MUC14-Related ncRNA-mRNA Network in Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. starBase Analysis
2.2. GEPIA Analysis
2.3. HPA Analysis
2.4. UALCAN Analysis
2.5. ROC Curve Analysis
2.6. Kaplan-Meier Plotter Analysis
2.7. bc-GenExMiner Analysis
2.8. miRNA Prediction
2.9. miRNet Analysis
3. Results
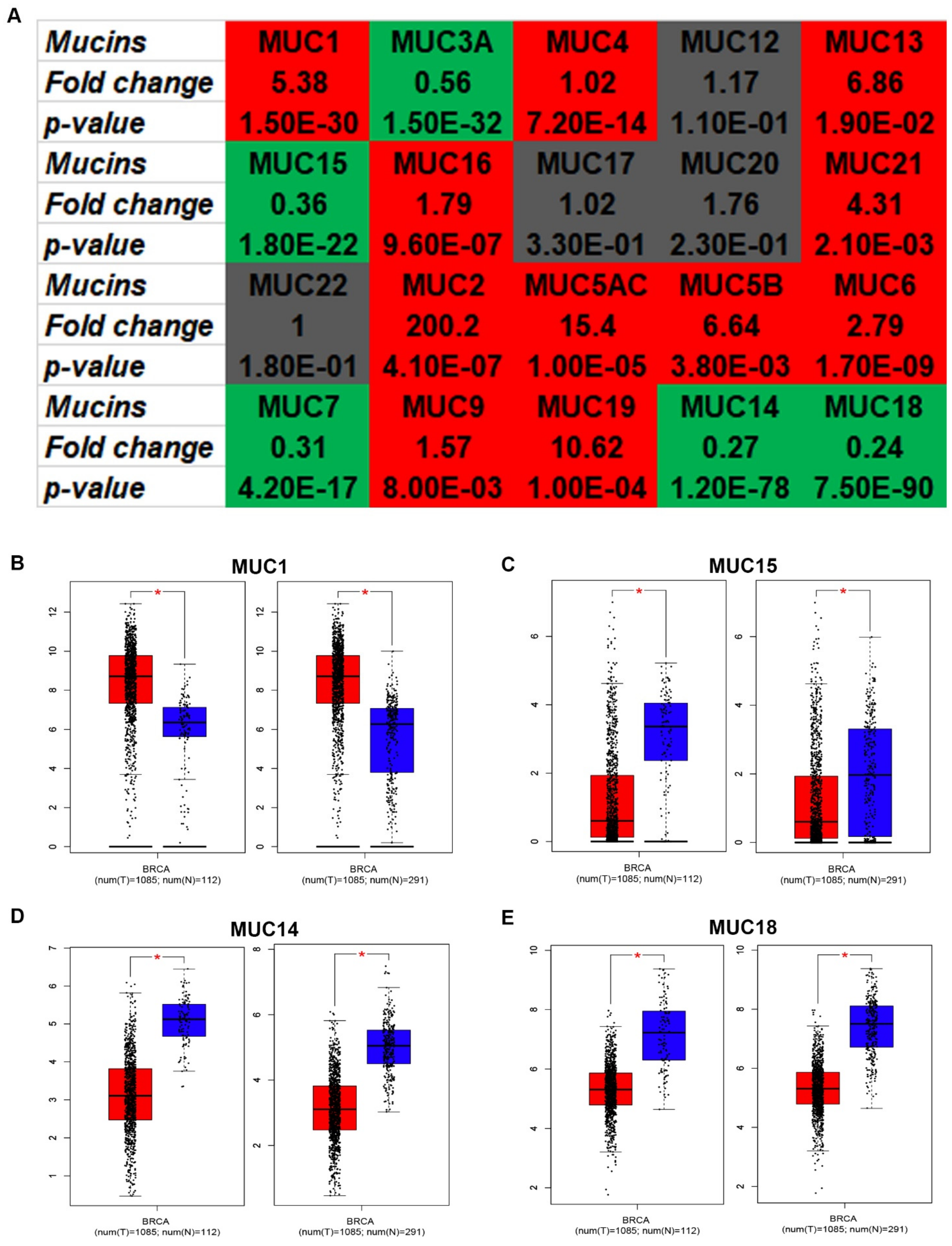
3.1. The Expression of MUCs in Breast Cancer
3.2. The Diagnostic and Prognostic Values of Candidate MUCs in Breast Cancer
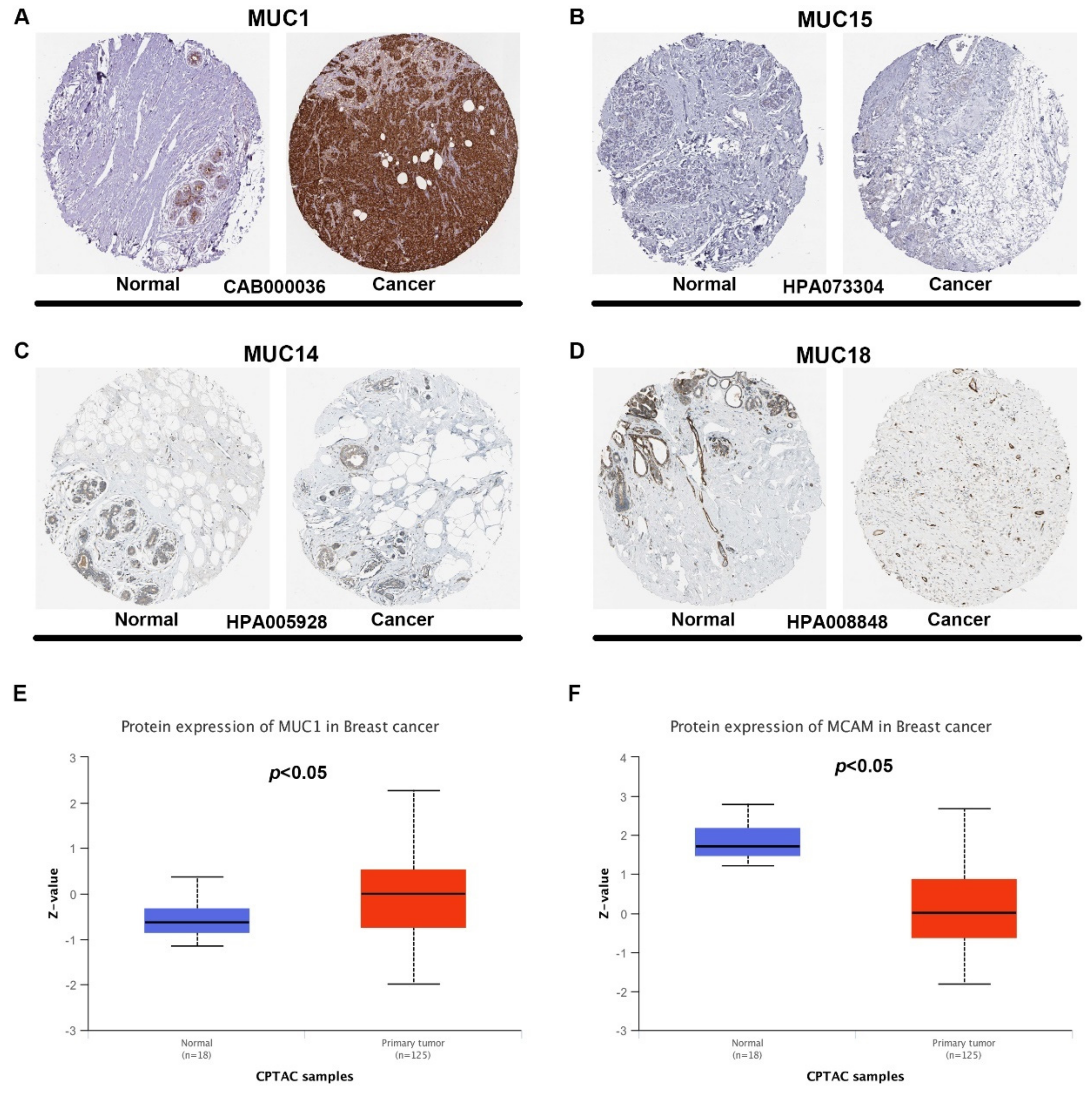
3.3. Identification of MUC14 as the Most Potential Protective Regulator in Breast Cancer
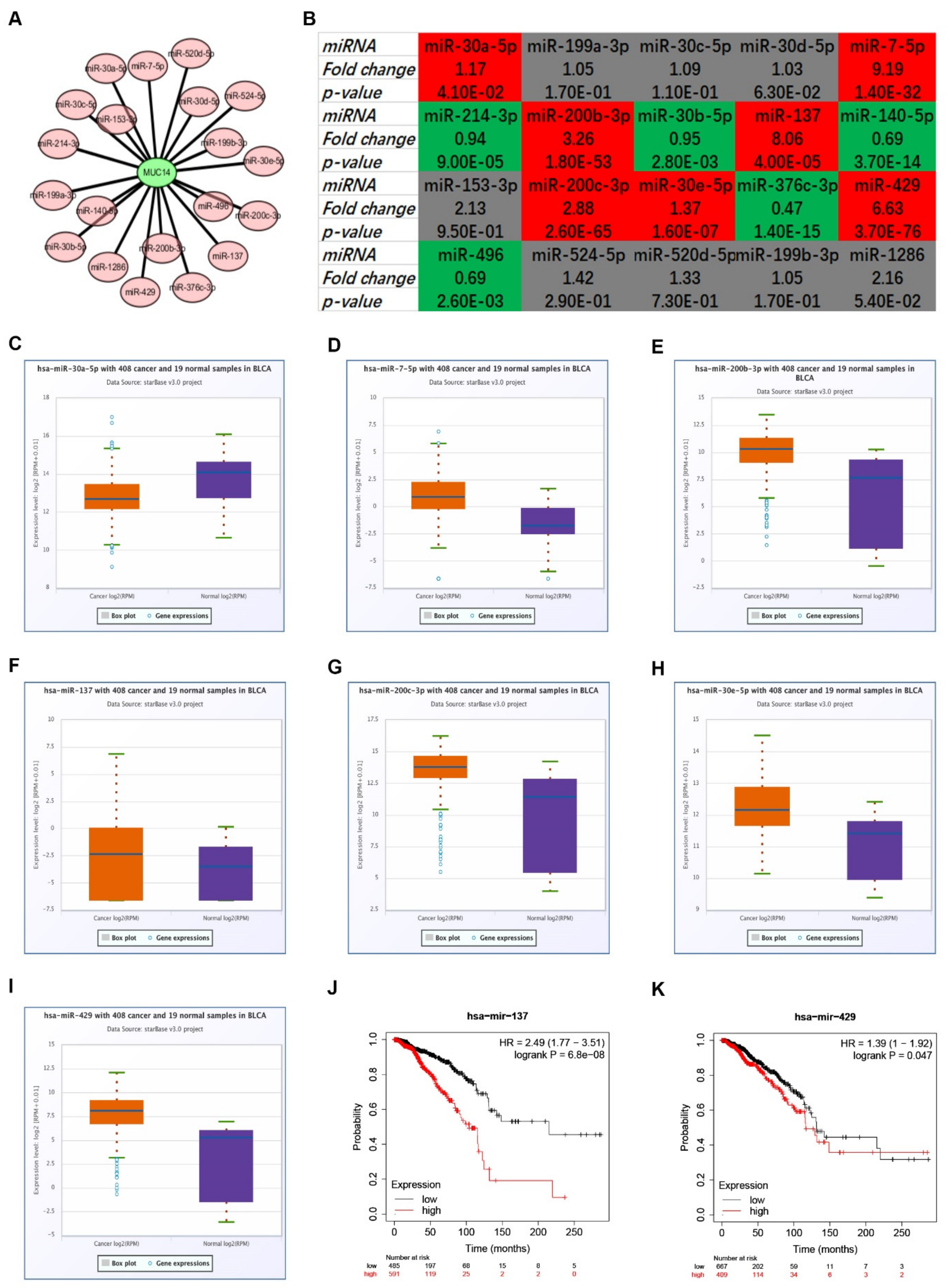
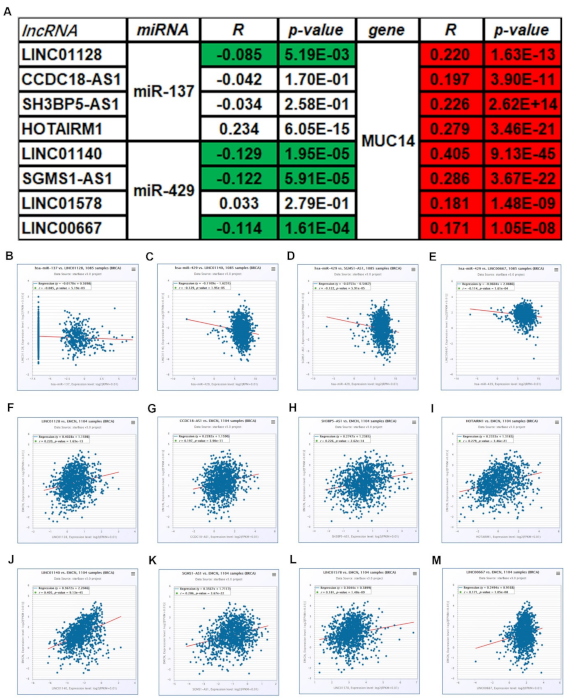
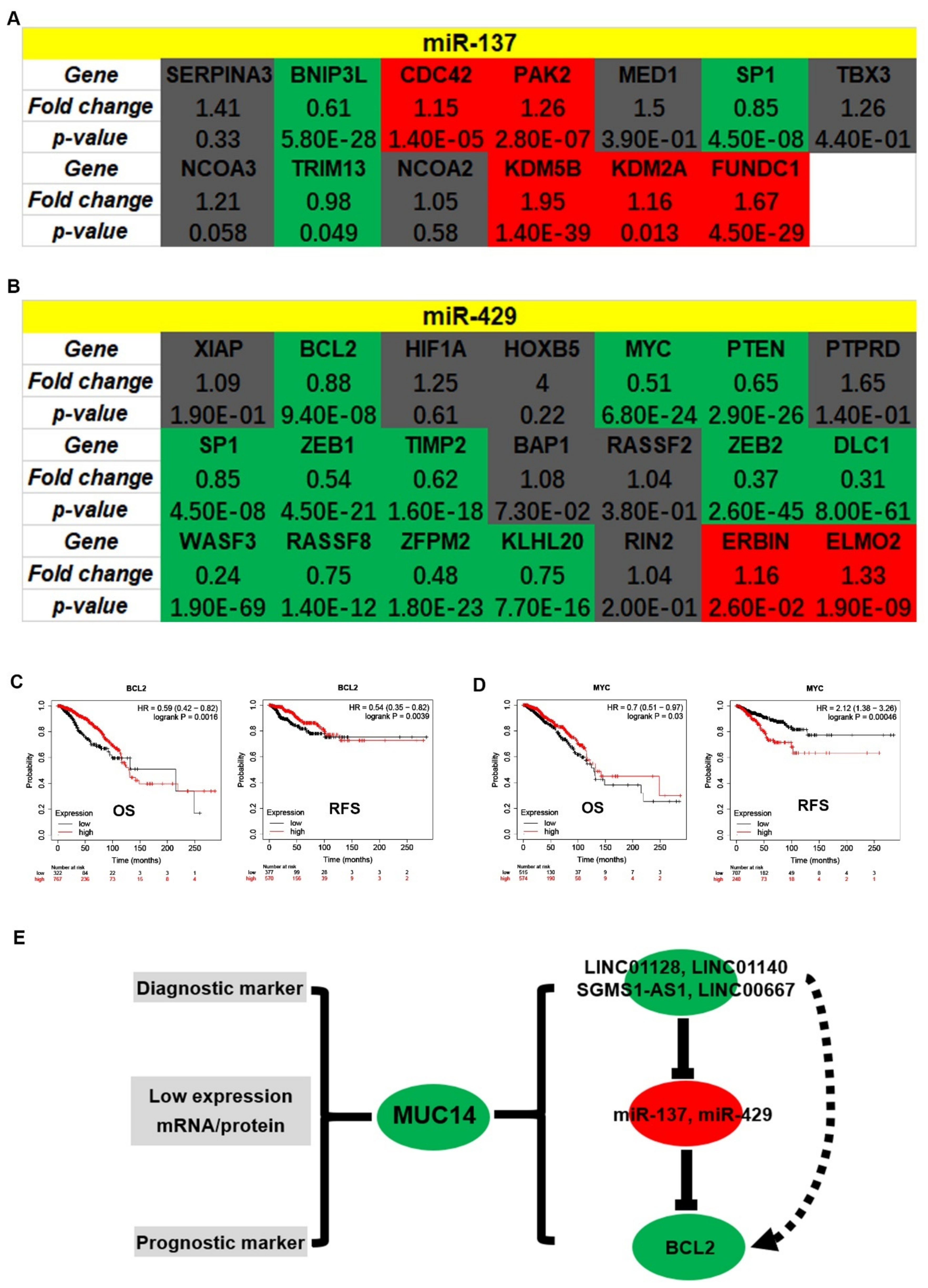
3.4. Prediction and Analysis of Upstream miRNAs of MUC14 in Breast Cancer
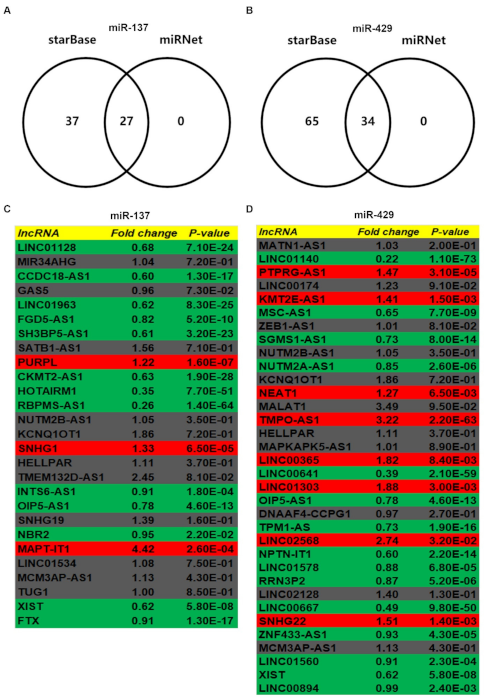
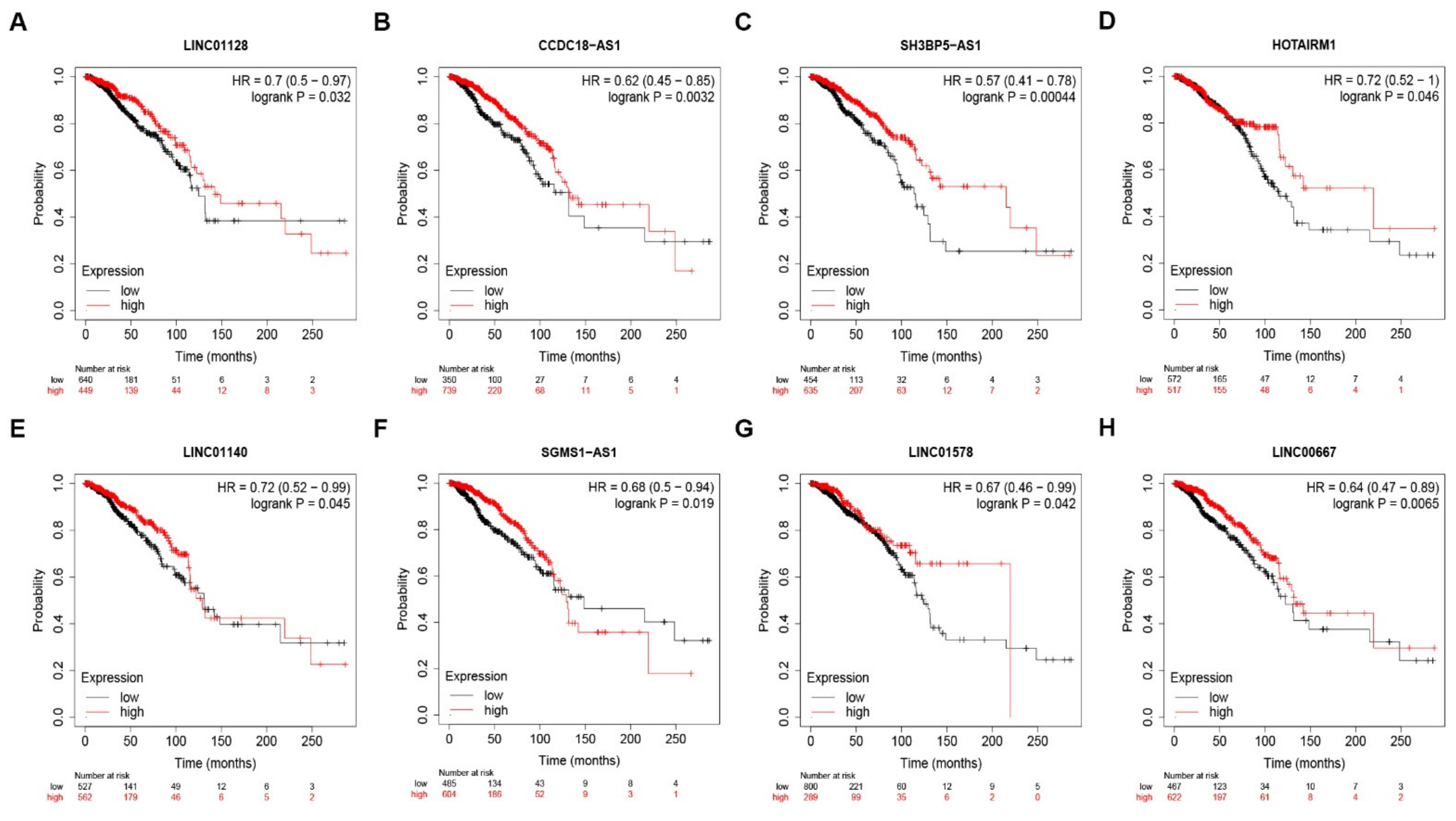
3.5. Prediction and Analysis of Upstream lncRNAs of MUC14-miR-137/miR-429 Axis in Breast Cancer
3.6. Identification of Potential Downstream Target Genes of MUC14-miR-137/miR-429 Axis in Breast Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2016, 389, 1134–1150. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, X.F.; Wang, Y.; Huang, B.F.; Hu, G.N.; Shao, J.K.; Wang, Q.; Tang, C.A.-O.; Wang, C.A.-O. Downregulated METTL14 Expression Correlates with Breast Cancer Tumor Grade and Molecular Classification. BioMed Res. Int. 2020, 2020, 8823270. [Google Scholar] [CrossRef] [PubMed]
- Lou, W.; Ding, B.; Zhong, G.; Yao, J.; Fan, W.; Fu, P. RP11-480I12.5-004 Promotes Growth and Tumorigenesis of Breast Cancer by Relieving miR-29c-3p-Mediated AKT3 and CDK6 Degradation. Mol. Ther.-Nucleic Acids 2020, 21, 916–931. [Google Scholar] [CrossRef] [PubMed]
- Lou, W.A.-O.; Ding, B.; Wang, S.; Fu, P. Overexpression of GPX3, a potential biomarker for diagnosis and prognosis of breast cancer, inhibits progression of breast cancer cells in vitro. Cancer Cell Int. 2020, 20, 1–15. [Google Scholar] [CrossRef]
- Jonckheere, N.; Auwercx, J.; Bachir, E.H.; Coppin, L.; Boukrout, N.; Vincent, A.; Neve, B.; Gautier, M.; Treviño, V.; Van Seuningen, I. Unsupervised Hierarchical Clustering of Pancreatic Adenocarcinoma Dataset from TCGA Defines a Mucin Expression Profile That Impacts Overall Survival. Cancers 2020, 12, 3309. [Google Scholar] [CrossRef]
- Ganguly, K.; Krishn, S.R.; Rachagani, S.; Jahan, R.; Shah, A.; Nallasamy, P.; Rauth, S.; Atri, P.; Cox, J.L.; Pothuraju, R.; et al. Secretory mucin 5AC promotes neoplastic progression by augmenting KLF4-mediated pancreatic cancer cell stemness. Cancer Res. 2020, 81, 91–102. [Google Scholar] [CrossRef]
- Xu, H.; Gao, H.; Li, H.; Li, D.; Yuan, W.; Zhang, L.; Cheng, P.; Su, X.; Li, Z.; Wang, G.; et al. Downregulated Mucin 1 alleviates paclitaxel resistance in non-small cell lung cancer cells. Mol. Med. Rep. 2020, 22, 2966–2972. [Google Scholar] [CrossRef]
- Gao, S.A.-O.; Yin, R.; Zhang, L.F.; Wang, S.M.; Chen, J.S.; Wu, X.Y.; Yue, C.; Zuo, L.; Tang, M. The oncogenic role of MUC12 in RCC progression depends on c-Jun/TGF-β signalling. J. Cell. Mol. Med. 2020, 24, 8789–8802. [Google Scholar] [CrossRef]
- Tiemin, P.; Fanzheng, M.; Peng, X.; Jihua, H.; Ruipeng, S.; Yaliang, L.; Yan, W.; Junlin, X.; Qingfu, L.; Zhefeng, H.; et al. MUC13 promotes intrahepatic cholangiocarcinoma progression via EGFR/PI3K/AKT pathways. J. Hepatol. 2019, 72, 761–773. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, H.; Wang, Y.; Huang, H.; Yang, J.; Gu, W.; Zhang, X.; Yang, J. Serum mucin 3A as a potential biomarker for extrahepatic cholangiocarcinoma. Saudi J. Gastroenterol. 2020, 26, 129. [Google Scholar] [PubMed]
- Niu, T.; Liu, Y.; Zhang, Y.; Fu, Q.; Liu, Z.; Wang, Z.; Fu, H.; Xu, J.; Liu, K. Increased expression of MUC3A is associated with poor prognosis in localized clear-cell renal cell carcinoma. Oncotarget 2016, 7, 50017–50026. [Google Scholar] [CrossRef] [Green Version]
- Abdelhady, A.S.; Abdel Hamid, F.F.; Hassan, N.M.; Ibrahim, D.M. Prognostic value of bone marrow MUC4 expression in acute myeloid leukaemia. Br. J. Biomed. Sci. 2020, 77, 202–207. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, J.; Liu, M.; Zhu, Q.; Li, Q.; Xie, C.; Han, C.; Wang, Y.; Gao, M.; Liu, J. Weighted correlation gene network analysis reveals a new stemness index-related survival model for prognostic prediction in hepatocellular carcinoma. Aging 2020, 12, 13502–13517. [Google Scholar] [CrossRef]
- Dai, W.; Liu, J.; Liu, B.; Li, Q.; Sang, Q.; Li, Y.Y. Systematical Analysis of the Cancer Genome Atlas Database Reveals EMCN/MUC15 Combination as a Prognostic Signature for Gastric Cancer. Front. Mol. Biosci. 2020, 7, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, B.; Zheng, C.; Yang, B.; Jin, Y.; Hou, K.; Li, Z.; Zheng, X.; Yu, S.; Zhang, X.; Fan, Y.; et al. Identification of Subtype-Specific Three-Gene Signature for Prognostic Prediction in Diffuse Type Gastric Cancer. Front. Oncol. 2019, 9, 1243. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Ding, G.; Ding, Q.; Wen, H. Overexpression of low density lipoprotein receptor-related protein 1 (LRP1) is associated with worsened prognosis and decreased cancer immunity in clear-cell renal cell carcinoma. Biochem. Biophys. Res. Commun. 2018, 503, 1537–1543. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Ding, B.; Lou, W. microRNA-Dependent Modulation of Genes Contributes to ESR1’s Effect on ERα Positive Breast Cancer. Front. Oncol. 2020, 10, 753. [Google Scholar] [CrossRef]
- Lou, W.; Ding, B.; Xu, L.; Fan, W. Construction of Potential Glioblastoma Multiforme-Related miRNA-mRNA Regulatory Network. Front. Mol. Neurosci. 2019, 12, 66. [Google Scholar] [CrossRef] [Green Version]
- Lou, W.; Liu, J.; Ding, B.; Xu, L.; Fan, W. Identification of chemoresistance-associated miRNAs in breast cancer. Cancer Manag. Res. 2018, 10, 4747–4757. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Jeong, J.-H.; Kang, S.H.; Kang, J.; Kim, E.A.; Lee, J.; Jung, J.H.; Park, H.Y.; Chae, Y.S. MicroRNA-137 Inhibits Cancer Progression by Targeting Del-1 in Triple-Negative Breast Cancer Cells. Int. J. Mol. Sci. 2019, 20, 6162. [Google Scholar] [CrossRef] [Green Version]
- Du, F.; Yu, L.; Wu, Y.; Wang, S.; Yao, J.; Zheng, X.; Xie, S.; Zhang, S.; Lu, X.; Liu, Y.; et al. miR-137 alleviates doxorubicin resistance in breast cancer through inhibition of epithelial-mesenchymal transition by targeting DUSP4. Cell Death Dis. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, Q.; Mu, Q.; Zhou, D.; Li, H.; Zhang, B.; Yin, C.A.-O. MiR-429 suppresses proliferation and invasion of breast cancer via inhibiting the Wnt/β-catenin signaling pathway. Thorac. Cancer 2020, 11, 3126–3138. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Zheng, J.; Liu, X.; Xue, Y.; Liu, L.; Ma, J.; He, Q.; Li, Z.; Cai, H.; Liu, Y. Knockdown of USF1 Inhibits the Vasculogenic Mimicry of Glioma Cells via Stimulating SNHG16/miR-212-3p and linc00667/miR-429 Axis. Mol. Ther.-Nucleic Acids 2019, 14, 465–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Li, L.; Cao, Y.; Chen, X. Downregulation of LINC01140 is associated with adverse features of breast cancer. Oncol. Lett. 2019, 19, 1157–1164. [Google Scholar] [CrossRef] [PubMed]
- Tsujimoto, Y.; Cossman, J.; Jaffe, E.; Croce, C.M. Involvement of the bcl-2 gene in human follicular lymphoma. Science 1985, 228, 1440–1443. [Google Scholar] [CrossRef]
- Tsujimoto, Y.; Ikegaki, N.; Croce, C.M. Characterization of the protein product of bcl-2, the gene involved in human follicular lymphoma. Oncogene 1987, 2, 3–7. [Google Scholar]
- Lee, K.M.; Lee, H.; Han, D.; Moon, W.K.; Kim, K.; Oh, H.J.; Choi, J.; Hwang, E.H.; Kang, S.E.; Im, S.A.; et al. Combined the SMAC mimetic and BCL2 inhibitor sensitizes neoadjuvant chemotherapy by targeting necrosome complexes in tyrosine aminoacyl-tRNA synthase-positive breast cancer. Breast Cancer Res. 2020, 22, 1–13. [Google Scholar] [CrossRef]
- Kim, J.; Shim, M.K.; Yang, S.; Moon, Y.; Song, S.; Choi, J.; Kim, J.; Kim, K. Combination of cancer-specific prodrug nanoparticle with Bcl-2 inhibitor to overcome acquired drug resistance. J. Control. Release 2020, 330, 920–932. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, F.; Javadirad, S.-M.; Amirmahani, F.; Fatehi, Z.; Tavassoli, M. Associations of BCL2 CA-Repeat Polymorphism and Breast Cancer Susceptibility in Isfahan Province of Iran. Biochem. Genet. 2020, 59, 506–515. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.H. StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pontén, F.; Schwenk, J.; Asplund, A.; Edqvist, P.H. The Human Protein Atlas as a proteomic resource for biomarker discovery. J. Intern. Med. 2011, 270, 428–446. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef] [Green Version]
- Jézéquel, P.; Frénel, J.; Campion, L.; Guérin-Charbonnel, C.; Gouraud, W.; Ricolleau, G.; Campone, M. Bc-GenExMiner 3.0: New mining module computes breast cancer gene expression correlation analyses. Database 2013, 2013, bas060. [Google Scholar] [CrossRef]
- Lou, W.; Liu, J.; Ding, B.; Jin, L.; Xu, L.; Li, X.; Chen, J.; Fan, W. Five miRNAs-mediated PIEZO2 downregulation, accompanied with activation of Hedgehog signaling pathway, predicts poor prognosis of breast cancer. Aging 2019, 11, 2628. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J.; Fan, Y.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology miRNet-Functional Analysis and Visual Exploration of miRNA-Target Interactions in a Network Context. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]


| ID | Target | R | P |
|---|---|---|---|
| miR-137 | SERPINA3 | −0.061 | 4.37 × 10−2 |
| miR-137 | AKT2 | −0.029 | 3.37 × 10−1 |
| miR-137 | BMP7 | 0.089 | 3.30 × 10−3 |
| miR-137 | BNIP3L | −0.080 | 8.31 × 10−3 |
| miR-137 | CDC42 | −0.154 | 3.52 × 10−7 |
| miR-137 | CDK6 | 0.230 | 1.94 × 10−14 |
| miR-137 | CSE1L | −0.043 | 1.57 × 10−1 |
| miR-137 | CTBP1 | 0.066 | 3.06 × 10−2 |
| miR-137 | E2F6 | −0.047 | 1.22 × 10−1 |
| miR-137 | EGFR | 0.216 | 6.73 × 10−13 |
| miR-137 | ESRRA | 0.171 | 1.49 × 10−8 |
| miR-137 | EZH2 | 0.100 | 9.51 × 10−4 |
| miR-137 | KIT | 0.029 | 3.34 × 10−1 |
| miR-137 | MET | 0.204 | 1.32 × 10−11 |
| miR-137 | MITF | 0.005 | 8.67 × 10−1 |
| miR-137 | MSI1 | 0.073 | 1.66 × 10−2 |
| miR-137 | COX2 | 0.000 | 1.00 |
| miR-137 | MYO1C | −0.034 | 2.59 × 10−1 |
| miR-137 | YBX1 | 0.215 | 8.68 × 10−13 |
| miR-137 | PAK2 | −0.118 | 1.04 × 10−4 |
| miR-137 | MED1 | −0.095 | 1.82 × 10−3 |
| miR-137 | PRKAA1 | 0.058 | 5.83 × 10−2 |
| miR-137 | PTGS2 | 0.131 | 1.44 × 10−5 |
| miR-137 | PXN | 0.049 | 1.04 × 10−1 |
| miR-137 | RASGRF1 | 0.130 | 1.74 × 10−5 |
| miR-137 | RORA | −0.059 | 5.21 × 10−2 |
| miR-137 | CXCL12 | −0.023 | 4.52 × 10−1 |
| miR-137 | SLC6A3 | 0.108 | 3.55 × 10−4 |
| miR-137 | SP1 | −0.201 | 2.25 × 10−11 |
| miR-137 | TBX3 | −0.196 | 8.10 × 10−11 |
| miR-137 | TGFB2 | 0.095 | 1.71 × 10−3 |
| miR-137 | NCOA3 | −0.121 | 6.42 × 10−5 |
| miR-137 | CUL4A | −0.010 | 7.33 × 10−1 |
| miR-137 | DCLK1 | −0.055 | 7.22 × 10−2 |
| miR-137 | KLF4 | −0.005 | 8.77 × 10−1 |
| miR-137 | KDM4A | 0.029 | 3.40 × 10−1 |
| miR-137 | TRIM13 | −0.094 | 1.98 × 10−3 |
| miR-137 | NCOA2 | −0.142 | 2.86 × 10−6 |
| miR-137 | KDM5B | −0.124 | 3.95 × 10−5 |
| miR-137 | GLIPR1 | 0.139 | 4.31 × 10−6 |
| miR-137 | KLF12 | −0.002 | 9.55 × 10−1 |
| miR-137 | Nr1i3 | 0.100 | 9.75 × 10−4 |
| miR-137 | KDM2A | −0.121 | 6.52 × 10−5 |
| miR-137 | KDM1A | 0.037 | 2.23 × 10−1 |
| miR-137 | KDM7A | −0.020 | 5.20 × 10−1 |
| miR-137 | CALN1 | 0.048 | 1.18 × 10−1 |
| miR-137 | ZNF804A | 0.010 | 7.44 × 10−1 |
| miR-137 | MTDH | −0.045 | 1.35 × 10−1 |
| miR-137 | FMNL2 | 0.158 | 1.83 × 10−7 |
| miR-137 | FUNDC1 | −0.127 | 2.63 × 10−5 |
| ID | Target | R | P |
|---|---|---|---|
| miR-429 | XIAP | −0.069 | 2.31 × 10−2 |
| miR-429 | RERE | 0.062 | 4.10 × 10−2 |
| miR-429 | BCL2 | −0.071 | 1.98 × 10−2 |
| miR-429 | CRKL | −0.022 | 4.65 × 10−1 |
| miR-429 | DNMT1 | 0.079 | 9.56 × 10−3 |
| miR-429 | EZH2 | 0.215 | 8.50 × 10−13 |
| miR-429 | HIF1A | −0.072 | 1.77 × 10−2 |
| miR-429 | HOXB5 | −0.082 | 6.84 × 10−3 |
| miR-429 | IL4 | −0.03 | 3.28 × 10−1 |
| miR-429 | MYB | 0.04 | 1.87 × 10−1 |
| miR-429 | MYC | −0.066 | 3.06 × 10−2 |
| miR-429 | PTEN | −0.165 | 4.99 × 10−8 |
| miR-429 | PTPRD | −0.213 | 1.24 × 10−12 |
| miR-429 | RBBP4 | 0.194 | 1.16 × 10−10 |
| miR-429 | SHC1 | −0.153 | 4.32 × 10−7 |
| miR-429 | FSCN1 | −0.054 | 7.76 × 10−2 |
| miR-429 | SOX2 | −0.005 | 8.63 × 10−1 |
| miR-429 | SP1 | −0.068 | 2.48 × 10−2 |
| miR-429 | ZEB1 | −0.316 | 1.38 × 10−26 |
| miR-429 | TIMP2 | −0.308 | 2.73 × 10−25 |
| miR-429 | VEGFA | 0.058 | 5.59 × 10−2 |
| miR-429 | BAP1 | −0.104 | 6.18 × 10−4 |
| miR-429 | KLF11 | −0.056 | 6.35 × 10−2 |
| miR-429 | ONECUT2 | 0.01 | 7.51 × 10−1 |
| miR-429 | RASSF2 | −0.199 | 3.83 × 10−11 |
| miR-429 | ZEB2 | −0.306 | 6.76 × 10−25 |
| miR-429 | DLC1 | −0.283 | 2.18 × 10−21 |
| miR-429 | WASF3 | −0.145 | 1.58 × 10−6 |
| miR-429 | RASSF8 | −0.144 | 1.77 × 10−6 |
| miR-429 | WDR37 | 0.046 | 1.33 × 10−1 |
| miR-429 | ZFPM2 | −0.291 | 1.18 × 10−22 |
| miR-429 | OSTF1 | 0.051 | 9.35 × 10−2 |
| miR-429 | KLHL20 | −0.201 | 2.61 × 10−11 |
| miR-429 | RIN2 | −0.175 | 6.67 × 10−9 |
| miR-429 | VAC14 | 0.053 | 8.28 × 10−2 |
| miR-429 | ERBIN | −0.151 | 5.96 × 10−7 |
| miR-429 | ELMO2 | −0.092 | 2.29 × 10−3 |
| miR-429 | TCF7L1 | −0.016 | 5.98 × 10−1 |
| miR-429 | MALAT1 | −0.02 | 5.03 × 10−1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Jin, J.; Chen, J.; Lou, W. MUC14-Related ncRNA-mRNA Network in Breast Cancer. Genes 2021, 12, 1677. https://doi.org/10.3390/genes12111677
Wang S, Jin J, Chen J, Lou W. MUC14-Related ncRNA-mRNA Network in Breast Cancer. Genes. 2021; 12(11):1677. https://doi.org/10.3390/genes12111677
Chicago/Turabian StyleWang, Shuqian, Jing Jin, Jing Chen, and Weiyang Lou. 2021. "MUC14-Related ncRNA-mRNA Network in Breast Cancer" Genes 12, no. 11: 1677. https://doi.org/10.3390/genes12111677
APA StyleWang, S., Jin, J., Chen, J., & Lou, W. (2021). MUC14-Related ncRNA-mRNA Network in Breast Cancer. Genes, 12(11), 1677. https://doi.org/10.3390/genes12111677






