Transcriptomic Markers of Recombinant Human Erythropoietin Micro-Dosing in Thoroughbred Horses
Abstract
:1. Introduction
2. Material and Methods
2.1. Horses
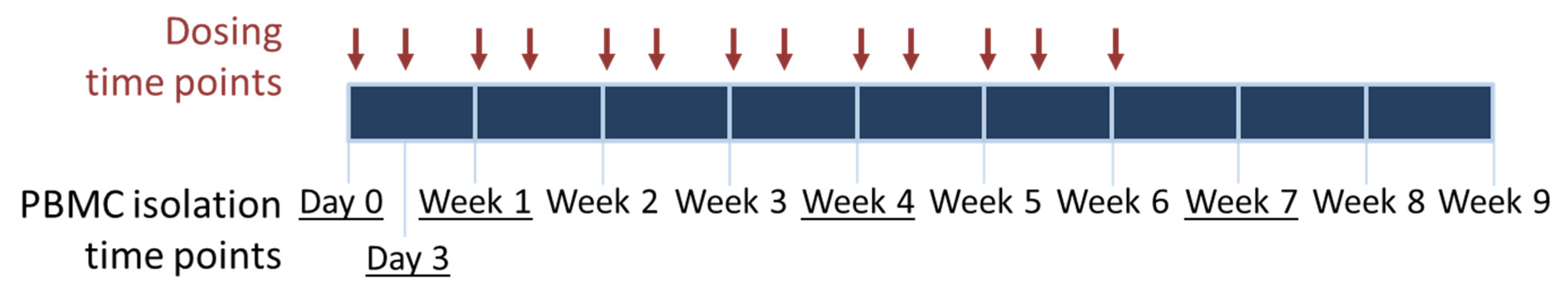
2.2. Sample Collection
2.3. PBMC and RNA Isolations
2.4. RNA Sequencing and Analysis
2.5. RT-q PCR Validation
3. Results
3.1. Horses
3.2. RNA Sequencing and Differential Transcript Analysis
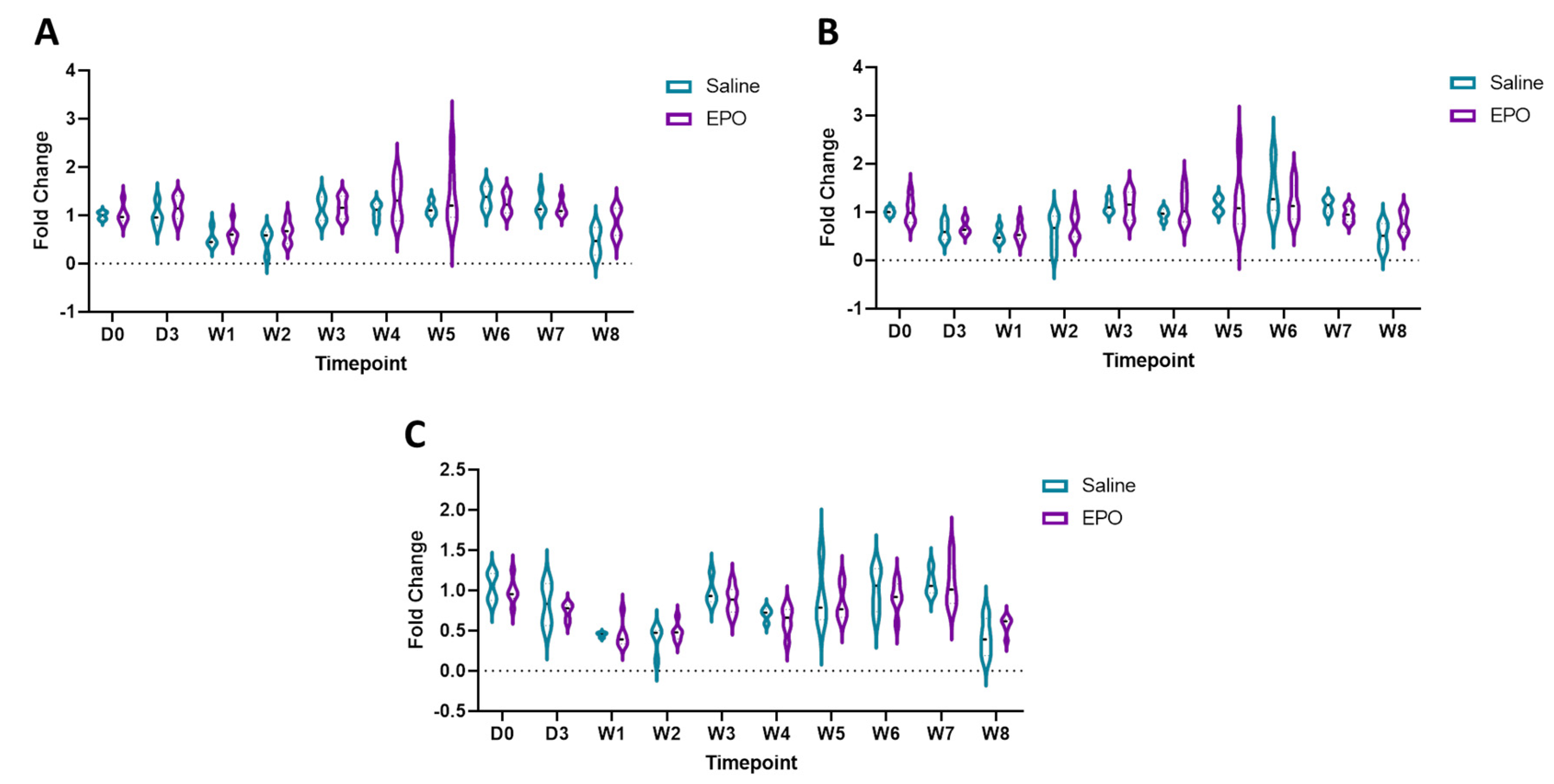
3.3. RT-qPCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jacob, J.; John, M.; Jaison, V.; Jain, K.; Kakkar, N. Erythropoietin Use and Abuse. Indian J. Endocrinol. Metab. 2012, 16, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.; Marx, G.; Littlewood, T.; Macdougall, I. Recombinant Erythropoietin in Clinical Practice. Postgrad. Med. J. 2003, 79, 367–376. [Google Scholar] [CrossRef] [Green Version]
- Ekblom, B.; Berglund, B. Effect of Erythropoietin Administration on Mammal Aerobic Power. Scand. J. Med. Sci. Sports 1991, 1, 88–93. [Google Scholar] [CrossRef]
- Thomsen, J.J.; Rentsch, R.L.; Robach, P.; Calbet, J.A.L.; Boushel, R.; Rasmussen, P.; Juel, C.; Lundby, C. Prolonged Administration of Recombinant Human Erythropoietin Increases Submaximal Performance More than Maximal Aerobic Capacity. Eur. J. Appl. Physiol. 2007, 101, 481–486. [Google Scholar] [CrossRef]
- Durussel, J.; Daskalaki, E.; Anderson, M.; Chatterji, T.; Wondimu, D.H.; Padmanabhan, N.; Patel, R.K.; McClure, J.D.; Pitsiladis, Y.P. Haemoglobin Mass and Running Time Trial Performance after Recombinant Human Erythropoietin Administration in Trained Men. PLoS ONE 2013, 8, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Siri, W.E.; Van Dyke, D.C.; Winchell, H.S.; Pollycove, M.; Parker, H.G.; Cleveland, A.S. Blood, and Physiological Responses To Severe Hypoxia in Man. J. Appl. Physiol. 1966, 21, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Wold-Anti Doping Agency. The 2021 WADA Prohibited List; World Anti-Doping: Montreal, QC, Canada, 2021; pp. 1–24. [Google Scholar]
- Parisotto, R.; Gore, C.J.; Emslie, K.R.; Ashenden, M.J.; Brugnara, C.; Howe, C.; Martin, D.T.; Trout, G.J.; Hahn, A.G. A Novel Method Utilizing Markers of Altered Erythropoiesis for the Detection of Recombinant Human Erythropoietin Abuse in Athletes. Haematologica 2000, 85, 564–572. [Google Scholar]
- Lundby, C.; Achman-Andersen, N.J.; Thomsen, J.J.; Norgaard, A.M.; Robach, P. Testing for Recombinant Human Erythropoietin in Urine: Problems Associated with Current Anti-Doping Testing. J. Appl. Physiol. 2008, 105, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Martin, L.; Audran, M.; Marchand, A. Combined Immuno-Purification and Detection of Recombinant Erythropoietins and Activin Receptor Type II-Fc Fusion Proteins by Isoelectric Focusing for Application in Doping Control. Drug Test. Anal. 2019, 11, 168–172. [Google Scholar] [CrossRef] [Green Version]
- Schwenke, D. Improved Detection of EPO in Blood and Urine Based on Novel Velum SAR Precast Horizontal Gels Optimized for Routine Analysis. Chemistry 2015, 1–6. Available online: https://www.semanticscholar.org/paper/Improved-detection-of-EPO-in-blood-and-urine-based-Schwenke/aa7c2b44892836f2b1f45268fc1561fdb67984ef (accessed on 25 March 2021).
- Ashenden, M.; Gough, C.E.; Garnham, A.; Gore, C.J.; Sharpe, K. Current Markers of the Athlete Blood Passport Do Not Flag Microdose EPO Doping. Eur. J. Appl. Physiol. 2011, 111, 2307–2314. [Google Scholar] [CrossRef]
- Reichel, C.; Erceg, D.; Lorenc, B.; Scheiblhofer, V.; Farmer, L.; Zanitzer, K.; Geisendorfer, T.; Gmeiner, G.; Thevis, M. Data from a Microdosed Recombinant Human Erythropoietin Administration Study Applying the New Biotinylated Clone AE7A5 Antibody and a Further Optimized Sarcosyl Polyacrylamide Gel Electrophoresis Protocol. Drug Test. Anal. 2021, 1–10. [Google Scholar] [CrossRef]
- Wang, G.; Durussel, J.; Shurlock, J.; Mooses, M.; Fuku, N.; Bruinvels, G.; Pedlar, C.; Burden, R.; Murray, A.; Yee, B.; et al. Validation of Whole-Blood Transcriptome Signature during Microdose Recombinant Human Erythropoietin (RHuEpo) Administration. BMC Genom. 2017, 18 (Suppl. 8), 817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKeever, K.H.; McNally, B.A.; Hinchcliff, K.W.; Lehnhard, R.A.; Poole, D.C. Effects of Erythropoietin on Systemic Hematocrit and Oxygen Transport in the Splenectomized Horse. Respir. Physiol. Neurobiol. 2016, 225, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Persson, S.G.B.; Ekman, L.; Lydin, G.; Tufvesson, G. Circulatory Effects of Splenectomy in the Horse: III. Effect on Pulse-work Relationship. Zent. für Veterinärmedizin Reihe A 1973, 20, 521–530. [Google Scholar] [CrossRef]
- Fragkaki, A.G.; Kioukia-Fougia, N.; Kiousi, P.; Kioussi, M.; Tsivou, M. Challenges in Detecting Substances for Equine Anti-Doping. Drug Test. Anal. 2017, 9, 1291–1303. [Google Scholar] [CrossRef] [Green Version]
- Guan, F.; Uboh, C.E.; Soma, L.R.; Birks, E.; Chen, J.; Mitchell, J.; You, W.; Rudy, J.; Xu, F.; Li, X.; et al. LC-MS/MS Method for Confirmation of Recombinant Human Erythropoietin and Darbepoetin α in Equine Plasma. Anal. Chem. 2007, 79, 4627–4635. [Google Scholar] [CrossRef]
- Souillard, A.; Audran, M.; Bressolle, F.; Jaussaud, P.; Gareau, R. Pharmacokinetics and Haematological Parameters of Recombinant Human Erythropoietin after Subcutaneous Administrations in Horses. Biopharm. Drug Dispos. 1996, 17, 805–815. [Google Scholar] [CrossRef]
- Lasne, F.; Popot, M.A.; Varlet-Marie, E.; Martin, L.; Martin, J.A.; Bonnaire, Y.; Audran, M.; De Ceaurriz, J. Detection of Recombinant Epoetin and Darbepoetin Alpha after Subcutaneous Administration in the Horse. J. Anal. Toxicol. 2005, 29, 835–837. [Google Scholar] [CrossRef] [Green Version]
- Lonnberg, M.; Bondesson, U.; Cormant, F.; Garcia, P.; Bonnaire, Y.; Carlsson, J.; Popot, M.-A.; Rollborn, N.; Rasbo, K.; Bailly-Chouriberry, L. Detection of Recombinant Human EPO Administered to Horses Using MAIIA Lateral Flow Isoform Test. Anal. Bioanal. Chem. 2012, 403, 1619–1628. [Google Scholar] [CrossRef]
- Tozaki, T.; Ohnuma, A.; Takasu, M.; Kikuchi, M.; Kakoi, H.; Hirota, K.I.; Kusano, K.; Nagata, S.I. Droplet Digital Pcr Detection of the Erythropoietin Transgene from Horse Plasma and Urine for Gene-Doping Control. Genes 2019, 10, 243. [Google Scholar] [CrossRef] [Green Version]
- Tozaki, T.; Ohnuma, A.; Kikuchi, M.; Ishige, T.; Kakoi, H.; Hirota, K.I.; Kusano, K.; Nagata, S.I. Microfluidic Quantitative PCR Detection of 12 Transgenes from Horse Plasma for Gene Doping Control. Genes 2020, 11, 457. [Google Scholar] [CrossRef] [Green Version]
- Cavalcanti, R.T.C.; Teixeira, P.A.C.; Levy, R.S.; Pereira, H.M.G.; Aquino Neto, F.R. Detection of ESAs in Equine Urine and Blood by SAR-PAGE. Drug Test. Anal. 2019, 11, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Bailly-Chouriberry, L.; Cormant, F.; Garcia, P.; Lönnberg, M.; Szwandt, S.; Bondesson, U.; Popot, M.A.; Bonnaire, Y. A New Analytical Method Based on Anti-EPO Monolith Column and LC-FAIMS-MS/MS for the Detection of RHuEPOs in Horse Plasma and Urine Samples. Analyst 2012, 137, 2445–2453. [Google Scholar] [CrossRef] [PubMed]
- Tozaki, T.; Ohnuma, A.; Takasu, M.; Nakamura, K.; Kikuchi, M.; Ishige, T.; Kakoi, H.; Hirora, K.-I.; Tamura, N.; Kusano, K.; et al. Detection of Non-Targeted Transgenes by Whole-Genome Resequencing for Gene-Doping Control. Gene Ther. 2021, 28, 199–205. [Google Scholar] [CrossRef]
- Lisowska, K.A.; Debska-Slizień, A.; Bryl, E.; Rutkowski, B.; Witkowski, J.M. Erythropoietin Receptor Is Expressed on Human Peripheral Blood T and B Lymphocytes and Monocytes and Is Modulated by Recombinant Human Erythropoietin Treatment. Artif. Organs 2010, 34, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Bailly-Chouriberry, L.; Noguier, F.; Manchon, L.; Piquemal, D.; Garcia, P.; Popot, M.A.; Bonnaire, Y. Blood Cells RNA Biomarkers as a First Long-Term Detection Strategy for EPO Abuse in Horseracing. Drug Test. Anal. 2010, 2, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Genome Analysis Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision Weights Unlock Linear Model Analysis Tools for RNA-Seq Read Counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Xiang-Hong, J.; Yan-Hong, Y.; Han-Jin, X.; Li-long, A.; Ying-Mei, X.; Pei-Rong, J.; Ming, L. Selection of Reference Genes for Gene Expression Studies in PBMC from Bama Miniature Pig under Heat Stress. Vet. Immunol. Immunopathol. 2011, 144, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Milhem, C.; Ingelaere, C.; Mordon, S.; Morales, O.; Delhem, N. Beta-2 Microglobulin And Ubiquitin C Identified as Two Robust Housekeeping Genes for RNA Expression Normalization in Real Time PCR on Human Leukocytes and Regulatory T Cells. Biomed. J. Sci. Tech. Res. 2020, 31, 24425–24430. [Google Scholar] [CrossRef]
- He, R.; Oliveira, J.L.; Hoyer, J.D.; Viswanatha, D.S. 24—Molecular Hematopathology. In Foundations in Diagnostic Pathology; Hsi, E.D.B.T.-H., Third, E., Eds.; Elsevier: Philadelphia, PA, USA, 2018; pp. 712–760.e18. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Naudin, C.; Hattabi, A.; Michelet, F.; Miri-Nezhad, A.; Benyoucef, A.; Pflumio, F.; Guillonneau, F.; Fichelson, S.; Vigon, I.; Dusanter-Fourt, I.; et al. PUMILIO/FOXP1 Signaling Drives Expansion of Hematopoietic Stem/Progenitor and Leukemia Cells. Blood 2017, 129, 2493–2506. [Google Scholar] [CrossRef] [Green Version]
- Beutler, E.; Lichtman, M.A.; Coller, B.S.; Kipps, T.J.; Seligsohn, U. Williams Hematology, 6th ed.; McGraw-Hill: New York, NY, USA, 2001. [Google Scholar]
- Van Dijk, T.B.; Gillemans, N.; Pourfarzad, F.; Van Lom, K.; Von Lindern, M.; Grosveld, F.; Philipsen, S. Fetal Globin Expression Is Regulated by Friend of Prmt1. Blood 2010, 116, 4349–4352. [Google Scholar] [CrossRef] [Green Version]
- Bourantas, K.L.; Georgiou, I.; Seferiadis, K. Fetal Globin Stimulation during a Short Term Trial of Erythropoietin in HbS/Beta-Thalassemia Patients. Acta Haematol. 1994, 92, 79–82. [Google Scholar] [CrossRef]
- Perrine, S.P.; Castaneda, S.A.; Boosalis, M.S.; White, G.L.; Jones, B.M.; Bohacek, R. Induction of Fetal Globin in β-Thalassemia: Cellular Obstacles and Molecular Progress Susan. Ann. N. Y. Acad. Sci. 2005, 1054, 257–265. [Google Scholar] [CrossRef]
- Bustin, S.A.; Nolan, T. Pitfalls of Quantitative Real-Time Reverse-Transcription Polymerase Chain Reaction. J. Biomol. Tech. 2004, 15, 155–166. [Google Scholar]
- Cooper, C.; Sears, W.; Bienzle, D. Reticulocyte Changes after Experimental Anemia and Erythropoietin Treatment of Horses. J. Appl. Physiol. 2005, 99, 915–921. [Google Scholar] [CrossRef] [Green Version]
- Finno, C.J.; Bordbari, M.H.; Valberg, S.J.; Lee, D.; Herron, J.; Hines, K.; Monsour, T.; Scott, E.; Bannasch, D.L.; Mickelson, J.; et al. Transcriptome Profiling of Equine Vitamin E Deficient Neuroaxonal Dystrophy Identifies Upregulation of Liver X Receptor Target Genes. Free Radic. Biol. Med. 2016, 101, 261–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus QPCR for Gene Expression Analysis with Low Abundant Targets: From Variable Nonsense to Publication Quality Data. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Scott, E.Y.; Woolard, K.D.; Finno, C.J.; Penedo, M.C.T.; Murray, J.D. Variation in MUTYH Expression in Arabian Horses with Cerebellar Abiotrophy. Brain Res. 2018, 1678, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Loup, B.; André, F.; Avignon, J.; Lhuaire, M.; Delcourt, V.; Barnabé, A.; Garcia, P.; Popot, M.A.; Bailly-Chouriberry, L. MiRNAs Detection in Equine Plasma by Quantitative Polymerase Chain Reaction for Doping Control: Assessment of Blood Sampling and Study of Eca-MiR-144 as Potential Erythropoiesis Stimulating Agent Biomarker. Drug Test. Anal. 2021. [Google Scholar] [CrossRef]
- Jung, M.; Schaefer, A.; Steiner, I.; Kempkensteffen, C.; Stephan, C.; Erbersdobler, A.; Jung, K. Robust MicroRNA Stability in Degraded RNA Preparations from Human Tissue and Cell Samples. Clin. Chem. 2010, 56, 998–1006. [Google Scholar] [CrossRef] [Green Version]
- Piercy, R.; Swardson, C.; Hinchcliff, K. Erythroid Hypoplasia and Anemia Following Administration of Recombinant Human Erythropoietin to Two Horses. J. Am. Vet. Med. Assoc. 1998, 212, 244–247. [Google Scholar]
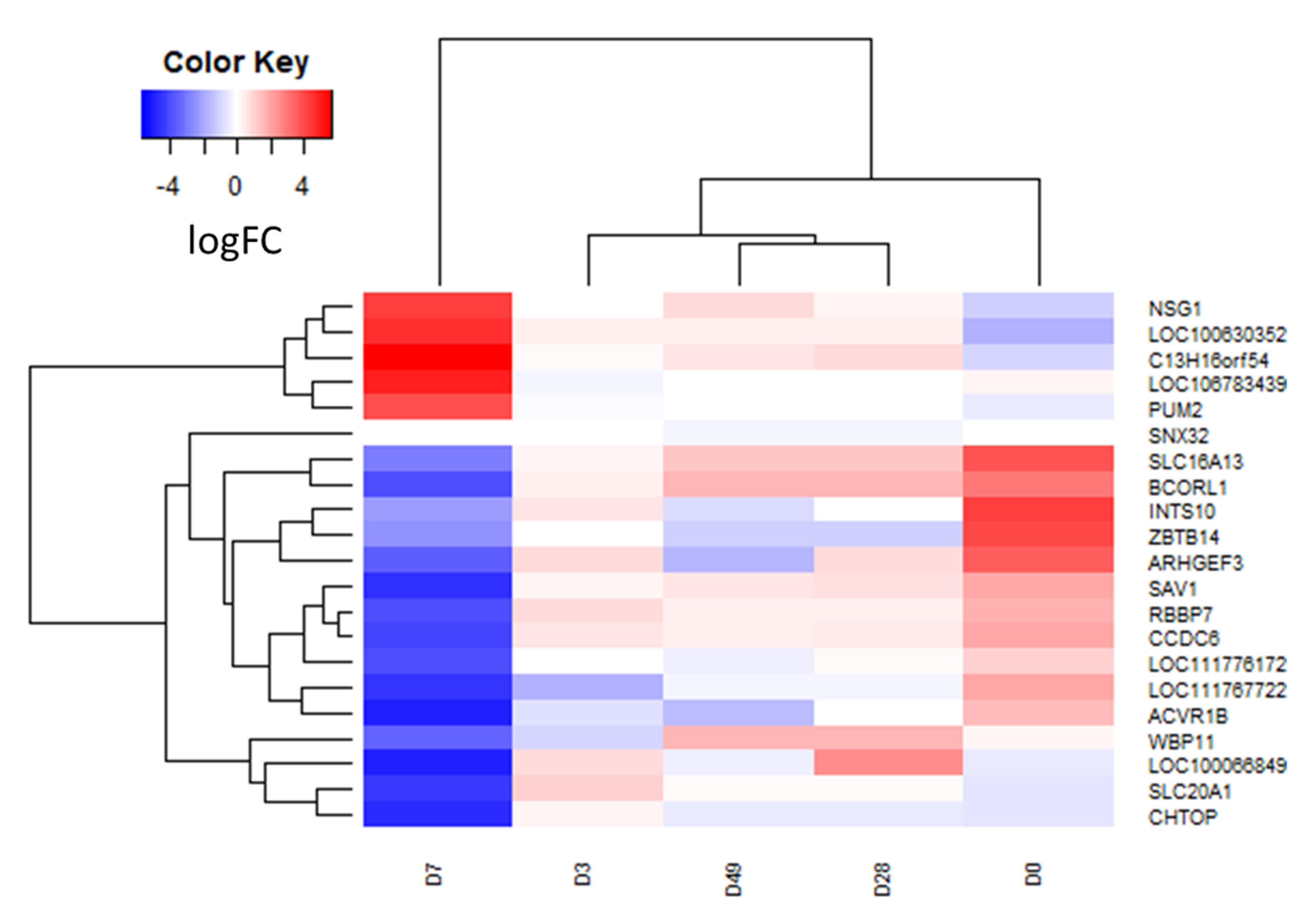
| Time Point | Gene Name | Log2FC | Ave Expr | p Value | adj. p. Val |
|---|---|---|---|---|---|
| D7 v D0 | BCORL1 | −6.91 | 3.58 | 1.04 × 10−6 | 7.90 × 10−3 |
| C13H16orf54 | 6.68 | 3.98 | 1.48 × 10−6 | 7.90 × 10−3 | |
| LOC100630352 | 6.32 | 5.13 | 2.25 × 10−6 | 8.04 × 10−3 | |
| SLC16A13 | −6.61 | 4.45 | 7.80 × 10−6 | 1.53 × 10-2 | |
| ARHGEF3 | −7.18 | 3.39 | 1.06 × 10−5 | 1.53 × 10−2 | |
| LOC111776172 | −4.96 | 5.93 | 1.11 × 10−5 | 1.53 × 10−2 | |
| LOC106783439 | 4.70 | 5.05 | 1.14 × 10−5 | 1.53 × 10−2 | |
| LOC111767722 | −6.38 | 2.94 | 1.15 × 10−5 | 1.53 × 10-2 | |
| PUM2 | 4.40 | 5.19 | 1.28 × 10−5 | 1.53 × 10−2 | |
| CCDC6 | −6.17 | 2.92 | 1.43 × 10−5 | 1.53 × 10−2 | |
| INTS10 | −6.56 | 2.85 | 1.61 × 10−5 | 1.57 × 10−2 | |
| SAV1 | −6.48 | 3.80 | 2.74 × 10−5 | 2.45 × 10−2 | |
| RBBP7 | −5.70 | 4.16 | 3.41 × 10−5 | 2.80 × 10−2 | |
| ACVR1B | −6.41 | 3.24 | 3.66 × 10−5 | 2.80 × 10-2 | |
| CHTOP | −4.03 | 4.68 | 4.42 × 10−5 | 3.15 × 10−2 | |
| NSG1 | 5.34 | 3.92 | 7.67 × 10−5 | 4.98 × 10−2 | |
| ZBTB14 | −6.47 | 1.41 | 7.91 × 10−5 | 4.98 × 10−2 | |
| D7 v D3 | LOC106783439 | 5.31 | 5.05 | 2.76 × 10−6 | 2.42 × 10−3 |
| CHTOP | −5.00 | 4.68 | 4.53 × 10−7 | 2.42 × 10−3 | |
| PUM2 | 4.11 | 5.19 | 1.21 × 10−5 | 4.32 × 10-2 | |
| C13H16orf54 | 5.55 | 3.98 | 2.08 × 10−5 | 4.32 × 10−2 | |
| SLC20A1 | −5.43 | 3.42 | 2.17 × 10−5 | 4.32 × 10−2 | |
| D28 v D0 | SNX32 | −0.39 | 13.85 | 9.48 × 10−7 | 1.01 × 10−2 |
| D28 v D7 | LOC106783439 | −5.08 | 5.05 | 2.90 × 10−7 | 3.10 × 10−3 |
| LOC100066849 | 7.49 | 2.30 | 1.32 × 10−6 | 7.08 × 10−3 | |
| WBP11 | 5.08 | 5.08 | 3.27 × 10−6 | 1.07 × 10−2 | |
| CHTOP | 4.24 | 4.68 | 1.25 × 10−5 | 3.35 × 10−2 | |
| D49 v D7 | LOC106783439 | −5.90 | 5.05 | 7.55 × 10−9 | 8.08 × 10−5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dahlgren, A.R.; Knych, H.K.; Arthur, R.M.; Durbin-Johnson, B.P.; Finno, C.J. Transcriptomic Markers of Recombinant Human Erythropoietin Micro-Dosing in Thoroughbred Horses. Genes 2021, 12, 1874. https://doi.org/10.3390/genes12121874
Dahlgren AR, Knych HK, Arthur RM, Durbin-Johnson BP, Finno CJ. Transcriptomic Markers of Recombinant Human Erythropoietin Micro-Dosing in Thoroughbred Horses. Genes. 2021; 12(12):1874. https://doi.org/10.3390/genes12121874
Chicago/Turabian StyleDahlgren, Anna R., Heather K. Knych, Rick M. Arthur, Blythe P. Durbin-Johnson, and Carrie J. Finno. 2021. "Transcriptomic Markers of Recombinant Human Erythropoietin Micro-Dosing in Thoroughbred Horses" Genes 12, no. 12: 1874. https://doi.org/10.3390/genes12121874
APA StyleDahlgren, A. R., Knych, H. K., Arthur, R. M., Durbin-Johnson, B. P., & Finno, C. J. (2021). Transcriptomic Markers of Recombinant Human Erythropoietin Micro-Dosing in Thoroughbred Horses. Genes, 12(12), 1874. https://doi.org/10.3390/genes12121874






