Drosophila Models for Charcot–Marie–Tooth Neuropathy Related to Aminoacyl-tRNA Synthetases
Abstract
1. Charcot–Marie–Tooth Neuropathy: The Most Common Genetic Affliction of the Peripheral Nervous System
2. Aminoacyl-tRNA Synthetases Causing Peripheral Neuropathies
3. Modeling aaRSCMT in D. melanogaster: Why and How
4. Mechanistic Insights Derived from D. melanogaster aaRSCMT Models
4.1. CMT Mutants Active for Aminoacylation Induce Neurodegeneration In Vivo
4.2. CMT-Mutations in YARS1 and GARS1 Reduce Global Protein Translation
4.3. CMT Mutations Do Not Impair the Subcellular Localization of aaRS in D. melanogaster
4.4. Nuclear YARS1 as a Transcriptional Regulator
4.5. GARS1 Is Secreted to Accumulate at the Synaptic Membrane and Interacts with the Plexin-Semaphorin Signaling
4.6. GARS1-Induced Neurotoxicity Is Rescued by Inhibition of Sirt2 Deacetylase Activity
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Catala, M.; Kubis, N. Gross anatomy and development of the peripheral nervous system. Handb. Clin. Neurol. 2013, 115, 29–41. [Google Scholar] [CrossRef]
- Tooth, H.H. The Peroneal Type of Progressive Muscular Atrophy; University of Cambridge: London, UK, 1886. [Google Scholar]
- Charcot, J.M. Sur une forme paticuliere d’atrophie musuculaire progressive souvent familial, debutante par les pieds et les jambes et atteignant plus tard les mains. Rev. Med. Fr. 1886, 6, 97–138. [Google Scholar]
- Skre, H. Genetic and clinical aspects of Charcot-Marie-Tooth’s disease. Clin. Genet. 1974, 6, 98–118. [Google Scholar] [CrossRef]
- Barreto, L.C.; Oliveira, F.S.; Nunes, P.S.; de Franca Costa, I.M.; Garcez, C.A.; Goes, G.M.; Neves, E.L.; de Souza Siqueira Quintans, J.; de Souza Araujo, A.A. Epidemiologic Study of Charcot-Marie-Tooth Disease: A Systematic Review. Neuroepidemiology 2016, 46, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Mladenovic, J.; Milic Rasic, V.; Keckarevic Markovic, M.; Romac, S.; Todorovic, S.; Rakocevic Stojanovic, V.; Kisic Tepavcevic, D.; Hofman, A.; Pekmezovic, T. Epidemiology of Charcot-Marie-Tooth disease in the population of Belgrade, Serbia. Neuroepidemiology 2011, 36, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Pareyson, D.; Saveri, P.; Pisciotta, C. New developments in Charcot-Marie-Tooth neuropathy and related diseases. Curr. Opin. Neurol. 2017, 30, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Pareyson, D.; Scaioli, V.; Laura, M. Clinical and electrophysiological aspects of Charcot-Marie-Tooth disease. Neuromol. Med. 2006, 8, 3–22. [Google Scholar] [CrossRef]
- Garcia, C.A.; Malamut, R.E.; England, J.D.; Parry, G.S.; Liu, P.; Lupski, J.R. Clinical variability in two pairs of identical twins with the Charcot-Marie-Tooth disease type 1A duplication. Neurology 1995, 45, 2090–2093. [Google Scholar] [CrossRef]
- Nagappa, M.; Sharma, S.; Taly, A.B. Charcot Marie Tooth. In StatPearls; StatPearls: Treasure Island, FL, USA, 2020. [Google Scholar]
- Reilly, M.M.; Shy, M.E. Diagnosis and new treatments in genetic neuropathies. J. Neurol. Neurosurg. Psychiatry 2009, 80, 1304–1314. [Google Scholar] [CrossRef] [PubMed]
- Schroder, J.M. Neuropathology of Charcot-Marie-Tooth and related disorders. Neuromolecular Med. 2006, 8, 23–42. [Google Scholar] [CrossRef]
- Bienfait, H.M.; Baas, F.; Koelman, J.H.; de Haan, R.J.; van Engelen, B.G.; Gabreels-Festen, A.A.; Ongerboer de Visser, B.W.; Meggouh, F.; Weterman, M.A.; De Jonghe, P.; et al. Phenotype of Charcot-Marie-Tooth disease Type 2. Neurology 2007, 68, 1658–1667. [Google Scholar] [CrossRef]
- Berciano, J.; Garcia, A.; Gallardo, E.; Peeters, K.; Pelayo-Negro, A.L.; Alvarez-Paradelo, S.; Gazulla, J.; Martinez-Tames, M.; Infante, J.; Jordanova, A. Intermediate Charcot-Marie-Tooth disease: An electrophysiological reappraisal and systematic review. J. Neurol. 2017, 264, 1655–1677. [Google Scholar] [CrossRef]
- Saporta, A.S.; Sottile, S.L.; Miller, L.J.; Feely, S.M.; Siskind, C.E.; Shy, M.E. Charcot-Marie-Tooth disease subtypes and genetic testing strategies. Ann. Neurol. 2011, 69, 22–33. [Google Scholar] [CrossRef]
- DiVincenzo, C.; Elzinga, C.D.; Medeiros, A.C.; Karbassi, I.; Jones, J.R.; Evans, M.C.; Braastad, C.D.; Bishop, C.M.; Jaremko, M.; Wang, Z.; et al. The allelic spectrum of Charcot-Marie-Tooth disease in over 17,000 individuals with neuropathy. Mol. Genet. Genomic Med. 2014, 2, 522–529. [Google Scholar] [CrossRef]
- Rossor, A.M.; Polke, J.M.; Houlden, H.; Reilly, M.M. Clinical implications of genetic advances in Charcot-Marie-Tooth disease. Nat. Rev. Neurol. 2013, 9, 562–571. [Google Scholar] [CrossRef]
- Antonellis, A.; Ellsworth, R.E.; Sambuughin, N.; Puls, I.; Abel, A.; Lee-Lin, S.Q.; Jordanova, A.; Kremensky, I.; Christodoulou, K.; Middleton, L.T.; et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am. J. Hum. Genet. 2003, 72, 1293–1299. [Google Scholar] [CrossRef]
- Jordanova, A.; Irobi, J.; Thomas, F.P.; Van Dijck, P.; Meerschaert, K.; Dewil, M.; Dierick, I.; Jacobs, A.; De Vriendt, E.; Guergueltcheva, V.; et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat. Genet. 2006, 38, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Latour, P.; Thauvin-Robinet, C.; Baudelet-Mery, C.; Soichot, P.; Cusin, V.; Faivre, L.; Locatelli, M.C.; Mayencon, M.; Sarcey, A.; Broussolle, E.; et al. A major determinant for binding and aminoacylation of tRNA(Ala) in cytoplasmic Alanyl-tRNA synthetase is mutated in dominant axonal Charcot-Marie-Tooth disease. Am. J. Hum. Genet. 2010, 86, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Vester, A.; Velez-Ruiz, G.; McLaughlin, H.M.; Program, N.C.S.; Lupski, J.R.; Talbot, K.; Vance, J.M.; Zuchner, S.; Roda, R.H.; Fischbeck, K.H.; et al. A loss-of-function variant in the human histidyl-tRNA synthetase (HARS) gene is neurotoxic in vivo. Hum. Mutat. 2013, 34, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.; McLaughlin, H.; Houlden, H.; Guo, M.; Yo-Tsen, L.; Hadjivassilious, M.; Speziani, F.; Yang, X.L.; Antonellis, A.; Reilly, M.M.; et al. Exome sequencing identifies a significant variant in methionyl-tRNA synthetase (MARS) in a family with late-onset CMT2. J. Neurol. Neurosurg. Psychiatry 2013, 84, 1247–1249. [Google Scholar] [CrossRef] [PubMed]
- Tsai, P.C.; Soong, B.W.; Mademan, I.; Huang, Y.H.; Liu, C.R.; Hsiao, C.T.; Wu, H.T.; Liu, T.T.; Liu, Y.T.; Tseng, Y.T.; et al. A recurrent WARS mutation is a novel cause of autosomal dominant distal hereditary motor neuropathy. Brain 2017, 140, 1252–1266. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, H.M.; Sakaguchi, R.; Liu, C.; Igarashi, T.; Pehlivan, D.; Chu, K.; Iyer, R.; Cruz, P.; Cherukuri, P.F.; Hansen, N.F.; et al. Compound heterozygosity for loss-of-function lysyl-tRNA synthetase mutations in a patient with peripheral neuropathy. Am. J. Hum. Genet. 2010, 87, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Guo, M. Structural characterization of human aminoacyl-tRNA synthetases for translational and nontranslational functions. Methods 2017, 113, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R.; Olsen, G.J.; Ibba, M.; Soll, D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000, 64, 202–236. [Google Scholar] [CrossRef]
- Jiang, L.; Jones, J.; Yang, X.L. Human diseases linked to cytoplasmic aminoacyl-tRNA synthetases. Enzymes 2020, 48, 277–319. [Google Scholar] [CrossRef]
- Motley, W.W.; Griffin, L.B.; Mademan, I.; Baets, J.; De Vriendt, E.; De Jonghe, P.; Antonellis, A.; Jordanova, A.; Scherer, S.S. A novel AARS mutation in a family with dominant myeloneuropathy. Neurology 2015, 84, 2040–2047. [Google Scholar] [CrossRef]
- McLaughlin, H.M.; Sakaguchi, R.; Giblin, W.; Program, N.C.S.; Wilson, T.E.; Biesecker, L.; Lupski, J.R.; Talbot, K.; Vance, J.M.; Zuchner, S.; et al. A recurrent loss-of-function alanyl-tRNA synthetase (AARS) mutation in patients with Charcot-Marie-Tooth disease type 2N (CMT2N). Hum. Mutat. 2012, 33, 244–253. [Google Scholar] [CrossRef]
- Royer-Bertrand, B.; Tsouni, P.; Mullen, P.; Campos Xavier, B.; Mittaz Crettol, L.; Lobrinus, A.J.; Ghika, J.; Baumgartner, M.R.; Rivolta, C.; Superti-Furga, A.; et al. Peripheral neuropathy and cognitive impairment associated with a novel monoallelic HARS variant. Ann. Clin. Transl. Neurol. 2019, 6, 1072–1080. [Google Scholar] [CrossRef]
- Blocquel, D.; Sun, L.; Matuszek, Z.; Li, S.; Weber, T.; Kuhle, B.; Kooi, G.; Wei, N.; Baets, J.; Pan, T.; et al. CMT disease severity correlates with mutation-induced open conformation of histidyl-tRNA synthetase, not aminoacylation loss, in patient cells. Proc. Natl. Acad. Sci. USA 2019, 116, 19440–19448. [Google Scholar] [CrossRef]
- Abbott, J.A.; Meyer-Schuman, R.; Lupo, V.; Feely, S.; Mademan, I.; Oprescu, S.N.; Griffin, L.B.; Alberti, M.A.; Casasnovas, C.; Aharoni, S.; et al. Substrate interaction defects in histidyl-tRNA synthetase linked to dominant axonal peripheral neuropathy. Hum. Mutat. 2018, 39, 415–432. [Google Scholar] [CrossRef]
- Storkebaum, E.; Leitao-Goncalves, R.; Godenschwege, T.; Nangle, L.; Mejia, M.; Bosmans, I.; Ooms, T.; Jacobs, A.; Van Dijck, P.; Yang, X.L.; et al. Dominant mutations in the tyrosyl-tRNA synthetase gene recapitulate in Drosophila features of human Charcot-Marie-Tooth neuropathy. Proc. Natl. Acad. Sci. USA 2009, 106, 11782–11787. [Google Scholar] [CrossRef] [PubMed]
- Griffin, L.B.; Sakaguchi, R.; McGuigan, D.; Gonzalez, M.A.; Searby, C.; Zuchner, S.; Hou, Y.M.; Antonellis, A. Impaired function is a common feature of neuropathy-associated glycyl-tRNA synthetase mutations. Hum. Mutat. 2014, 35, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Safka Brozkova, D.; Deconinck, T.; Griffin, L.B.; Ferbert, A.; Haberlova, J.; Mazanec, R.; Lassuthova, P.; Roth, C.; Pilunthanakul, T.; Rautenstrauss, B.; et al. Loss of function mutations in HARS cause a spectrum of inherited peripheral neuropathies. Brain 2015, 138, 2161–2172. [Google Scholar] [CrossRef] [PubMed]
- Weterman, M.A.J.; Kuo, M.; Kenter, S.B.; Gordillo, S.; Karjosukarso, D.W.; Takase, R.; Bronk, M.; Oprescu, S.; Ruissen, F.; Witteveen, R.J.W.; et al. Hypermorphic and hypomorphic AARS alleles in patients with CMT2N expand clinical and molecular heterogeneities. Hum. Mol. Genet. 2018, 27, 4036–4050. [Google Scholar] [CrossRef] [PubMed]
- Bervoets, S.; Wei, N.; Erfurth, M.L.; Yusein-Myashkova, S.; Ermanoska, B.; Mateiu, L.; Asselbergh, B.; Blocquel, D.; Kakad, P.; Penserga, T.; et al. Transcriptional dysregulation by a nucleus-localized aminoacyl-tRNA synthetase associated with Charcot-Marie-Tooth neuropathy. Nat. Commun. 2019, 10, 5045. [Google Scholar] [CrossRef]
- Greenberg, Y.; King, M.; Kiosses, W.B.; Ewalt, K.; Yang, X.; Schimmel, P.; Reader, J.S.; Tzima, E. The novel fragment of tyrosyl tRNA synthetase, mini-TyrRS, is secreted to induce an angiogenic response in endothelial cells. FASEB J. 2008, 22, 1597–1605. [Google Scholar] [CrossRef]
- Otani, A.; Slike, B.M.; Dorrell, M.I.; Hood, J.; Kinder, K.; Ewalt, K.L.; Cheresh, D.; Schimmel, P.; Friedlander, M. A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 178–183. [Google Scholar] [CrossRef]
- Wakasugi, K.; Schimmel, P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science 1999, 284, 147–151. [Google Scholar] [CrossRef]
- Achilli, F.; Bros-Facer, V.; Williams, H.P.; Banks, G.T.; AlQatari, M.; Chia, R.; Tucci, V.; Groves, M.; Nickols, C.D.; Seburn, K.L.; et al. An ENU-induced mutation in mouse glycyl-tRNA synthetase (GARS) causes peripheral sensory and motor phenotypes creating a model of Charcot-Marie-Tooth type 2D peripheral neuropathy. Dis. Model. Mech. 2009, 2, 359–373. [Google Scholar] [CrossRef]
- Seburn, K.L.; Nangle, L.A.; Cox, G.A.; Schimmel, P.; Burgess, R.W. An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron 2006, 51, 715–726. [Google Scholar] [CrossRef]
- Ermanoska, B.; Motley, W.W.; Leitao-Goncalves, R.; Asselbergh, B.; Lee, L.H.; De Rijk, P.; Sleegers, K.; Ooms, T.; Godenschwege, T.A.; Timmerman, V.; et al. CMT-associated mutations in glycyl- and tyrosyl-tRNA synthetases exhibit similar pattern of toxicity and share common genetic modifiers in Drosophila. Neurobiol. Dis. 2014, 68, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Niehues, S.; Bussmann, J.; Steffes, G.; Erdmann, I.; Kohrer, C.; Sun, L.; Wagner, M.; Schafer, K.; Wang, G.; Koerdt, S.N.; et al. Impaired protein translation in Drosophila models for Charcot-Marie-Tooth neuropathy caused by mutant tRNA synthetases. Nat. Commun. 2015, 6, 7520. [Google Scholar] [CrossRef] [PubMed]
- Mullen, P.; Abbott, J.A.; Wellman, T.; Aktar, M.; Fjeld, C.; Demeler, B.; Ebert, A.M.; Francklyn, C.S. Neuropathy-associated histidyl-tRNA synthetase variants attenuate protein synthesis in vitro and disrupt axon outgrowth in developing zebrafish. FEBS J. 2020, 288, 142–159. [Google Scholar] [CrossRef]
- Charng, W.L.; Yamamoto, S.; Bellen, H.J. Shared mechanisms between Drosophila peripheral nervous system development and human neurodegenerative diseases. Curr. Opin. Neurobiol. 2014, 27, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.D.; Celniker, S.E.; Holt, R.A.; Evans, C.A.; Gocayne, J.D.; Amanatides, P.G.; Scherer, S.E.; Li, P.W.; Hoskins, R.A.; Galle, R.F.; et al. The genome sequence of Drosophila melanogaster. Science 2000, 287, 2185–2195. [Google Scholar] [CrossRef]
- Reiter, L.T.; Potocki, L.; Chien, S.; Gribskov, M.; Bier, E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001, 11, 1114–1125. [Google Scholar] [CrossRef]
- Yamamoto, S.; Jaiswal, M.; Charng, W.L.; Gambin, T.; Karaca, E.; Mirzaa, G.; Wiszniewski, W.; Sandoval, H.; Haelterman, N.A.; Xiong, B.; et al. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 2014, 159, 200–214. [Google Scholar] [CrossRef]
- Chien, S.; Reiter, L.T.; Bier, E.; Gribskov, M. Homophila: Human disease gene cognates in Drosophila. Nucleic Acids Res. 2002, 30, 149–151. [Google Scholar] [CrossRef]
- Hopkins, A.L.; Groom, C.R. The druggable genome. Nat. Rev. Drug Discov. 2002, 1, 727–730. [Google Scholar] [CrossRef]
- Roote, J.; Prokop, A. How to design a genetic mating scheme: A basic training package for Drosophila genetics. G3 2013, 3, 353–358. [Google Scholar] [CrossRef]
- Piper, M.D.W.; Partridge, L. Drosophila as a model for ageing. Biochim Biophys Acta Mol. Basis Dis. 2018, 1864, 2707–2717. [Google Scholar] [CrossRef]
- Bhat, M.A. Molecular organization of axo-glial junctions. Curr. Opin. Neurobiol. 2003, 13, 552–559. [Google Scholar] [CrossRef]
- Larkin, A.; Marygold, S.J.; Antonazzo, G.; Attrill, H.; Dos Santos, G.; Garapati, P.V.; Goodman, J.L.; Gramates, L.S.; Millburn, G.; Strelets, V.B.; et al. FlyBase: Updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2021, 49, D899–D907. [Google Scholar] [CrossRef] [PubMed]
- Ugur, B.; Chen, K.; Bellen, H.J. Drosophila tools and assays for the study of human diseases. Dis. Model. Mech. 2016, 9, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Venken, K.J.; Bellen, H.J. Transgenesis upgrades for Drosophila melanogaster. Development 2007, 134, 3571–3584. [Google Scholar] [CrossRef] [PubMed]
- Kanca, O.; Bellen, H.J.; Schnorrer, F. Gene Tagging Strategies to Assess Protein Expression, Localization, and Function in Drosophila. Genetics 2017, 207, 389–412. [Google Scholar] [CrossRef] [PubMed]
- Senturk, M.; Bellen, H.J. Genetic strategies to tackle neurological diseases in fruit flies. Curr. Opin. Neurobiol. 2018, 50, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Ewen-Campen, B.; Ni, X.; Housden, B.E.; Perrimon, N. In Vivo Transcriptional Activation Using CRISPR/Cas9 in Drosophila. Genetics 2015, 201, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.T.; Zirin, J.; Kanca, O.; Lin, W.W.; Schulze, K.L.; Li-Kroeger, D.; Tao, R.; Devereaux, C.; Hu, Y.; Chung, V.; et al. A gene-specific T2A-GAL4 library for Drosophila. eLife 2018, 7, e35574. [Google Scholar] [CrossRef]
- Deisseroth, K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 2015, 18, 1213–1225. [Google Scholar] [CrossRef]
- Lu, J.; Marygold, S.J.; Gharib, W.H.; Suter, B. The aminoacyl-tRNA synthetases of Drosophila melanogaster. Fly 2015, 9, 53–61. [Google Scholar] [CrossRef][Green Version]
- Grice, S.J.; Sleigh, J.N.; Motley, W.W.; Liu, J.L.; Burgess, R.W.; Talbot, K.; Cader, M.Z. Dominant, toxic gain-of-function mutations in gars lead to non-cell autonomous neuropathology. Hum. Mol. Genet. 2015, 24, 4397–4406. [Google Scholar] [CrossRef]
- Chihara, T.; Luginbuhl, D.; Luo, L. Cytoplasmic and mitochondrial protein translation in axonal and dendritic terminal arborization. Nat. Neurosci. 2007, 10, 828–837. [Google Scholar] [CrossRef]
- Leitao-Goncalves, R.; Ermanoska, B.; Jacobs, A.; De Vriendt, E.; Timmerman, V.; Lupski, J.R.; Callaerts, P.; Jordanova, A. Drosophila as a platform to predict the pathogenicity of novel aminoacyl-tRNA synthetase mutations in CMT. Amino Acids 2012, 42, 1661–1668. [Google Scholar] [CrossRef]
- Zhao, Y.; Xie, L.; Shen, C.; Qi, Q.; Qin, Y.; Xing, J.; Zhou, D.; Qi, Y.; Yan, Z.; Lin, X.; et al. SIRT2-knockdown rescues GARS-induced Charcot-Marie-Tooth neuropathy. Aging Cell 2021, 20, e13391. [Google Scholar] [CrossRef]
- Zuko, A.; Mallik, M.; Thompson, R.; Spaulding, E.L.; Wienand, A.R.; Been, M.; Tadenev, A.L.D.; van Bakel, N.; Sijlmans, C.; Santos, L.A.; et al. tRNA overexpression rescues peripheral neuropathy caused by mutations in tRNA synthetase. Science 2021, 373, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.J.; Godenschwege, T.A.; Tanouye, M.A.; Phelan, P. Making an escape: Development and function of the Drosophila giant fibre system. Semin. Cell Dev. Biol. 2006, 17, 31–41. [Google Scholar] [CrossRef]
- Iyer, J.; Wang, Q.; Le, T.; Pizzo, L.; Gronke, S.; Ambegaokar, S.S.; Imai, Y.; Srivastava, A.; Troisi, B.L.; Mardon, G.; et al. Quantitative Assessment of Eye Phenotypes for Functional Genetic Studies Using Drosophila melanogaster. G3 2016, 6, 1427–1437. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.P. Building an ommatidium one cell at a time. Dev. Dyn. 2012, 241, 136–149. [Google Scholar] [CrossRef]
- Grueber, W.B.; Ye, B.; Yang, C.H.; Younger, S.; Borden, K.; Jan, L.Y.; Jan, Y.N. Projections of Drosophila multidendritic neurons in the central nervous system: Links with peripheral dendrite morphology. Development 2007, 134, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Babcock, D.T.; Landry, C.; Galko, M.J. Cytokine signaling mediates UV-induced nociceptive sensitization in Drosophila larvae. Curr. Biol. 2009, 19, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Hwang, R.Y.; Zhong, L.; Xu, Y.; Johnson, T.; Zhang, F.; Deisseroth, K.; Tracey, W.D. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr. Biol. 2007, 17, 2105–2116. [Google Scholar] [CrossRef]
- Xiang, Y.; Yuan, Q.; Vogt, N.; Looger, L.L.; Jan, L.Y.; Jan, Y.N. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature 2010, 468, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Jefferis, G.S.; Potter, C.J.; Chan, A.M.; Marin, E.C.; Rohlfing, T.; Maurer, C.R., Jr.; Luo, L. Comprehensive maps of Drosophila higher olfactory centers: Spatially segregated fruit and pheromone representation. Cell 2007, 128, 1187–1203. [Google Scholar] [CrossRef] [PubMed]
- Marin, E.C.; Jefferis, G.S.; Komiyama, T.; Zhu, H.; Luo, L. Representation of the glomerular olfactory map in the Drosophila brain. Cell 2002, 109, 243–255. [Google Scholar] [CrossRef]
- Wong, A.M.; Wang, J.W.; Axel, R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell 2002, 109, 229–241. [Google Scholar] [CrossRef]
- Storkebaum, E. Peripheral neuropathy via mutant tRNA synthetases: Inhibition of protein translation provides a possible explanation. Bioessays 2016, 38, 818–829. [Google Scholar] [CrossRef]
- Erdmann, I.; Marter, K.; Kobler, O.; Niehues, S.; Abele, J.; Muller, A.; Bussmann, J.; Storkebaum, E.; Ziv, T.; Thomas, U.; et al. Cell-selective labelling of proteomes in Drosophila melanogaster. Nat. Commun. 2015, 6, 7521. [Google Scholar] [CrossRef]
- Holt, C.E.; Martin, K.C.; Schuman, E.M. Local translation in neurons: Visualization and function. Nat. Struct Mol. Biol. 2019, 26, 557–566. [Google Scholar] [CrossRef]
- Lin, J.Q.; van Tartwijk, F.W.; Holt, C.E. Axonal mRNA translation in neurological disorders. RNA Biol. 2021, 18, 936–961. [Google Scholar] [CrossRef]
- Antonellis, A.; Lee-Lin, S.Q.; Wasterlain, A.; Leo, P.; Quezado, M.; Goldfarb, L.G.; Myung, K.; Burgess, S.; Fischbeck, K.H.; Green, E.D. Functional analyses of glycyl-tRNA synthetase mutations suggest a key role for tRNA-charging enzymes in peripheral axons. J. Neurosci. 2006, 26, 10397–10406. [Google Scholar] [CrossRef]
- Nangle, L.A.; Zhang, W.; Xie, W.; Yang, X.L.; Schimmel, P. Charcot-Marie-Tooth disease-associated mutant tRNA synthetases linked to altered dimer interface and neurite distribution defect. Proc. Natl. Acad. Sci. USA 2007, 104, 11239–11244. [Google Scholar] [CrossRef]
- Lund, E.; Dahlberg, J.E. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 1998, 282, 2082–2085. [Google Scholar] [CrossRef]
- Wei, N.; Shi, Y.; Truong, L.N.; Fisch, K.M.; Xu, T.; Gardiner, E.; Fu, G.; Hsu, Y.S.; Kishi, S.; Su, A.I.; et al. Oxidative stress diverts tRNA synthetase to nucleus for protection against DNA damage. Mol. Cell 2014, 56, 323–332. [Google Scholar] [CrossRef]
- Cao, X.; Li, C.; Xiao, S.; Tang, Y.; Huang, J.; Zhao, S.; Li, X.; Li, J.; Zhang, R.; Yu, W. Acetylation promotes TyrRS nuclear translocation to prevent oxidative damage. Proc. Natl. Acad. Sci. USA 2017, 114, 687–692. [Google Scholar] [CrossRef]
- He, W.; Bai, G.; Zhou, H.; Wei, N.; White, N.M.; Lauer, J.; Liu, H.; Shi, Y.; Dumitru, C.D.; Lettieri, K.; et al. CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature 2015, 526, 710–714. [Google Scholar] [CrossRef]
- Sleigh, J.N.; Dawes, J.M.; West, S.J.; Wei, N.; Spaulding, E.L.; Gomez-Martin, A.; Zhang, Q.; Burgess, R.W.; Cader, M.Z.; Talbot, K.; et al. Trk receptor signaling and sensory neuron fate are perturbed in human neuropathy caused by Gars mutations. Proc. Natl. Acad. Sci. USA 2017, 114, E3324–E3333. [Google Scholar] [CrossRef]
- Grice, S.J.; Sleigh, J.N.; Zameel Cader, M. Plexin-Semaphorin Signaling Modifies Neuromuscular Defects in a Drosophila Model of Peripheral Neuropathy. Front. Mol. Neurosci. 2018, 11, 55. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, X.J. Tubulin acetylation: Responsible enzymes, biological functions and human diseases. Cell Mol. Life Sci. 2015, 72, 4237–4255. [Google Scholar] [CrossRef] [PubMed]
- Janssens, K.; Goethals, S.; Atkinson, D.; Ermanoska, B.; Fransen, E.; Jordanova, A.; Auer-Grumbach, M.; Asselbergh, B.; Timmerman, V. Human Rab7 mutation mimics features of Charcot-Marie-Tooth neuropathy type 2B in Drosophila. Neurobiol. Dis. 2014, 65, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Jordanova, A.; Thomas, F.P.; Guergueltcheva, V.; Tournev, I.; Gondim, F.A.; Ishpekova, B.; De Vriendt, E.; Jacobs, A.; Litvinenko, I.; Ivanova, N.; et al. Dominant intermediate Charcot-Marie-Tooth type C maps to chromosome 1p34-p35. Am. J. Hum. Genet. 2003, 73, 1423–1430. [Google Scholar] [CrossRef]
- Thomas, F.P.; Guergueltcheva, V.; Gondim, F.A.; Tournev, I.; Rao, C.V.; Ishpekova, B.; Kinsella, L.J.; Pan, Y.; Geller, T.J.; Litvinenko, I.; et al. Clinical, neurophysiological and morphological study of dominant intermediate Charcot-Marie-Tooth type C neuropathy. J. Neurol. 2016, 263, 467–476. [Google Scholar] [CrossRef]
- Del Bo, R.; Locatelli, F.; Corti, S.; Scarlato, M.; Ghezzi, S.; Prelle, A.; Fagiolari, G.; Moggio, M.; Carpo, M.; Bresolin, N.; et al. Coexistence of CMT-2D and distal SMA-V phenotypes in an Italian family with a GARS gene mutation. Neurology 2006, 66, 752–754. [Google Scholar] [CrossRef]
- Wei, N.; Cui, H.; Shi, Y.; Fu, G.; Rauniyar, N.; Yates, J.R.; Yang, X.-L. Nucleus translocation of tRNA synthetase mediates late integrated stress response. bioRxiv 2020. [Google Scholar] [CrossRef]
- Garin, S.; Levi, O.; Forrest, M.E.; Antonellis, A.; Arava, Y.S. Comprehensive characterization of mRNAs associated with yeast cytosolic aminoacyl-tRNA synthetases. RNA Biol. 2021, 1–12. [Google Scholar] [CrossRef]
- Levi, O.; Arava, Y. mRNA association by aminoacyl tRNA synthetase occurs at a putative anticodon mimic and autoregulates translation in response to tRNA levels. PLoS Biol. 2019, 17, e3000274. [Google Scholar] [CrossRef]
- Levi, O.; Garin, S.; Arava, Y. RNA mimicry in post-transcriptional regulation by aminoacyl tRNA synthetases. Wiley Interdiscip. Rev. RNA 2020, 11, e1564. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.J.; Park, S.; Nguyen, L.T.; Hwang, J.; Lee, E.Y.; Giong, H.K.; Lee, J.S.; Yoon, I.; Lee, J.H.; Kim, J.H.; et al. A threonyl-tRNA synthetase-mediated translation initiation machinery. Nat. Commun. 2019, 10, 1357. [Google Scholar] [CrossRef] [PubMed]
- Spaulding, E.L.; Hines, T.J.; Bais, P.; Tadenev, A.L.D.; Schneider, R.; Jewett, D.; Pattavina, B.; Pratt, S.L.; Morelli, K.H.; Stum, M.G.; et al. The integrated stress response contributes to tRNA synthetase-associated peripheral neuropathy. Science 2021, 373, 1156–1161. [Google Scholar] [CrossRef] [PubMed]
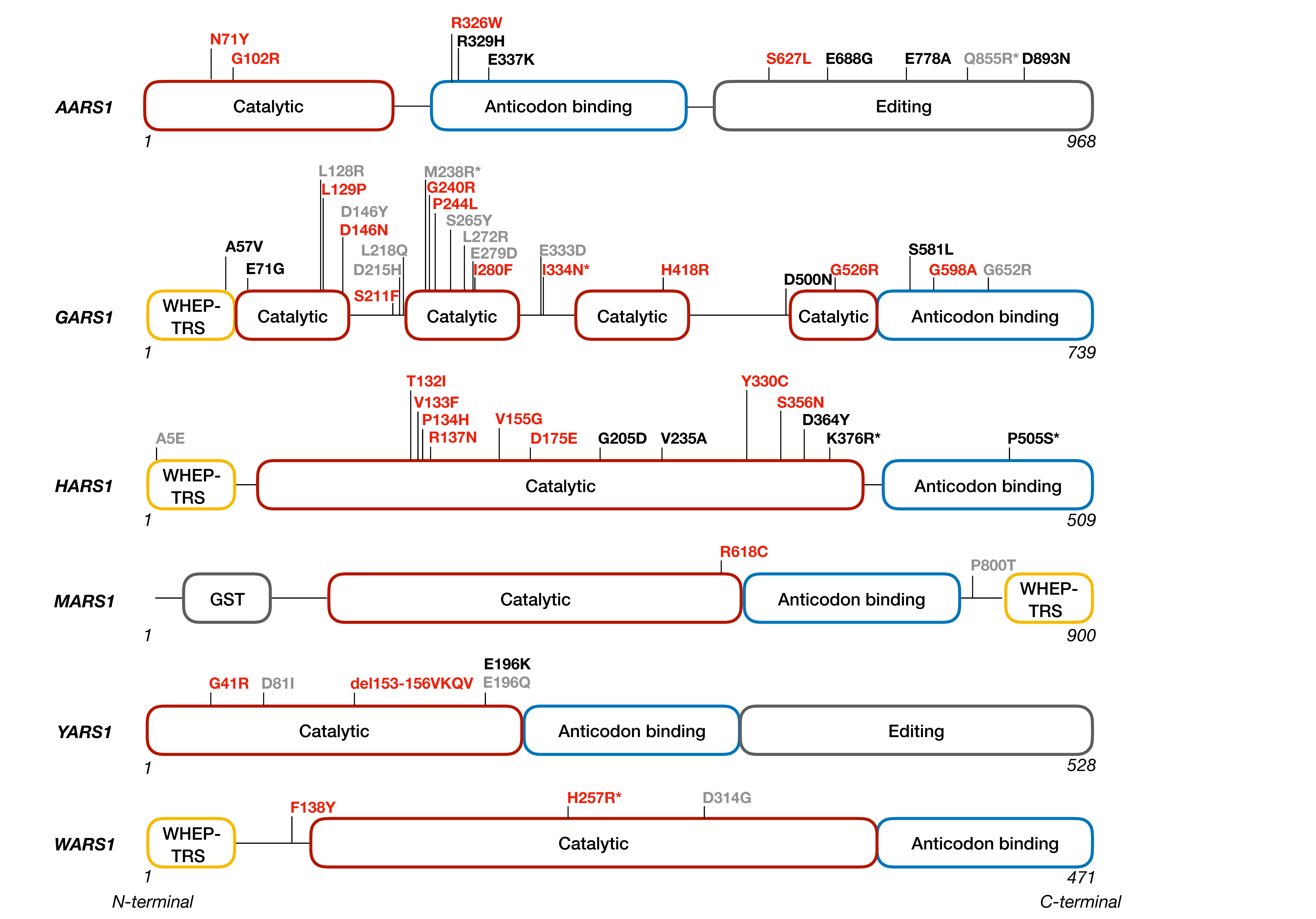
| aaRS Gene | OMIM * | Associated Clinical Phenotype | Nomenclature |
|---|---|---|---|
| AARS1 | 613287 | Axonal Charcot–Marie–Tooth neuropathy type 2N | CMT2N |
| GARS1 | 601472 | Axonal Charcot–Marie–Tooth neuropathy type 2D Distal hereditary motor neuropathy type VA Distal spinal muscular atrophy type V | CMT2D dHMN-VA dSMA-V |
| HARS1 | 616625 | Axonal Charcot–Marie–Tooth neuropathy type 2W | CMT2W |
| MARS1 | 616280 | Axonal Charcot–Marie–Tooth neuropathy type 2U | CMT2U |
| YARS1 | 608323 | Dominant intermediate Charcot–Marie–Tooth neuropathy type C | DICMTC |
| WARS1 | 617721 | Distal hereditary motor neuropathy type IX | dHMN-IX |
| Human Gene | D. melanogaster Orthologue % Identity at Protein Level | Modeled CMT Mutation | Reference |
|---|---|---|---|
| hGARS1 | GlyRS (dGARS) 54 | E71G | [44,65] |
| L219P | [65] | ||
| P234KY * | [43,64] | ||
| G240R | [43,44,64] | ||
| G526R | [44] | ||
| hYARS1 | TyrRS (dYARS) 67 | G41R | [33,43,44] |
| E196K | |||
| 153–156delVKQV | |||
| K265N | [66] |
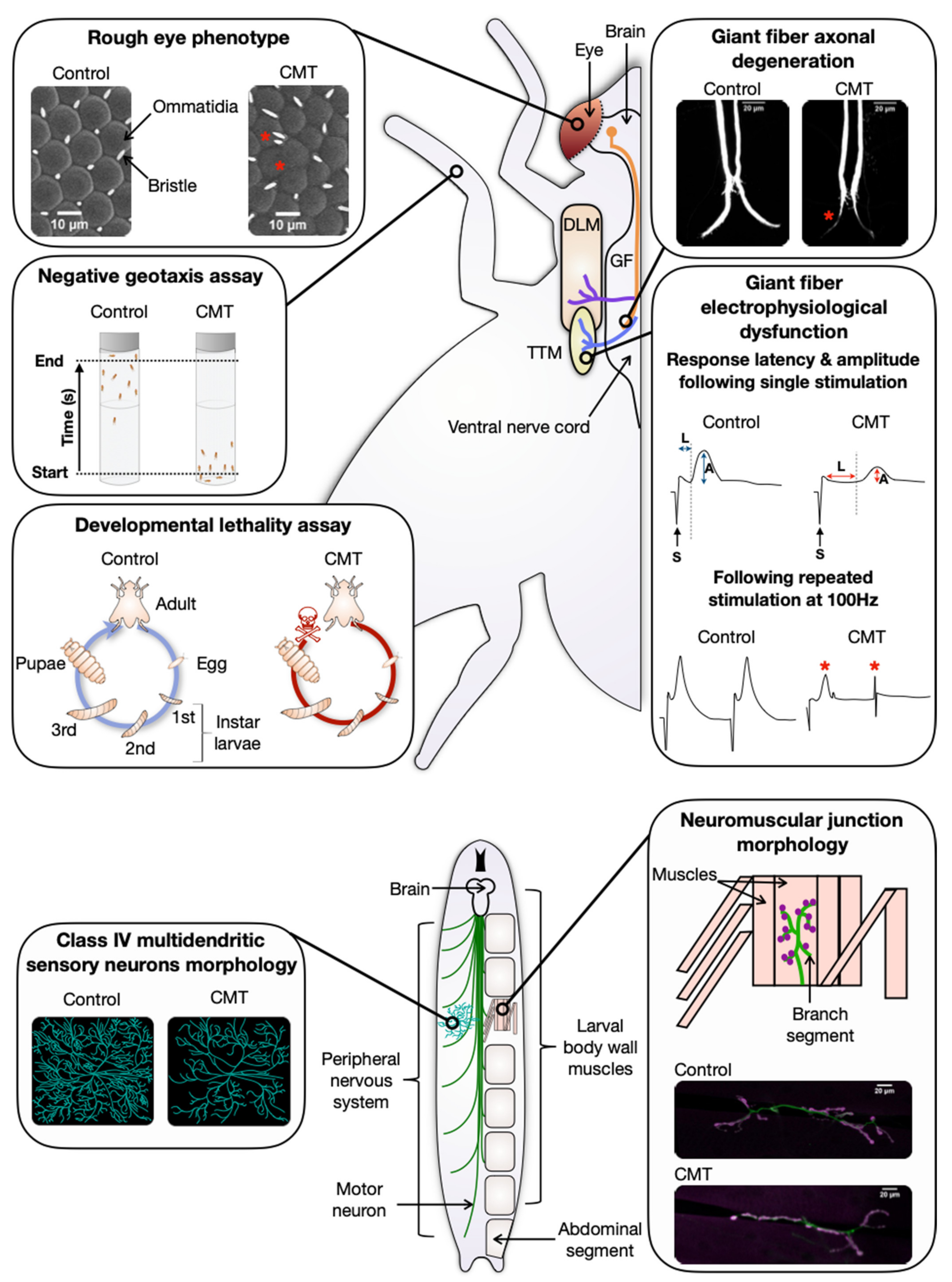
| Phenotype Observed | Transgene | CMT Associated Mutations | Spatial Expression (Driver Used) | References |
|---|---|---|---|---|
| Age-dependent locomotor deficits | hYARS1 | G41R, del153-156VKQV, E196K | Ubiquitous (Actin-5c-GAL4) | [33] |
| Nervous system (elav-GAL4, nSyb-GAL4) | [33,37] | |||
| dGARS | G240R | Nervous system (nSyb-GAL4) | [43] | |
| hGARS1 | E71G, G240R, G526R | Glutamatergic neurons (OK371-GAL4) | [44,67,68] | |
| GFS morphological and electrophysiological deficits | hYARS1 | G41R, del153-156VKQV | Giant fiber (A307-GAL4) | [33] |
| dGARS | P234KY, G240R | [43] | ||
| hYARS1 | del153-156VKQV | Pre-synaptic expression in GF (C17-GAL4, R91H05-GAL4) | [37] | |
| dGARS | P234KY | Pre-synaptically TTMn (C42.2-GAL4) or Post-synaptically TTMn (ShakB-GAL4) | [43] | |
| Neuromuscular junction defects * | dGARS | P234KY | Ubiquitous (1032-GAL4) | [64] |
| Mesoderm (how-GAL4) | ||||
| Muscle (MHC-GAL4) | ||||
| hYARS1 | E196K | Nervous system (nSyb-GAL4) | [37] | |
| hGARS1 | E71G, G240R, G526R | Glutamatergic neurons (OK371-GAL4) | [44,67,68] | |
| Muscle Denervation * | hGARS1 | G240R | Motor neuron (D42-GAL4) | [44,68] |
| hGARS1 | E71G, G240R, G526R | Glutamatergic neurons (OK371-GAL4) | ||
| Muscle contractions defect * | dGARS | P234KY, G240R | Ubiquitous (Tubulin-GAL4) | [64] |
| Muscle (MHC-GAL4) | ||||
| P234KY | Mesoderm (how-GAL4) | |||
| Nervous system (elav-GAL4) | ||||
| Reduction of dendritic coverage * | hGARS1 | E71G, G240R, G526R | Class IV multidendritic sensory neurons (ppk-GAL4) | [44,68] |
| Rough eye | dGARS | P234KY | Eye photoreceptor cell (GMR-GAL4) | [43] |
| hYARS1 | E196K | [33] | ||
| Developmental lethality | hYARS1 | G41R, del153-156VKQV, E196K | Ubiquitous (Actin5c-GAL4, Tubulin-GAL4) | [33] |
| hGARS1 | E71G, G240R, G526R | [44,68] | ||
| hYARS1 | E196K | Nervous system (nSyb-GAL4) | [37] | |
| hGARS1 | E71G, G240R, G526R | [44,68] | ||
| dGARS | P234KY | [33,43] | ||
| dGARS | P234KY | Nervous system (elav-GAL4) | [64] | |
| Mesoderm (how-GAL4) | ||||
| Muscle (MHC-GAL4) | ||||
| Short lifespan | hGARS1 | E71G, G240R, G526R | Ubiquitous (Tubulin-GAL4) | [44,67,68] |
| Inhibition of global protein translation * | hGARS1 | E71G, G240R, G526R | Class IV multidendritic sensory neurons (ppk-GAL4) | [44,68] |
| Ubiquitous (Tubulin-GAL4) | ||||
| Glutamatergic neurons (OK371-GAL4) | ||||
| hYARS1 | G41R, del153-156VKQV, E196K | Class IV multidendritic sensory neurons (ppk-GAL4) | ||
| Glutamatergic neurons (OK371-GAL4) | ||||
| Transcriptional dysregulation | hYARS1 | E196K | Nervous system (nSyb-GAL4) | [37] |
| GARS1CMT accumulation at the synapse * | dGARS | P234KY | Ubiquitous (1032-GAL4) | [64] |
| Mesoderm (how-GAL4) | ||||
| Muscle (MHC-GAL4) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morant, L.; Erfurth, M.-L.; Jordanova, A. Drosophila Models for Charcot–Marie–Tooth Neuropathy Related to Aminoacyl-tRNA Synthetases. Genes 2021, 12, 1519. https://doi.org/10.3390/genes12101519
Morant L, Erfurth M-L, Jordanova A. Drosophila Models for Charcot–Marie–Tooth Neuropathy Related to Aminoacyl-tRNA Synthetases. Genes. 2021; 12(10):1519. https://doi.org/10.3390/genes12101519
Chicago/Turabian StyleMorant, Laura, Maria-Luise Erfurth, and Albena Jordanova. 2021. "Drosophila Models for Charcot–Marie–Tooth Neuropathy Related to Aminoacyl-tRNA Synthetases" Genes 12, no. 10: 1519. https://doi.org/10.3390/genes12101519
APA StyleMorant, L., Erfurth, M.-L., & Jordanova, A. (2021). Drosophila Models for Charcot–Marie–Tooth Neuropathy Related to Aminoacyl-tRNA Synthetases. Genes, 12(10), 1519. https://doi.org/10.3390/genes12101519






