Tracing Key Molecular Regulators of Lipid Biosynthesis in Tuber Development of Cyperus esculentus Using Transcriptomics and Lipidomics Profiling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Measurement of Oil Content
2.3. Nile Red Staining
2.4. Lipid Extraction
2.5. Lipidomic Analysis by LC/MS
2.6. Differential Expression Analysis
2.7. Cloning and Functional Identification of CePAH1 Gene
2.8. Measurement of DAG and Total Oil Content
2.9. Data Analysis
3. Results
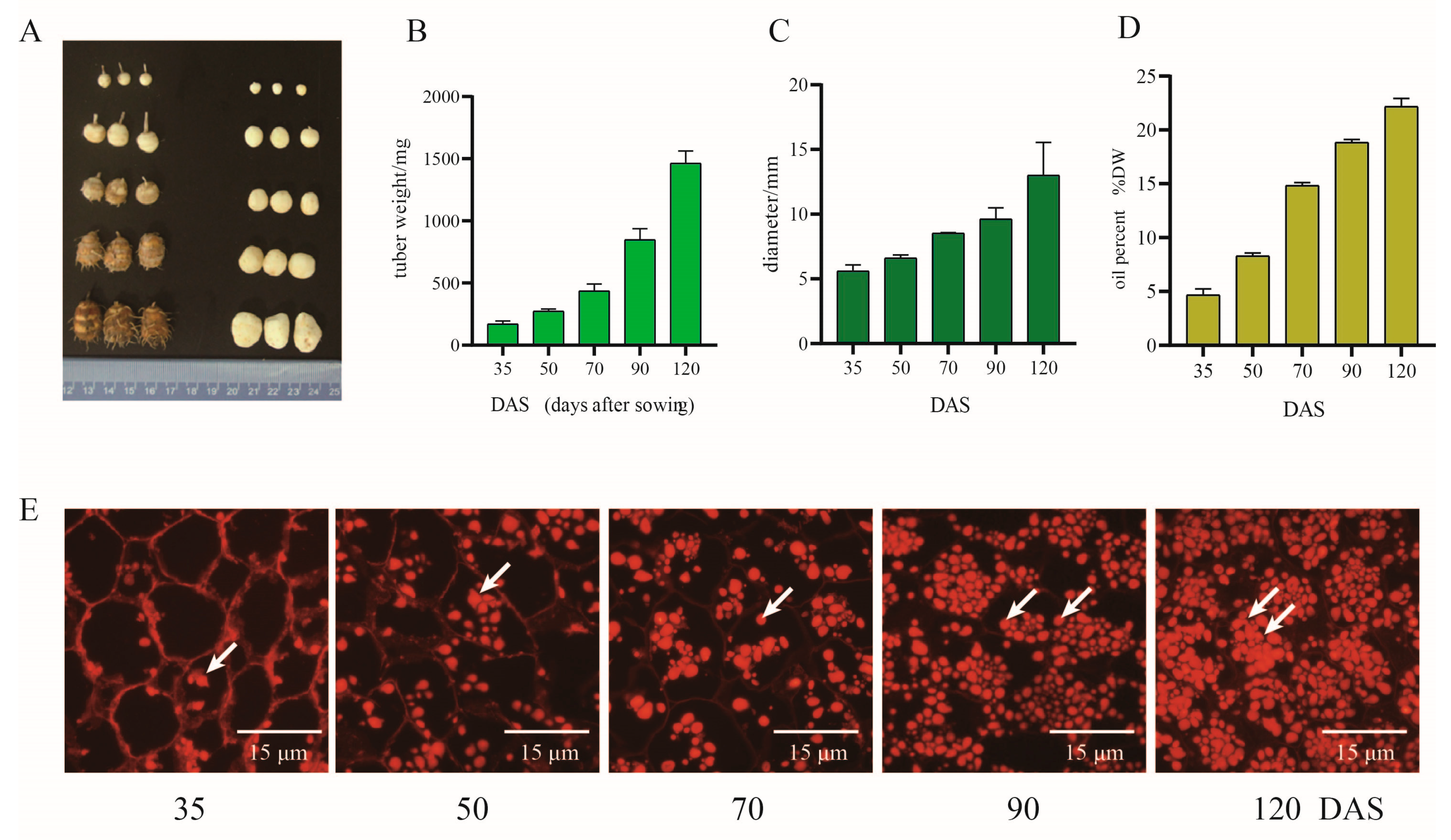
3.1. Raw Oil Contents in Developing Tiger Nut Tubers
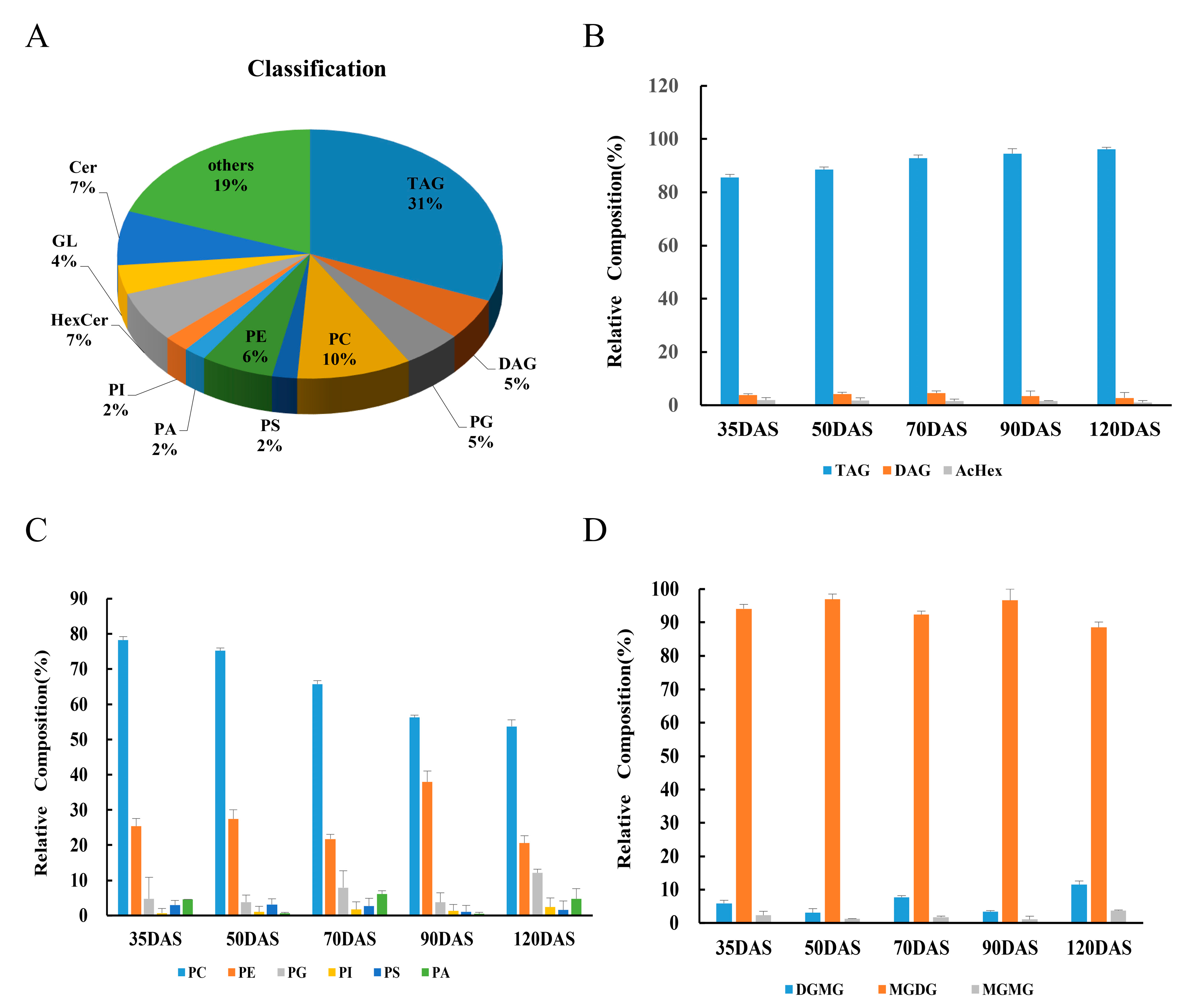
3.2. Lipid Composition and Variation
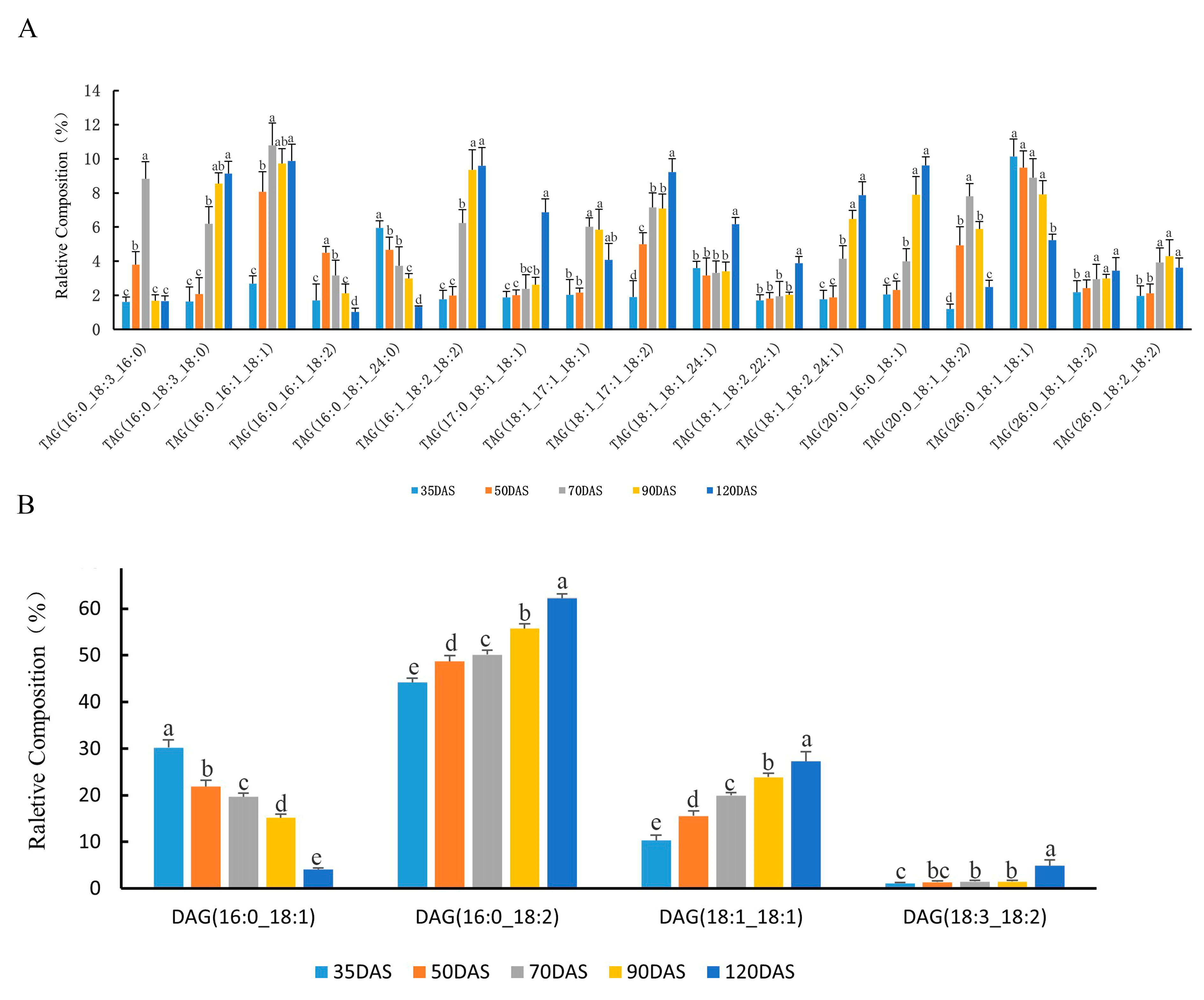
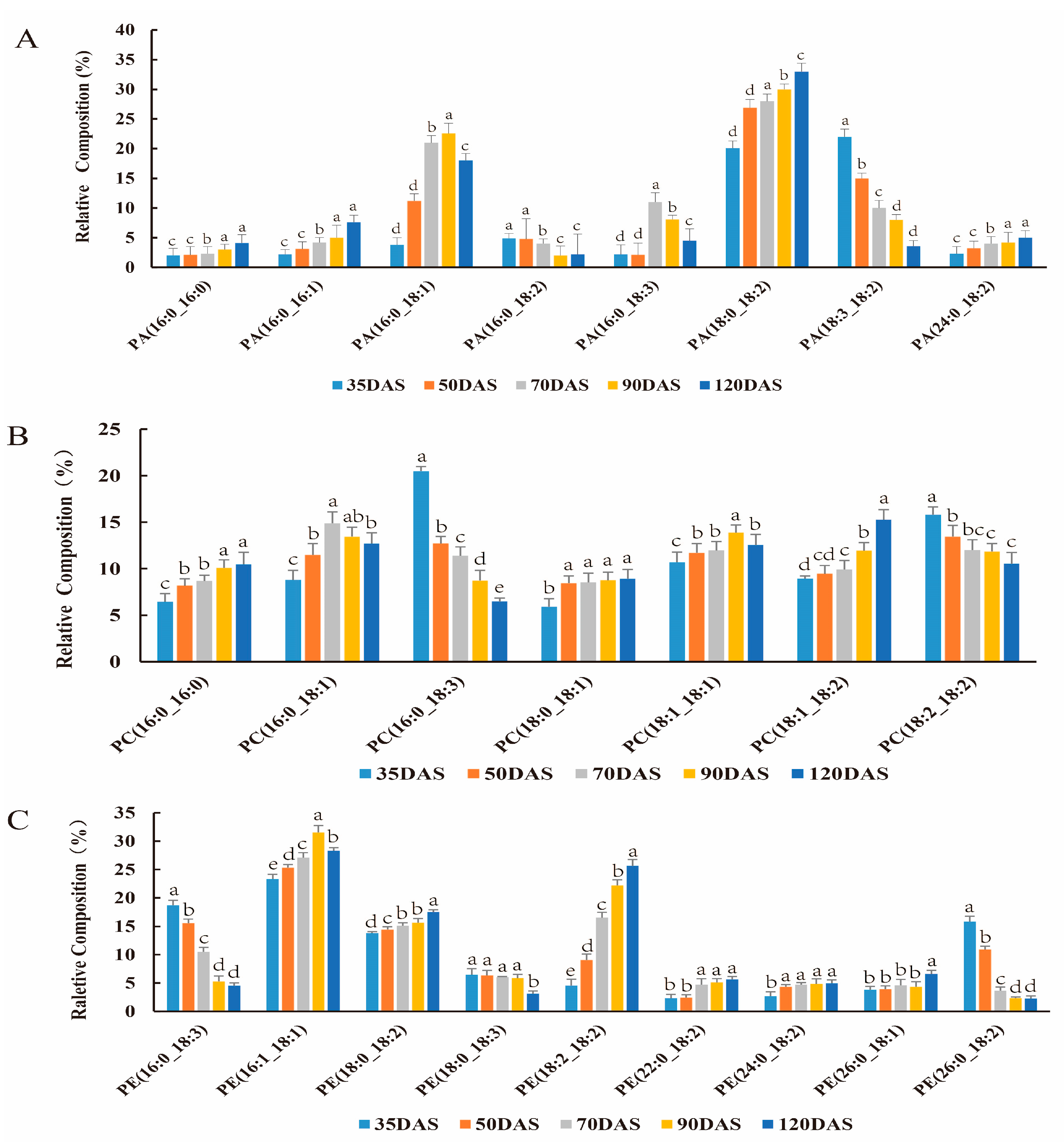
3.3. Changes in Lipids Accumulation during Tuber Development
3.4. Transcriptome Analysis of C. esculentus during Tuber Development
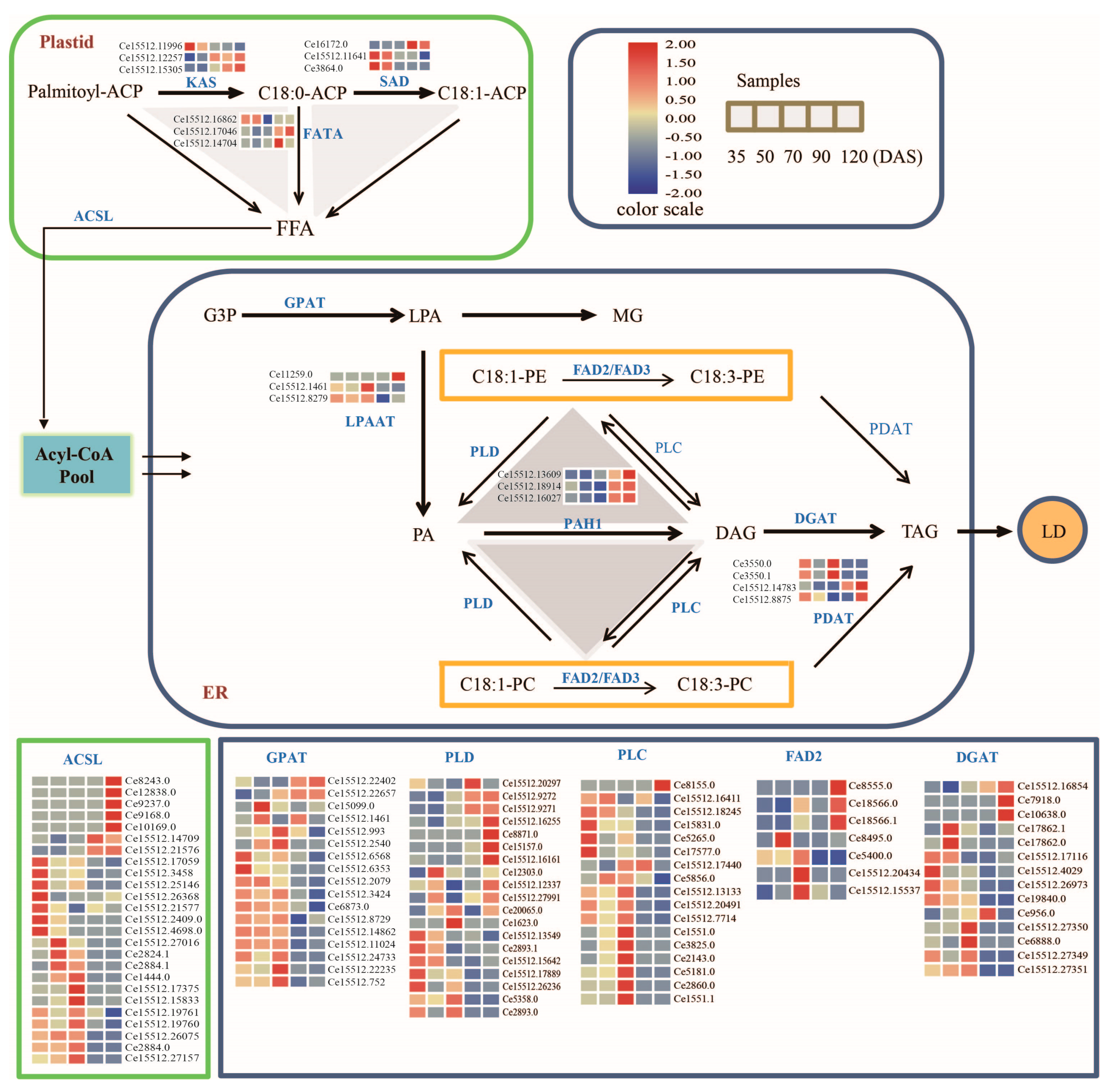
3.5. Majors Genes Are Responsible for the Molecular Types Composition of TAG
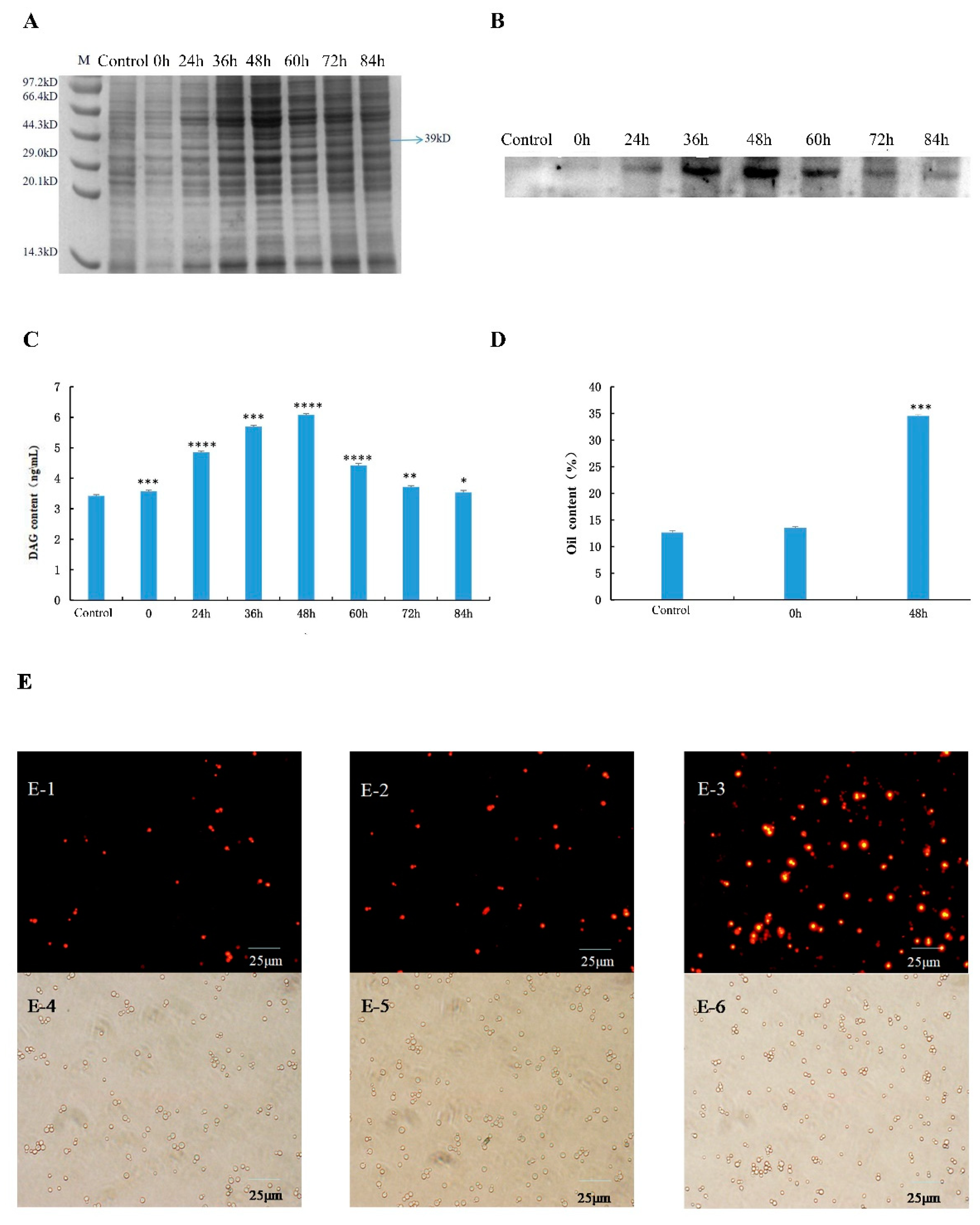
3.6. Expression and Verification of Recombinant Protein
3.7. Detection of DAG Content and Oil Content in Recombinant Saccharomyces cerevisiae Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TAG | triacylglycerol |
| ESI-MS | electrospray ionization mass spectrometry |
| PC | phosphatidylcholines |
| PE | Phosphatidyl ethanolamine |
| DAG | diacylglycerol |
| LPAAT | lysophosphatidic acid acyltransferase |
| DGAT | diglyceride acyltransferase |
| PDAT1 | phospholipid:diacylglycerol acyltransferase 1 |
| DAS | days after sowing |
| OBs | oil body |
| QC | quality control |
| SAD | stromal stearoyl-ACP desaturase |
| FA | fatty acid |
| LACS | long-chain acyl-CoA synthetase |
| G3P | glycerol-3-phosphate |
| GPAT | glycerol-3-phosphate acyltransferase |
| LPA | lysophosphatidic acid |
| AcHex | acetylhexosamine |
References
- Bado, S.; Bazongo, P.; Son, G.; Kyaw, M.T.; Forster, B.P.; Nielen, S.; Lykke, A.M.; Ouedraogo, A.; Bassole, I.H. Physicochemical characteristics and composition of three morphotypes of cyperus esculentus tubers and tuber oils. J. Anal. Methods Chem. 2015, 2015, 673547. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Gross, R.W. Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc. Natl. Acad. Sci. USA 1994, 91, 10635–10639. [Google Scholar] [CrossRef] [Green Version]
- Woodfield, H.K.; Cazenave-Gassiot, A.; Haslam, R.P.; Guschina, I.A.; Wenk, M.R.; Harwood, J.L. Using lipidomics to reveal details of lipid accumulation in developing seeds from oilseed rape (Brassica napus L.). Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2018, 1863, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Y.; Shi, D.Q.; Zhang, W.J.; Tang, Z.S.; Liu, J.; Yang, W.C. The Arabidopsis alkaline ceramidase TOD1 is a key turgor pressure regulator in plant cells. Nat. Commun. 2015, 6, 6030. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Bi, J.; Shui, G.; Liu, Z.; Xiang, Y.; Liu, Y.; Wenk, M.R.; Yang, H.; Huang, X. Tissue-autonomous function of Drosophila seipin in preventing ectopic lipid droplet formation. PLoS Genet. 2011, 7, e1001364. [Google Scholar] [CrossRef] [Green Version]
- Weselake, R.J.; Taylor, D.C.; Rahman, M.H.; Shah, S.; Laroche, A.; McVetty, P.B.; Harwood, J.L. Increasing the flow of carbon into seed oil. Biotechnol. Adv. 2009, 27, 866–878. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.D.; Browse, J. The significance of different diacylgycerol synthesis pathways on plant oil composition and bioengineering. Front. Plant Sci. 2012, 3, 147. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, K.B.; Cullerne, D.P.; El Tahchy, A.; Rolland, V.; Blanchard, C.L.; Wood, C.C.; Singh, S.P.; Petrie, J.R. Identification of genes involved in lipid biosynthesis through de novo transcriptome assembly from cocos nucifera developing endosperm. Plant Cell Physiol. 2019, 60, 945–960. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.D.; Xu, W.Z.; Shu, Q.Y.; Li, S.S.; Wu, Q.; Feng, C.Y.; Gu, Z.Y.; Wang, L.S. Fatty acid desaturase 3 (PsFAD3) from Paeonia suffruticosa reveals high alpha-linolenic acid accumulation. Plant Sci. 2018, 274, 212–222. [Google Scholar] [CrossRef] [PubMed]
- González-Mellado, D.; Salas, J.J.; Venegas-Calerón, M.; Moreno-Pérez, A.J.; Garcés, R.; Martínez-Force, E. Functional characterization and structural modelling of Helianthus annuus (sunflower) ketoacyl-CoA synthases and their role in seed oil composition. Planta 2019, 249, 1823–1836. [Google Scholar] [CrossRef] [Green Version]
- Khan, K.; Kumar, V.; Niranjan, A.; Shanware, A.; Sane, V.A. JcMYB1, a Jatropha R2R3MYB transcription factor gene, modulates lipid biosynthesis in transgenic plants. Plant Cell Physiol. 2019, 60, 462–475. [Google Scholar] [CrossRef]
- Tian, Y.; Lv, X.; Xie, G.; Wang, L.; Dai, T.; Qin, X. FAX2 mediates fatty acid export from plastids in developing arabidopsis seeds. Plant Cell Physiol. 2019, 60, 2231–2242. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Lv, X.; Xie, G.; Zhang, J.; Xu, Y.; Chen, F. Seed-specific overexpression of AtFAX1 increases seed oil content in Arabidopsis. Biochem. Biophys. Res. Commun. 2018, 500, 370–375. [Google Scholar] [CrossRef]
- Zhukovsky, M.A.; Filograna, A.; Luini, A.; Corda, D.; Valente, C. The structure and function of acylglycerophosphate acyltransferase 4/ lysophosphatidic acid acyltransferase delta (AGPAT4/LPAATdelta). Front. Cell Dev. Biol. 2019, 7, 147. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.H.; Wang, T.T.; Li, Q.Q.; Zhao, N.; Zhang, Y.; Liu, D.; Hu, Q.; Li, F.L. Two novel diacylglycerol acyltransferase genes from Xanthoceras sorbifolia are responsible for its seed oil content. Gene 2013, 527, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Seo, P.J. Interaction of DGAT1 and PDAT1 to enhance TAG assembly in Arabidopsis. Plant Signal. Behav. 2019, 14, 1554467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Ji, H.; Yang, Z. Functional characterization of three novel genes encoding diacylglycerol acyltransferase (DGAT) from oil-rich tubers of cyperus esculentus. Plant Cell Physiol. 2020, 61, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Yan, C.; Roston, R.; Shanklin, J.; Xu, C. Arabidopsis lipins, PDAT1 acyltransferase, and SDP1 triacylglycerol lipase synergistically direct fatty acids toward beta-oxidation, thereby maintaining membrane lipid homeostasis. Plant Cell 2014, 26, 4119–4134. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Shao, J.; Tang, S.; Shen, Q.; Wang, T.; Chen, W.; Hong, Y. Wrinkled1 accelerates flowering and regulates lipid homeostasis between oil accumulation and membrane lipid anabolism in brassica napus. Front. Plant Sci. 2015, 6, 1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.G.; Kim, H.; Suh, M.C.; Kim, H.U.; Seo, P.J. The MYB96 Transcription factor regulates triacylglycerol accumulation by activating DGAT1 and PDAT1 expression in arabidopsis seeds. Plant Cell Physiol. 2018, 59, 1432–1442. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, X.; Jia, Q.; Ohlrogge, J. FUSCA3 activates triacylglycerol accumulation in Arabidopsis seedlings and tobacco BY2 cells. Plant J. 2016, 88, 95–107. [Google Scholar] [CrossRef]
- Manan, S.; Ahmad, M.Z.; Zhang, G.; Chen, B.; Haq, B.U.; Yang, J.; Zhao, J. Soybean LEC2 regulates subsets of genes involved in controlling the biosynthesis and catabolism of seed storage substances and seed development. Front. Plant Sci. 2017, 8, 1604. [Google Scholar] [CrossRef] [Green Version]
- Triebl, A.; Trötzmüller, M.; Hartler, J.; Stojakovic, T.; Köfeler, H.C. Lipidomics by ultrahigh performance liquid chromatography-high resolution mass spectrometry and its application to complex biological samples. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017, 1053, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; McCarthy, D.J.; Chen, Y.; Okoniewski, M.; Smyth, G.K.; Huber, W.; Robinson, M.D. Count-based differential expression analysis of RNA sequencing data using R and bioconductor. Nat. Protoc. 2013, 8, 1765–1786. [Google Scholar] [CrossRef] [PubMed]
- Olzmann, J.A.; Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019, 20, 137–155. [Google Scholar] [CrossRef]
- Chen, G.; Woodfield, H.K.; Pan, X.; Harwood, J.L.; Weselake, R.J. Acyl-trafficking during plant oil accumulation. Lipids 2015, 50, 1057–1068. [Google Scholar] [CrossRef]
- Sembongi, H.; Miranda, M.; Han, G.S.; Fakas, S.; Grimsey, N.; Vendrell, J.; Carman, G.M.; Siniossoglou, S. Distinct roles of the phosphatidate phosphatases lipin 1 and 2 during adipogenesis and lipid droplet biogenesis in 3T3-L1 cells. J. Biol. Chem. 2013, 288, 34502–34513. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Peng, Z.; Yang, X.; Wang, W.; Fu, J.; Wang, J.; Han, Y.; Chai, Y.; Guo, T.; Yang, N.; et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013, 45, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Liu, K.; Zhang, Y.; Feng, Q.; Wang, L.; Zhao, Y.; Li, D.; Zhao, Q.; Zhu, X.; Zhu, X.; et al. Genetic discovery for oil production and quality in sesame. Nat. Commun. 2015, 6, 8609. [Google Scholar] [CrossRef] [Green Version]
- Vanhercke, T.; Dyer, J.M.; Mullen, R.T.; Kilaru, A.; Rahman, M.M.; Petrie, J.R.; Green, A.G.; Yurchenko, O.; Singh, S.P. Metabolic engineering for enhanced oil in biomass. Prog. Lipid Res. 2019, 74, 103–129. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Ji, H.; Liu, D. Oil biosynthesis in underground oil-rich storage vegetative tissue: Comparison of cyperus esculentus tuber with oil seeds and fruits. Plant Cell Physiol. 2016, 57, 2519–2540. [Google Scholar] [CrossRef] [PubMed]
- Kritioti, A.; Menexes, G.; Drouza, C. Chemometric characterization of virgin olive oils of the two major Cypriot cultivars based on their fatty acid composition. Food Res. Int. 2018, 103, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.L.; Chen, H.H.; Jiang, J.G. Implications of glycerol metabolism for lipid production. Prog. Lipid Res. 2017, 68, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Yan, C.; Zhang, X.; Xu, C. Dual role for phospholipid:diacylglycerol acyltransferase: Enhancing fatty acid synthesis and diverting fatty acids from membrane lipids to triacylglycerol in Arabidopsis leaves. Plant Cell 2013, 25, 3506–3518. [Google Scholar] [CrossRef] [Green Version]
- Bates, P.D. Understanding the control of acyl flux through the lipid metabolic network of plant oil biosynthesis. Biochim. Biophys. Acta 2016, 1861, 1214–1225. [Google Scholar] [CrossRef]
- Fisher, F.M.; Chui, P.C.; Nasser, I.A.; Popov, Y.; Cunniff, J.C.; Lundasen, T.; Kharitonenkov, A.; Schuppan, D.; Flier, J.S.; Maratos-Flier, E. Fibroblast growth factor 21 limits lipotoxicity by promoting hepatic fatty acid activation in mice on methionine and choline-deficient diets. Gastroenterology 2014, 147, 1073–1083.e6. [Google Scholar] [CrossRef] [Green Version]
- Jako, C.; Kumar, A.; Wei, Y.; Zou, J.; Barton, D.L.; Giblin, E.M.; Covello, P.S.; Taylor, D.C. Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 2001, 126, 861–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lardizabal, K.; Effertz, R.; Levering, C.; Mai, J.; Pedroso, M.C.; Jury, T.; Aasen, E.; Gruys, K.; Bennett, K. Expression of Umbelopsis ramanniana DGAT2A in seed increases oil in soybean. Plant Physiol. 2008, 148, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Zheng, P.; Allen, W.B.; Roesler, K.; Williams, M.E.; Zhang, S.; Li, J.; Glassman, K.; Ranch, J.; Nubel, D.; Solawetz, W.; et al. A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat. Genet. 2008, 40, 367–372. [Google Scholar] [CrossRef]
- Oakes, J.; Brackenridge, D.; Colletti, R.; Daley, M.; Hawkins, D.J.; Xiong, H.; Mai, J.; Screen, S.E.; Val, D.; Lardizabal, K.; et al. Expression of fungal diacylglycerol acyltransferase2 genes to increase kernel oil in maize. Plant Physiol. 2011, 155, 1146–1157. [Google Scholar] [CrossRef] [Green Version]
- Kroon, J.T.; Wei, W.; Simon, W.J.; Slabas, A.R. Identification and functional expression of a type 2 acyl-CoA:diacylglycerol acyltransferase (DGAT2) in developing castor bean seeds which has high homology to the major triglyceride biosynthetic enzyme of fungi and animals. Phytochemistry 2006, 67, 2541–2549. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Mao, X.; Zhao, K.; Ji, X.; Ji, C.; Xue, J.; Li, R. Characterisation of phospholipid: Diacylglycerol acyltransferases (PDATs) from Camelina sativa and their roles in stress responses. Biol. Open 2017, 6, 1024–1034. [Google Scholar] [CrossRef] [Green Version]
- Ducos, E.; Vergès, V.; Dugé de Bernonville, T.; Blanc, N.; Giglioli-Guivarc’h, N.; Dutilleul, C. Remarkable evolutionary conservation of antiobesity ADIPOSE/WDTC1 homologs in animals and plants. Genetics 2017, 207, 153–162. [Google Scholar] [CrossRef] [Green Version]
- Dahlqvist, A.; Ståhl, U.; Lenman, M.; Banas, A.; Lee, M.; Sandager, L. Phospholipid:diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 2000, 97, 6487–6492. [Google Scholar] [CrossRef] [Green Version]
- Han, G.S.; Wu, W.I.; Carman, G.M. The saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 2006, 281, 9210–9218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pascual, F.; Carman, G.M. Phosphatidate phosphatase, a key regulator of lipid homeostasis. Biochim. Biophys. Acta 2013, 1831, 514–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, S.A.; Kohlwein, S.D.; Carman, G.M. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 2012, 190, 317–349. [Google Scholar] [CrossRef] [Green Version]
- Carman, G.M.; Henry, S.A. Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 37293–37297. [Google Scholar] [CrossRef] [Green Version]
- Ukey, R.; Carmon, T.; Hardman, D.; Hill, N.T.; Fakas, S. The Yarrowia lipolytica PAH1 homologue contributes but is not required for triacylglycerol biosynthesis during growth on glucose. Yeast 2020, 37, 93–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarabia, L.D.; Boughton, B.A.; Rupasinghe, T.; Callahan, D.L.; Hill, C.B.; Roessner, U. Comparative spatial lipidomics analysis reveals cellular lipid remodelling in different developmental zones of barley roots in response to salinity. Plant Cell Environ. 2020, 4, 327–343. [Google Scholar] [CrossRef]
- Carrasco, S.; Mérida, I. Diacylglycerol, when simplicity becomes complex. Trends Biochem. Sci. 2007, 32, 27–36. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Jing, M.; Ahmad, N.; Wang, Y.; Wang, Y.; Li, J.; Li, X.; Liu, W.; Wang, N.; Wang, F.; et al. Tracing Key Molecular Regulators of Lipid Biosynthesis in Tuber Development of Cyperus esculentus Using Transcriptomics and Lipidomics Profiling. Genes 2021, 12, 1492. https://doi.org/10.3390/genes12101492
Wang L, Jing M, Ahmad N, Wang Y, Wang Y, Li J, Li X, Liu W, Wang N, Wang F, et al. Tracing Key Molecular Regulators of Lipid Biosynthesis in Tuber Development of Cyperus esculentus Using Transcriptomics and Lipidomics Profiling. Genes. 2021; 12(10):1492. https://doi.org/10.3390/genes12101492
Chicago/Turabian StyleWang, Liyan, Meiling Jing, Naveed Ahmad, Yifei Wang, Yijin Wang, Jia Li, Xiaowei Li, Weican Liu, Nan Wang, Fawei Wang, and et al. 2021. "Tracing Key Molecular Regulators of Lipid Biosynthesis in Tuber Development of Cyperus esculentus Using Transcriptomics and Lipidomics Profiling" Genes 12, no. 10: 1492. https://doi.org/10.3390/genes12101492
APA StyleWang, L., Jing, M., Ahmad, N., Wang, Y., Wang, Y., Li, J., Li, X., Liu, W., Wang, N., Wang, F., Dong, Y., & Li, H. (2021). Tracing Key Molecular Regulators of Lipid Biosynthesis in Tuber Development of Cyperus esculentus Using Transcriptomics and Lipidomics Profiling. Genes, 12(10), 1492. https://doi.org/10.3390/genes12101492




