google Scholar] [CrossRef]
- Tefferi, A.; Lasho, T.L.; Begna, K.H.; Patnaik, M.M.; Zblewski, D.L.; Finke, C.M.; Laborde, R.R.; Wassie, E.; Schimek, L.; Hanson, C.A.; et al. A Pilot Study of the Telomerase Inhibitor Imetelstat for Myelofibrosis. N. Engl. J. Med. 2015, 373, 908–919. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Zhang, J.; Yang, S.; Kuang, Z.; Tan, G.; Yang, G.; Wei, Q.; Guo, Z. Telomerase antagonist imetelstat increases radiation sensitivity in esophageal squamous cell carcinoma. Oncotarget 2017, 8, 13600–13619. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Hao, J.; Fan, W.; Li, Y.; Tang, R.; Tian, C.; Yang, Q.; Zhu, T.; Diao, C.; Hu, S.; Chen, M.; et al. Melatonin synergizes BRAF-targeting agent vemurafenib in melanoma treatment by inhibiting iNOS/hTERT signaling and cancer-stem cell traits. J. Exp. Clin. Cancer Res. CR 2019, 38, 48. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tarazón, E.; de Unamuno Bustos, B.; Murria Estal, R.; Pérez Simó, G.; Sahuquillo Torralba, A.; Simarro, J.; Palanca Suela, S.; Botella Estrada, R. MiR-138-5p Suppresses Cell Growth and Migration in Melanoma by Targeting Telomerase Reverse Transcriptase. Genes 2021, 12, 1931. https://doi.org/10.3390/genes12121931
Tarazón E, de Unamuno Bustos B, Murria Estal R, Pérez Simó G, Sahuquillo Torralba A, Simarro J, Palanca Suela S, Botella Estrada R. MiR-138-5p Suppresses Cell Growth and Migration in Melanoma by Targeting Telomerase Reverse Transcriptase. Genes. 2021; 12(12):1931. https://doi.org/10.3390/genes12121931
Chicago/Turabian StyleTarazón, Estefanía, Blanca de Unamuno Bustos, Rosa Murria Estal, Gema Pérez Simó, Antonio Sahuquillo Torralba, Javier Simarro, Sarai Palanca Suela, and Rafael Botella Estrada. 2021. "MiR-138-5p Suppresses Cell Growth and Migration in Melanoma by Targeting Telomerase Reverse Transcriptase" Genes 12, no. 12: 1931. https://doi.org/10.3390/genes12121931
APA StyleTarazón, E., de Unamuno Bustos, B., Murria Estal, R., Pérez Simó, G., Sahuquillo Torralba, A., Simarro, J., Palanca Suela, S., & Botella Estrada, R. (2021). MiR-138-5p Suppresses Cell Growth and Migration in Melanoma by Targeting Telomerase Reverse Transcriptase. Genes, 12(12), 1931. https://doi.org/10.3390/genes12121931





