DNA Methylation of Pig FUT3 Promoter Alters mRNA Expression to Regulate E. coli F18 Susceptibility
Abstract
:1. Introduction
2. Results
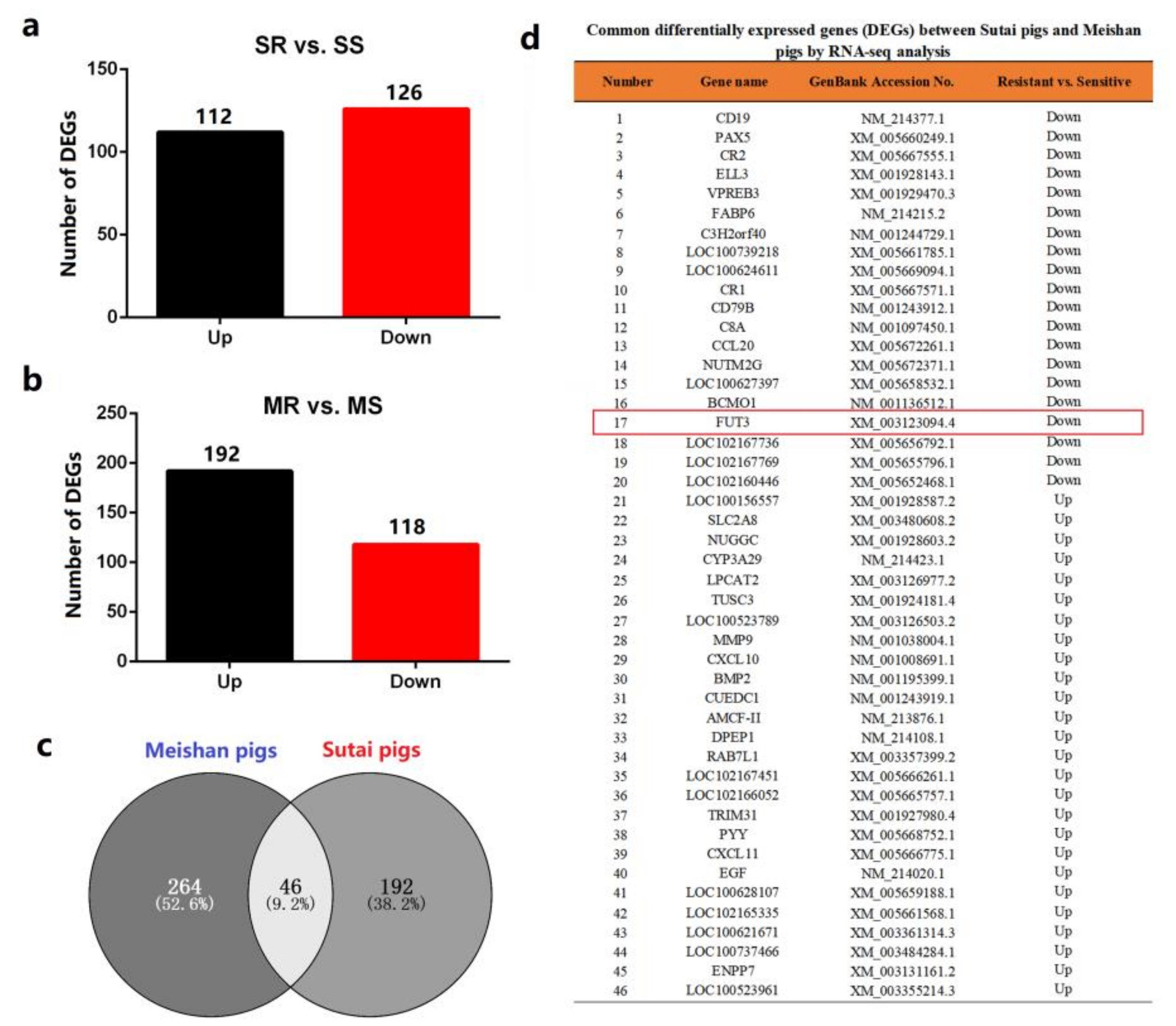
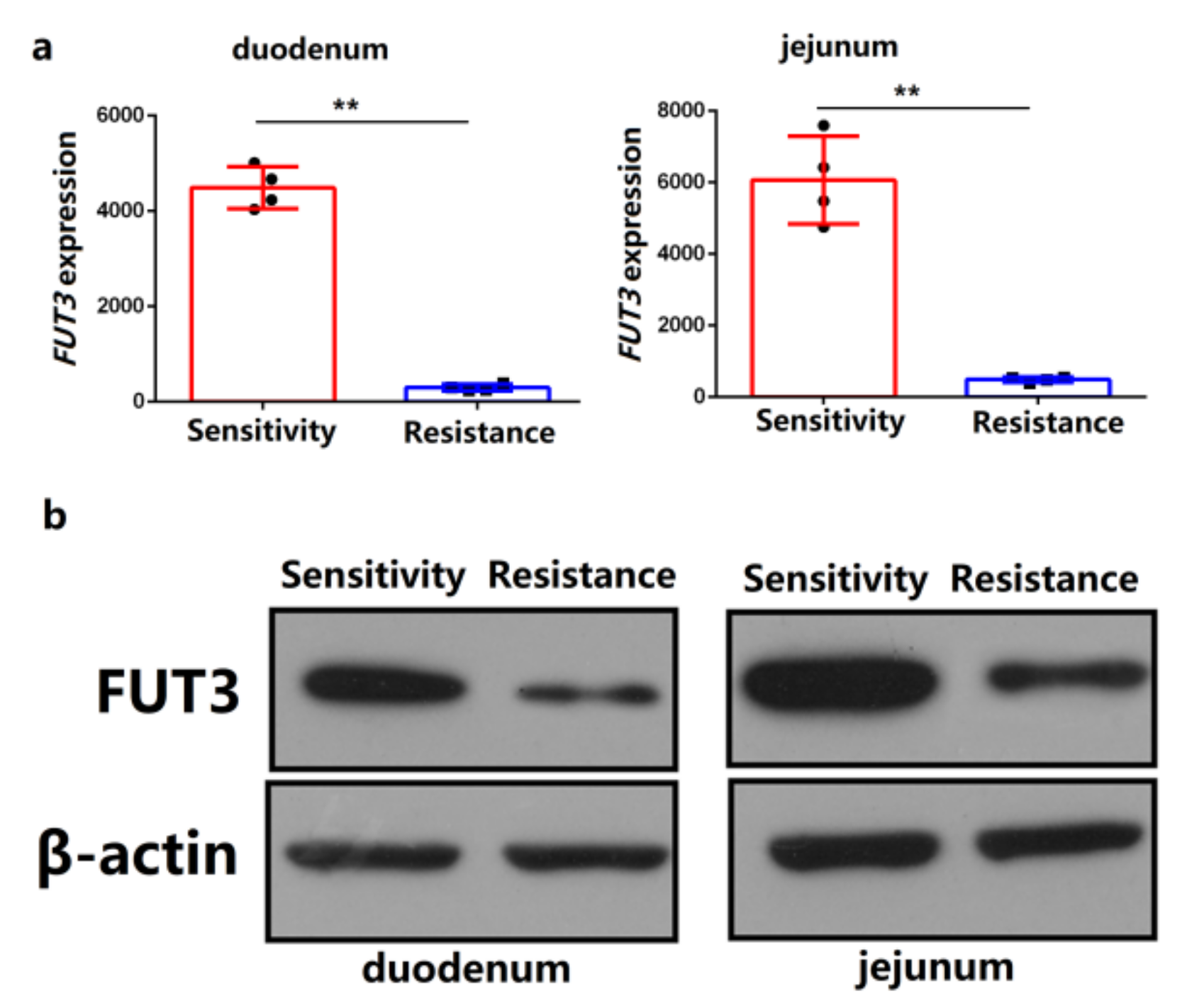
2.1. FUT3 Was Evaluated as a New Target to Combat E. coli F18 Infection on the Basis of Comparative Transcriptome Analysis
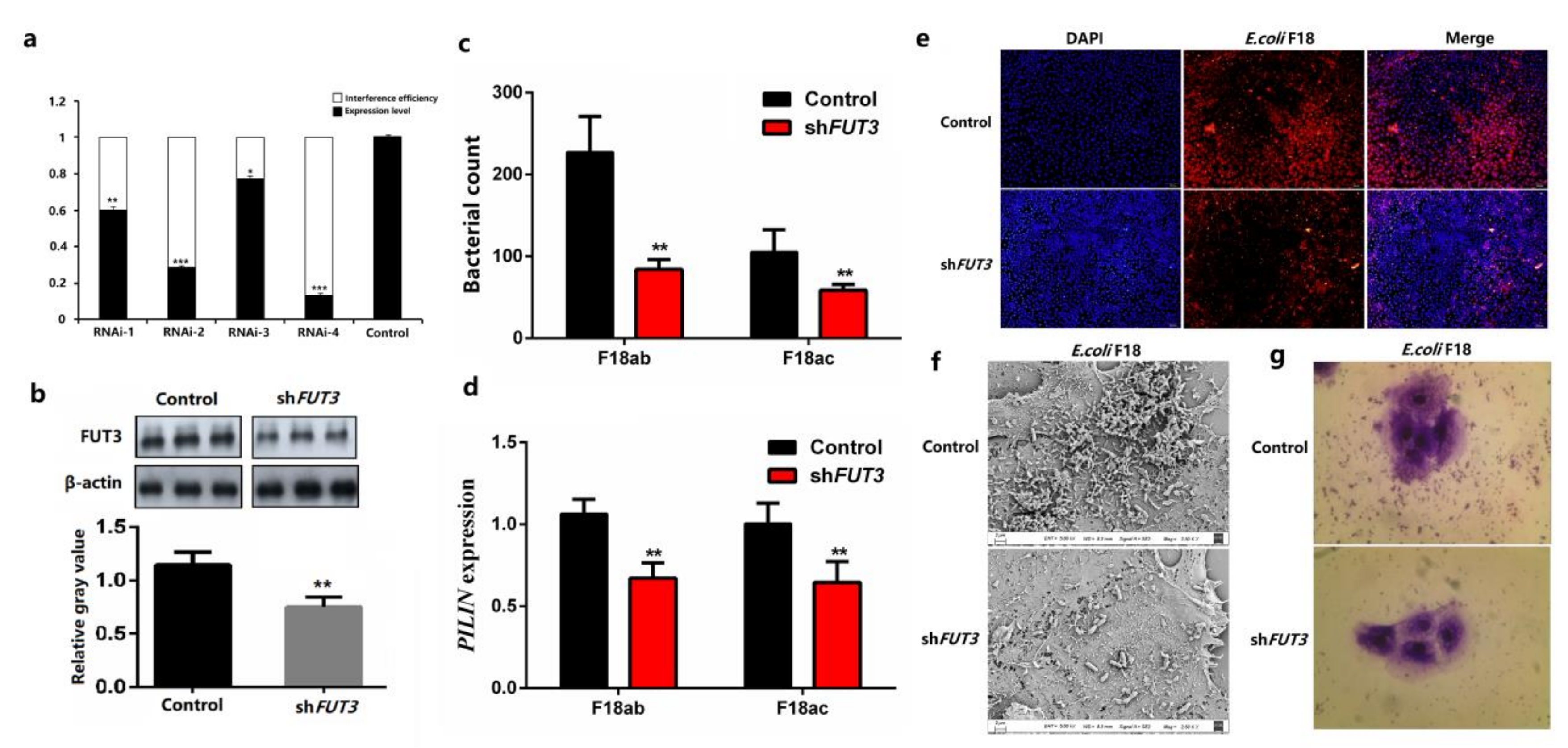
2.2. Knockdown of Pig FUT3 Enhances E. coli F18 Resistance
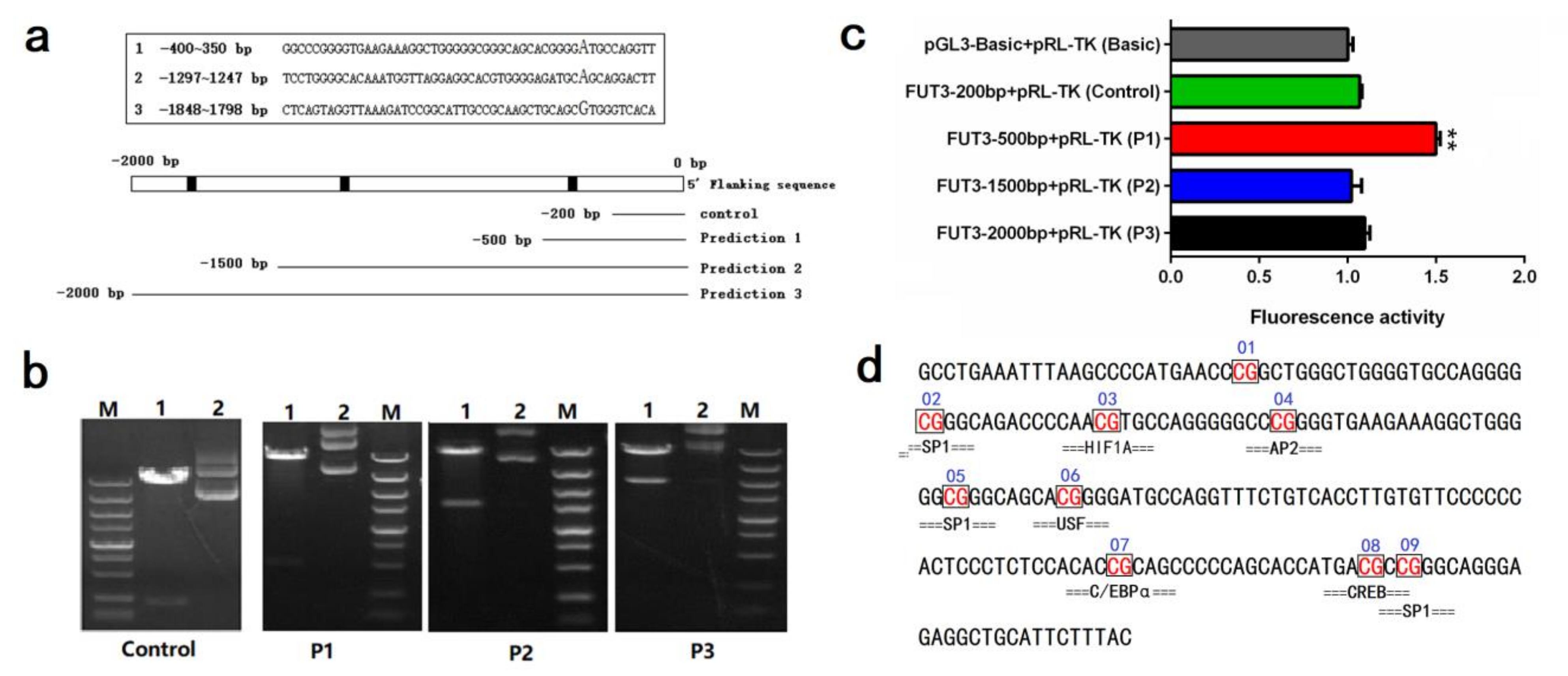
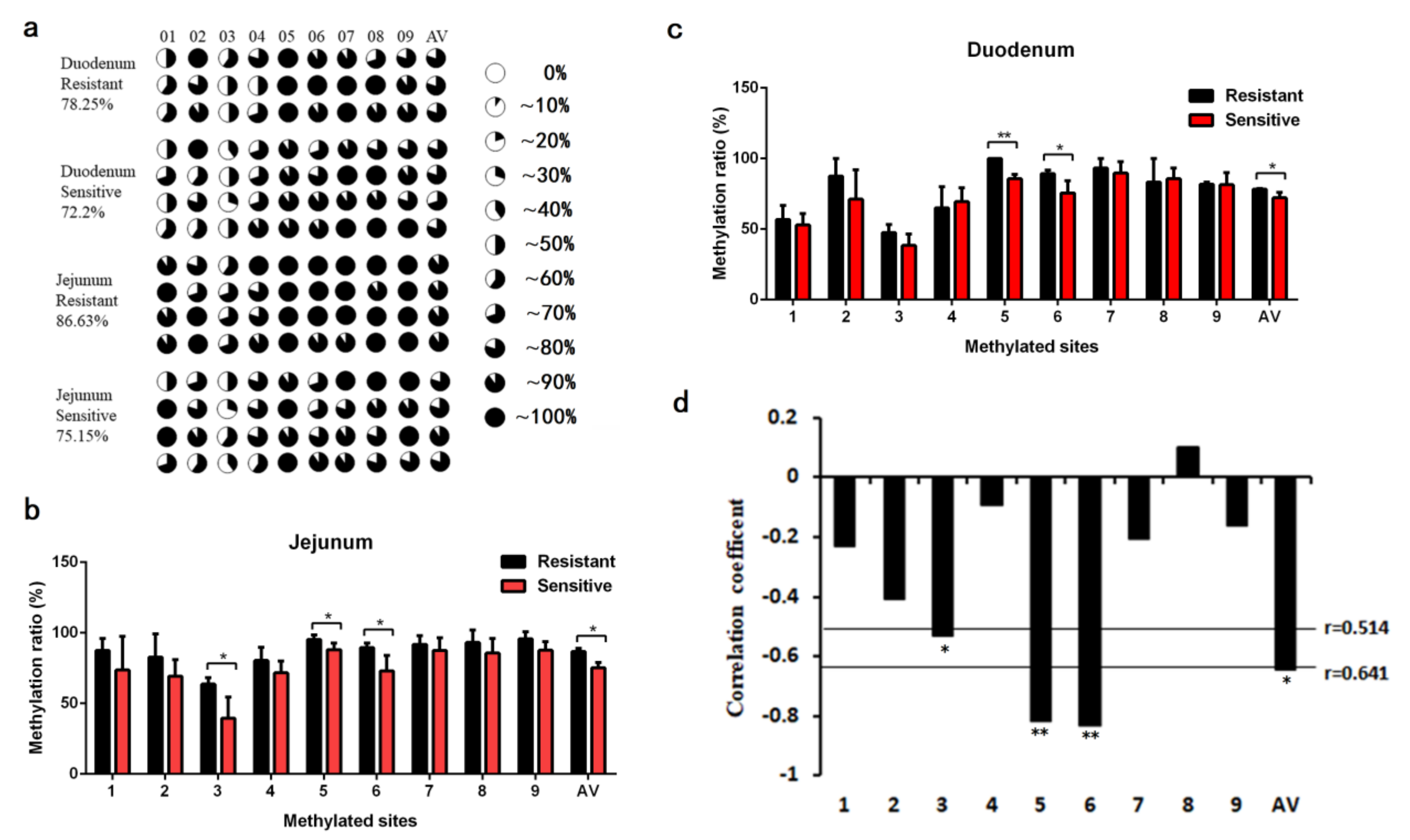
2.3. Impact of Pig FUT3 Promoter Methylation on Gene Expression
2.4. Evaluation of Key Transcription Factors in Pig FUT3 Core Promoter
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Ethics Statement
5.2. Experimental Sample
5.3. Transcriptome Sequencing (RNA-seq)
5.4. Knockdown Analysis
5.5. Adhesion Level Detection of E. coli F18 to IPEC-J2 In Vitro
5.6. RT-qPCR Analysis
5.7. Determination of Pig FUT3 Core Promoter Region
5.8. Detection of Methylation in Pig FUT3 Core Promoter Region
5.9. Identification of Key Transcription Factor in Pig FUT3 Core Promoter Region
5.10. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| E. coli | Escherichia coli |
| FUT3 | a-1,3-fucosyltransferase |
| PWD | Post-weaning diarrhea |
| TF | transcription factor |
| Sp1 | Specificity protein 1 |
| HIF1A | hypoxia-inducible factor 1-α |
| USF | upstream stimulatory factor |
| RNA-seq | RNA sequencing |
| DEGs | differentially expressed genes |
| SEM | Scanning electron microscopy |
| BSAS | bisulfite amplicon sequencing |
| IGV | Integrative Genomics Viewer |
| TFBS | transcription factor binding sites |
References
- Bender, J. DNA methylation and epigenetics. Annu. Rev. Plant Biol. 2004, 55, 41–68. [Google Scholar] [CrossRef] [PubMed]
- Flores, E.M.; Woeller, C.F.; Falsetta, M.L.; Susiarjo, M.; Phipps, R.P. Thy1 (CD90) expression is regulated by DNA methylation during adipogenesis. FASEB J. 2018, 33, 3353–3363. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Chen, T. DNA Methylation Reprogramming during Mammalian Development. Genes 2019, 10, 257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenberg, M.V.C.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Cao, Y.; Gao, Z.C.; Wu, Z.C.; Wang, H.F.; Bao, W.B. Tissue-specific expression and correlation with promoter DNA methylation of the LBP gene in pigs. J. Integr. Agric. 2020, 19, 1054–1063. [Google Scholar] [CrossRef]
- Sun, L.; Wang, J.; Yin, X.M.; Sun, S.Y.; Zi, C.; Zhu, G.Q.; Wu, S.L.; Bao, W.B. Identification of a 5-Methylcytosine Site that may Regulate C/EBPβ Binding and Determine Tissue-Specific Expression of the BPI Gene in Piglets. Sci. Rep. 2016, 6, 28506. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, X.; Yu, L.; Wang, Y.; Wang, H.; Wu, Z.; Wu, S.; Bao, W. SLC4A11 and MFSD3 Gene Expression Changes in Deoxynivalenol Treated IPEC-J2 Cells. Front. Genet. 2021, 12, 697883. [Google Scholar] [CrossRef]
- Wang, H.F.; Wu, J.Y.; Wu, S.; Wu, S.L.; Bao, W.B. DNA methylation differences of the BPI promoter among pig breeds and the regulation of gene expression. RSC Adv. 2017, 7, 48025–48030. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.F.; Feng, H.Y.; Sun, J.; Zhou, Y.J.; Zhu, G.Q.; Wu, S.L.; Bao, W.B. Age-associated changes in DNA methylation and expression of the TNFα gene in pigs. Gene Genet. Syst. 2018, 93, 191–198. [Google Scholar] [CrossRef] [Green Version]
- Dai, C.; Sun, L.; Xia, R.; Sun, S.; Zhu, G.; Wu, S.; Bao, W. Correlation between the methylation of the FUT1 promoter region and FUT1 expression in the duodenum of piglets from newborn to weaning. 3 Biotech 2017, 7, 247. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yin, X.M.; Sun, L.; Sun, S.Y.; Zi, C.; Zhu, G.Q.; Wu, S.L.; Bao, W.B. Correlation between BPI Gene Upstream CpG Island Methylation and mRNA Expression in Piglets. Int. J. Mol. Sci. 2014, 15, 10989–10998. [Google Scholar] [CrossRef]
- Zheng, Z.X.; Wan, C.; Mo, D.L.; Li, J.P.; Chen, Y.S.; Zhang, Z.W.; Cong, P. Promoter CpG methylation status in porcine Lyn is associated with its expression levels. Gene 2012, 511, 73–78. [Google Scholar] [CrossRef]
- Dong, W.H.; Yin, X.M.; Sun, L.; Wang, J.; Sun, S.Y.; Zhu, G.Q.; Wu, S.L.; Bao, W.B. Age-associated methylation change of TAP1 promoter in piglet. Gene 2015, 573, 70–74. [Google Scholar] [CrossRef]
- Boldin, B. Persistence and spread of gastro-intestinal infections: The case of enterotoxigenic Escherichia coli in piglets. Bull. Math. Biol. 2008, 70, 2077–2101. [Google Scholar] [CrossRef]
- Ye, L.; Su, X.M.; Wu, Z.C.; Zheng, X.R.; Wang, J.; Zi, C.; Wu, S.L.; Bao, W.B. Analysis of differential miRNA expression in the duodenum of Escherichia coli F18-sensitive and -resistant weaned piglets. PLoS ONE 2012, 7, e437412012. [Google Scholar] [CrossRef]
- Wu, Z.C.; Dong, W.H.; Liu, Y.; Yang, J.S.; Zhu, G.Q.; Wu, S.L.; Bao, W.B. Attack Experiment and Phenotype Analysis of Meishan piglets by E. coli F18 Strain. Acta Vet. Zootech. Sin. 2014, 45, 1608–1615. (In Chinese) [Google Scholar]
- Heggelund, J.E.; Varrot, A.; Imberty, A.; Krengel, U. Histo-blood group antigens as mediators of infections. Curr. Opin. Struct. Biol. 2017, 44, 190–200. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Liu, Y.; Tan, M. Histo-blood group antigens as receptors for rotavirus, new understanding on rotavirus epidemiology and vaccine strategy. Emerg. Microbes Infect. 2017, 6, e22. [Google Scholar] [CrossRef] [Green Version]
- Coddens, A.; Diswall, M.; Angström, J.; Breimer, M.E.; Goddeeris, B.; Cox, E.; Teneberg, S. Recognition of blood group ABH type 1 determinants by the FedF adhesin of F18-fimbriated Escherichia coli. J. Biol. Chem. 2009, 284, 9713–9726. [Google Scholar] [CrossRef] [Green Version]
- Moonens, K.; Bouckaert, J.; Coddens, A.; Tran, T.; Panjikar, S.; De Kerpel, M.; Cox, E.; Remaut, H.; De Greve, H. Structural insight in histo-blood group binding by the F18 fimbrial adhesin FedF. Mol. Microbiol. 2012, 86, 82–95. [Google Scholar] [CrossRef]
- Lonardi, E.; Moonens, K.; Buts, L.; de Boer, A.R.; Olsson, J.D.; Weiss, M.S.; Fabre, E.; Guérardel, Y.; Deelder, A.M.; Oscarson, S.; et al. Structural sampling of glycan interaction profiles reveals mucosal receptors for fimbrial adhesins of enterotoxigenic Escherichia coli. Biology 2013, 2, 894–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, C.H.; Yang, L.; Jin, J.; Wang, H.F.; Wu, S.L.; Bao, W.B. Regulation and Molecular Mechanism of TLR5 on Resistance to Escherichia coli F18 in Weaned Piglets. Animals 2019, 9, 735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Liu, Y.; Dong, W.; Zhu, G.Q.; Wu, S.; Bao, W. CD14 in the TLRs signaling pathway is associated with the resistance to E. coli F18 in Chinese domestic weaned piglets. Sci. Rep. 2016, 6, 24611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Q.S.; Xie, X.M.; Liu, X.C.; Huang, S.Q.; He, C.Q. Experimental results on enterotoxigenic E. coli F18 receptor genotypes. Hereditas 2002, 24, 656–658. (In Chinese) [Google Scholar]
- Marionneau, S.; Cailleau-Thomas, A.; Rocher, J.; Le Moullac-Vaidye, B.; Ruvoën, N.; Clément, M.; Le Pendu, J. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie 2001, 83, 565–573. [Google Scholar] [CrossRef]
- Hu, D.Y.; Zhang, D.G.; Zheng, S.Z.; Guo, M.D.; Lin, X.X.; Jiang, Y. Association of Ulcerative Colitis with FUT2 and FUT3 Polymorphisms in Patients from Southeast China. PLoS ONE 2016, 11, e0146557. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.L.; Yu, Y.; Yuan, Z.F.; Yang, J.; Ma, P.P.; Li, D.C.; Yu, S.K.; An, F.; Feng, X.J.; Zhang, Y. Comparative analysis on content and distribution of CpG sites in milk production traits and mastitis-related genes in dairy cattle. Hereditas 2012, 34, 437–444. (In Chinese) [Google Scholar] [CrossRef]
- Walczak-Drzewiecka, A.; Ratajewski, M.; Pułaski, Ł.; Dastych, J. DNA methylation-dependent suppression of HIF1A in an immature hematopoietic cell line HMC-1. Biochem. Biophys. Res. Commun. 2010, 391, 1028–1032. [Google Scholar] [CrossRef]
- Tian, H.P.; Lun, S.M.; Huang, H.J.; He, R.; Kong, P.Z.; Wang, Q.S.; Li, X.Q.; Feng, Y.M. DNA Methylation Affects the SP1-regulated Transcription of FOXF2 in Breast Cancer Cells. J. Biol. Chem. 2015, 290, 19173–19183. [Google Scholar] [CrossRef] [Green Version]
- Mancarelli, M.M.; Zazzeroni, F.; Ciccocioppo, L.; Capece, D.; Po, A.; Murgo, S.; Di Camillo, R.; Rinaldi, C.; Ferretti, E.; Gulino, A.; et al. The tumor suppressor gene KCTD11REN is regulated by Sp1 and methylation and its expression is reduced in tumors. Mol. Cancer 2010, 9, 172. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.P.; Leonard, W.J. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: A role for DNA methylation. J. Exp. Med. 2007, 204, 1543–1551. [Google Scholar] [CrossRef]
- Fujii, G.; Nakamura, Y.; Tsukamoto, D.; Ito, M.; Shiba, T.; Takamatsu, N. CpG methylation at the USF-binding site is important for the liver-specific transcription of the chipmunk HP-27 gene. Biochem. J. 2006, 395, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Wang, J.; Zhao, Q.; Zi, C.; Wu, Z.; Su, X.; Huo, Y.; Zhu, G.; Wu, S.; Bao, W. Genetic variation in exon 10 of the BPI gene is associated with Escherichia coli F18 susceptibility in Sutai piglets. Gene 2013, 523, 70–75. [Google Scholar] [CrossRef]
- Dai, C.H.; Gan, L.N.; Qin, W.U.; Zi, C.; Zhu, G.Q.; Wu, S.L.; Bao, W.B. Use of fluorescence quantitative polymerase chain reaction (PCR) for the detection of Escherichia coli adhesion to pig intestinal epithelial cells. Pol. J. Vet. Sci. 2016, 19, 303–308. [Google Scholar] [CrossRef] [Green Version]
- Kumaki, Y.; Oda, M.; Okano, M. QUMA: Quantification tool for methylation analysis. Nucleic Acids Res. 2008, 36, W170–W175. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

| Name. | Sequence of the Oligo (5’ → 3’) |
|---|---|
| R1F | GATCCggtctggttcagcatggaatcTTCAAGAGAgattccatgctgaaccagaccTTTTTTG |
| R1R | AATTCAAAAAAggtctggttcagcatggaatcTCTCTTGAAgattccatgctgaaccagaccG |
| R2F | GATCCgcagggactctgatatcttcaTTCAAGAGAtgaagatatcagagtccctgcTTTTTTG |
| R2R | AATTCAAAAAAgcagggactctgatatcttcaTCTCTTGAAtgaagatatcagagtccctgcG |
| R3F | GATCCgctcaacatctcggccaagaaTTCAAGAGAttcttggccgagatgttgagcTTTTTTG |
| R3R | AATTCAAAAAAgctcaacatctcggccaagaaTCTCTTGAAttcttggccgagatgttgagcG |
| R4F | GATCCgacctccaagtggacgtgtatgTTCAAGAGAcatacacgtccacttggaggtcTTTTTTG |
| R4R | AATTCAAAAAAgacctccaagtggacgtgtatgTCTCTTGAAcatacacgtccacttggaggtcG |
| NC-F | GATCCttctccgaacgtgtcacgtTTCAAGAGAagttagttgggactttgttgcTTTTTTG |
| NC-R | AATTCAAAAAAttctccgaacgtgtcacgtTCTCTTGAAagttagttgggactttgttgcG |
| Gene Name | GenBank Accession Number | Primer Sequence | Fragment Size |
|---|---|---|---|
| FUT3 | AF130972.1 | F: 5′-CCCGAAGCCTTCATCCACAT-3′ | 150 bp |
| R: 5′-CATCAAGGCCCAGCTGAAGA-3′ | |||
| PILIN | M25302.1 | F: 5′-AGGCCGAACCAAAGAAGCAT-3′ | 117 bp |
| R: 5′-TCACCATCAGGGTTTCTGAGT-3′ | |||
| β-actin | NC_010445.3 | F:GTCGTACTCCTGCTTGCTGAT R:CCTTCTCCTTCCAGATCATCGC | 119 bp |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Z.; Shi, D.; Jin, J.; Fan, H.; Bao, W.; Wu, S. DNA Methylation of Pig FUT3 Promoter Alters mRNA Expression to Regulate E. coli F18 Susceptibility. Genes 2021, 12, 1586. https://doi.org/10.3390/genes12101586
Wu Z, Shi D, Jin J, Fan H, Bao W, Wu S. DNA Methylation of Pig FUT3 Promoter Alters mRNA Expression to Regulate E. coli F18 Susceptibility. Genes. 2021; 12(10):1586. https://doi.org/10.3390/genes12101586
Chicago/Turabian StyleWu, Zhengchang, Dongfeng Shi, Jian Jin, Hairui Fan, Wenbin Bao, and Shenglong Wu. 2021. "DNA Methylation of Pig FUT3 Promoter Alters mRNA Expression to Regulate E. coli F18 Susceptibility" Genes 12, no. 10: 1586. https://doi.org/10.3390/genes12101586
APA StyleWu, Z., Shi, D., Jin, J., Fan, H., Bao, W., & Wu, S. (2021). DNA Methylation of Pig FUT3 Promoter Alters mRNA Expression to Regulate E. coli F18 Susceptibility. Genes, 12(10), 1586. https://doi.org/10.3390/genes12101586








