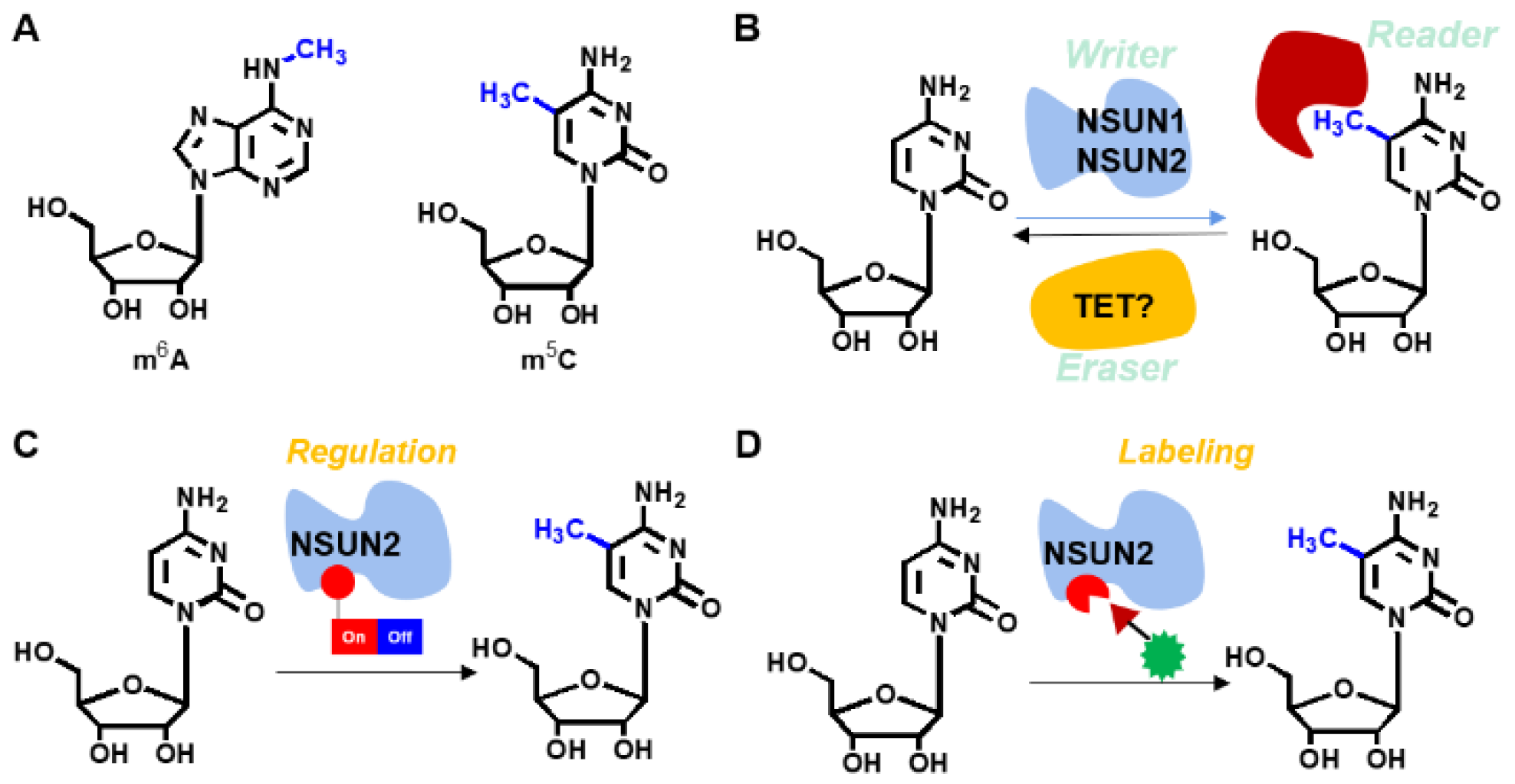
1. Introduction
In living organisms, ribonucleic acids regulate gene expression, cell migration, differentiation, and cell death [
1,
2]. Besides the four canonical nucleotides, over 150 chemical modifications in endogenous nucleic acids facilitate their diversified structures and functions [
3,
4,
5]. To achieve the regulation purpose, RNA modifying enzymes play essential roles, adding a wide range of chemical modifications into target RNAs. Methylation is heavily associated with intrinsic RNAs in various species [
6,
7,
8,
9,
10,
11,
12]. Compared to m
6A, 5-methylcytosine (m
5C) is a less common modification in mammalian animals that has increasingly been evaluated in recent years. The m
5C modification is performed by various enzymes, including
NOL1/NOP2/SUN domain (
NSUN) family [
13,
14,
15]. m
5C is a post-transcriptional RNA modification in various RNA species, including messenger RNAs and transfer RNA among others [
11]. The addition of m
5C to RNA is enabled using the
NSUN family of enzymes and the DNA methyltransferase,
DNMT2, in mammalian cells.
NSUN2 is a critical RNA methyltransferase for adding m
5C to mRNA. Yang et al. revealed that m
5C modification is enriched in the CG-rich motif [
14,
15], which is located down-stream of translation initiation sites and has conserved features across mammalian transcriptomes. Moreover, m
5C is recognized by the mRNA export adaptor,
ALYREF, as shown by in vitro and in vivo studies, where
NSUN2 modulated nuclear-cytoplasmic shuttling of
ALYREF, RNA-binding affinity, and associated mRNA export [
13]. Based on the roles of
NSUN2 enzymes in m
5C-associated RNA biological activities in living organisms, including cellular proliferation, senescence, migration, differentiation, mRNA nuclear export, enhanced mRNA translation, tRNA stabilization, and cleavage [
16], achieving site-specific labeling and modulation of
NSUN2 is, therefore, of high importance. However, precise regulation of nuclear-cytoplasmic shuttling of endogenous RNAs by manipulating the activity of
NSUN2 remains elusive.
The genetic encoding expansion technique developed by Schultz et al. has been successfully applied in mammals as a powerful tool in molecular biology. It has been used in identification of PPIs, regulation of proteins and RNAs as well as in drug discoveries among others [
17,
18,
19,
20,
21,
22]. In recent years, achievements have been made in protein regulation. For instance, Chen et al. used genetic encoding expansion techniques manipulate the functions of various enzymes, including
FTO,
luciferase, and
KRAS [
22]. Based on our previous study [
23], we installed the non-canonical PBBK into the critical site of Cas9 endonuclease through genetic encoding expansion, achieving a precise regulation of CRISPR-Cas9 gene editing. Therefore, we postulate that
NSUN2 can be labeled and regulated through the genetic encoding expansion technique (
Figure 1). Moreover, if the defined functionality is simultaneously installed, regulation of m
5C levels on specific RNAs in a spatiotemporal manner and further site-specific labeling of
NUSN2 can be achieved. Therefore, this study aimed at evaluating the intrinsic nature of the m
5C modifying enzyme,
NSUN2. Moreover, site-specific labeling and modulation of
NSUN2 besides the upregulated genes was also clarified.
2. Materials and Methods
All chemicals were purchased from commercially available sources otherwise stated including Innochem (Beijing, China), Aladdin (Shanghai, China), Ark (Shanghai, China), TCI (Shanghai, China), Sigma-Aldrich Inc. (Shanghai, China). All solvents were used directly purchased from Innochem (Shanghai, China) without further purification. Buffers including phosphate-buffered saline (PBS) and Tris(hydroxymethyl) aminomethane (Tris) were purchased from Innochem (Beijing, China). 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES, Cas # 7365-45-9), acrylamide and thiazolyl blue tetrazolium bromide (MTT, Cas # 298-93-1) were purchased from Sigma-Aldrich (Shanghai, China).
NMR were performed on a Bruker AM-400 spectrometer. Mass spectra was performed on Advion Expression L (Bohui Innovation Biotechnology, Beijing, China) using electrospray ionization (ESI). UV spectra was performed on Perkin Elmer Lambda 365 (Minden, German). Gel Imaging was performed using Pharos FX Molecular Imager (Bio-Rad, Hercules, CA, USA). Confocal microscope images were analyzed on Zeiss LSM780. Flow cytometry were analyzed on an LSR-II Flow Cytometer (BD Biosciences, Franklin Lakes, NJ, USA) and data were analyzed using FlowJo software (Tree Star, HongKong, China).
Synthesis of substrate S1. 2-Nitrobenzyl alcohol (500.0 mg, 3.3 mmol) was dissolved in THF (7.0 mL), containing Na2CO3 (345.5 mg, 3.3 mmol), and cooled to 0 °C. Triphosgene (967.4 mg, 3.3 mmol) was added to this solution and the reaction was continued to be stirred at r.t. for 12 h. The reaction was filtered, and the volatiles were subsequently evaporated without heating and the residue dried under vacuum to give carbamate intermediate in quantitative yield (702.8 mg, 3.3 mmol). Carbamate intermediate (702.8 mg, 3.3 mmol) was added to a solution of Boc-l-lysine (840.6 mg, 3.6 mmol) in a mixture of THF and 1.0 M NaOH (1: 4, 14.0 mL) at 0 °C. The reaction was stirred at r.t. for 12 h. The aqueous layer was washed with Et2O (5.0 mL) and subsequently acidified with ice-cold 1.0 M HCl (20.0 mL) to pH = 1 and was extracted with ethyl acetate (30 mL). The organic layer was dried over Na2SO4, filtered, and the volatiles were evaporated to give Boc-S1 (1.2 g) in 87% yield. 1H-NMR (400 MHz, CDCl3) δ 8.06 (d, J = 8.0 Hz, 1H), 7.73–7.53 (m, 2H), 7.53–7.35 (m, 1H), 5.43 (s, 2H), 4.31–4.27 (m, 1H), 3.21–3.17 (m, 2H), 1.85–1.80 (m, 2H), 1.71–1.67 (m, 2H), 1.57–1.52 (m, 2H), 1.42 (s, 9H). 13C-NMR (101 MHz, CDCl3) δ 176.15, 156.29, 147.04, 137.52, 133.84, 133.24, 128.42, 127.85, 124.77, 79.93, 63.07, 61.48, 40.65, 31.91, 28.23, 22.41. MS (ESI): [M + Na] + = 447.8; HRMS (EI): Calculated for C19H27N3O8 [M + Na] +: 448.1696; Found: 448.1754.
Overall protocol for unnatural lysine substitution.
- Step 1.1.
Screen specific MbPylRS.
- Step 1.2.
Construction of a dual fluorescence reporter system for unnatural amino acid substitution detection. Details are presented in
Supplementary Material.
- Step 1.3.
Replacement of unnatural amino acids in the active site of NSUN2.
Through the above experiments, the construction and screening of the unnatural amino acid substitution system for the above four lysine derivatives in eukaryotic cells have been completed, and then the unnatural amino acid substitution of the active site C271 in NSUN2 will be further implemented. C271 is an important active site of NSUN2, responsible for the separation of NSUN2 from the substrate after catalysis.
- Step 1.4.
Western blot detection of the effect of unnatural amino acid substitution at C271 of NSUN2 on protein expression.
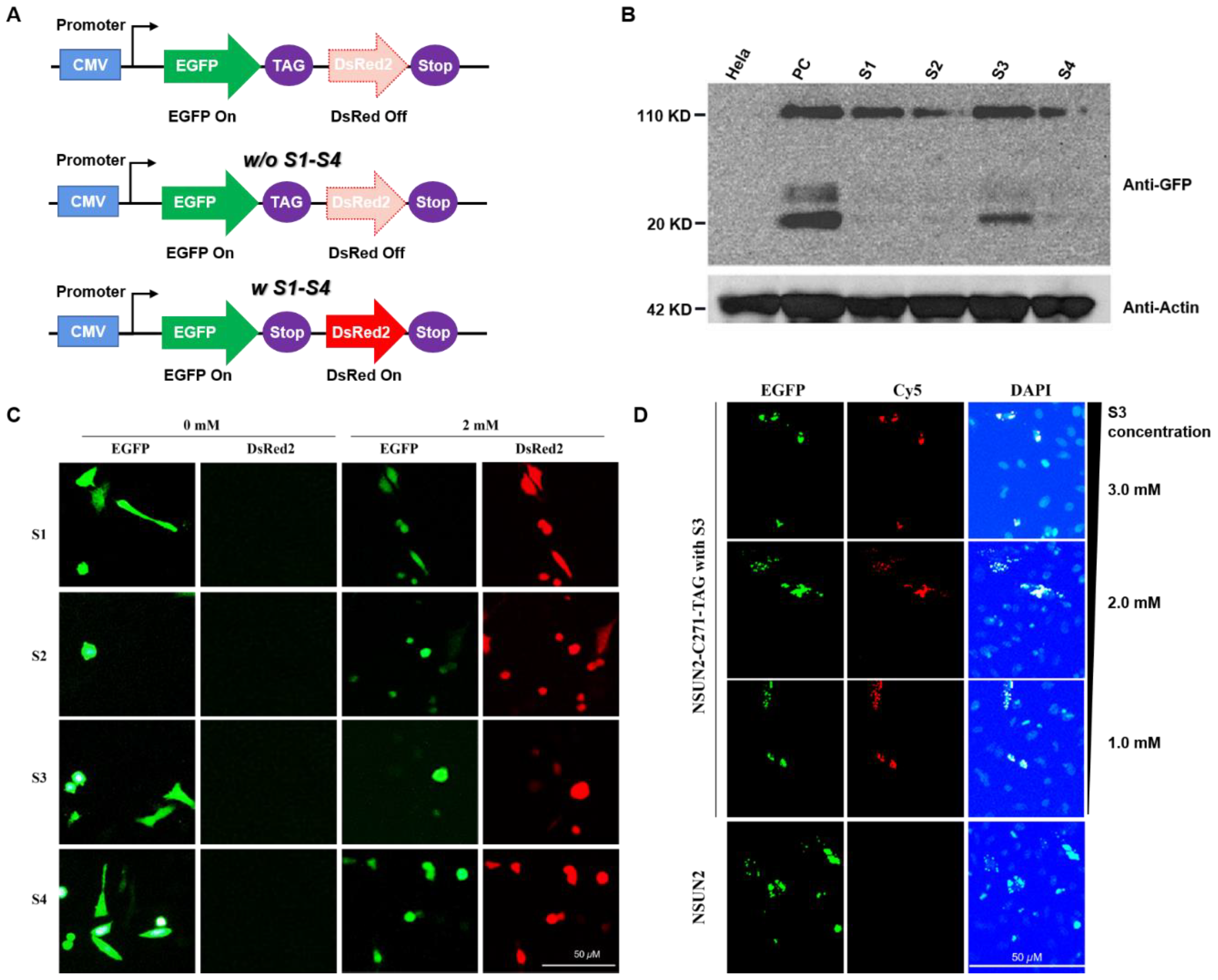
First, the eukaryotic expression system of NSUN2 was constructed as shown. The C271 site of the NSUN2 gene sequence was mutated to a TAG amber codon. The C-terminal of NSUN2 has an EGFP fluorescent protein tag. NSUN2-EGFP eukaryotic expression system was transfected into MbPylRS-tRNACUA expressing cells. The cells were cultured in a DMEM medium containing different lysine derivatives. After 24 h, the cells were lysed, and the total cell protein was extracted. The protein expression of NSUN2 was detected by Western blot. The experimental results show that the expression of NSUN2 C271 mutant protein can be detected in the presence of S1–S4.
In HeLa cells expressing the MbPylRS-tRNACUA system, they were cultured in a DMEM medium containing S1–S4 (2.0 mM) synthetic lysine. After 24 h, the cells were lysed, and the total protein was extracted. The protein expression was detected by Western blot. Wild-type HeLa cells were used as a negative control, and HeLa cells transfected with NSUN2 overexpression plasmid were used as a positive control.
Bio-orthogonal reaction to label NSUN2 and detect its subcellular localization.
Through Western blot detection, it is found that the unnatural amino acid substitution of C271 at the mutation site of NSUN2 can be efficiently realized in the presence of S3. Next, the click reaction was used to label S3 at the C271 site of NSUN2 with Cy5 fluorescent dye, and the subcellular localization of NSUN2 could be tracked. To further test whether this protein labeling method is accurate, the EGFP fusion protein of NSUN2 was constructed as described in the previous section. Detecting the co-localization of Cy5 (red fluorescence) and EGFP (green fluorescence) can detect the subcellular localization of NSUN2. In addition, in order to improve the expression efficiency of NSUN2, the concentration of S3 was further optimized. Cells were cultured in a DMEM medium containing 3.0 mM, 2.0 mM, and 1.0 mM S3, respectively, and then stained with Cy5 to detect protein localization under a fluorescence microscope. The experimental results show that with the continuous increase of Lys-N3 (S3) addition level, the replacement efficiency of unnatural amino acids in the process of NSUN2 protein expression also gradually increases, and the positioning of green fluorescent protein and red fluorescence shows that there are two kinds of fluorescence. The phenomenon of co-localization also shows that Lys-N3 (S3) has achieved amino acid substitution at position C271 of NSUN2.
In the HeLa cells expressing the MbPylRS-tRNACUA unnatural amino acid substitution system and the NSUN2-EGFP C271TAG mutation system, the cells were cultured in a DMEM medium containing different concentrations of S3 (1.0, 2.0, 3.0 mM) for 24 h, and then reacted with Cy5 and Lys-N3 (S3) via the click reaction to label NSUN2 with unnatural amino acids. NSUN2 itself has a EGFP label, and the subcellular localization of the two fluorescence could be detected by fluorescence microscope to reflect the unnatural amino acid replacement efficiency and subcellular localization of NSUN2.
3. Results
To establish the feasibility of our method, we synthesized four lysine substrates (
Supplementary Figures S1–S14, Schemes S1–S4). As shown in
Figure 2, the four lysine substrates were synthesized and evaluated by NMR and mass spectroscopy (
Figure 2A). S3 was obtained from commercially available Boc-protected
L-lysine through two linear steps. A yield of over 95% was obtained (
Figure 2B). The four lysine analogs can be transformed into their native forms using established methods [
23,
24,
25,
26]. For instance, substrate S1 was restored to lysine by UV (365 nm)-light within 5 min [
24]. Substrate S2 was transformed to its untreated form through basic hydrolysis [
25] while substrate S4 was rescued into lysine by the signaling molecule, hydrogen peroxide, as previously reported. [
23] Additionally, substrate S3 was restored to its native form through rapid addition of tricarboxyethyl phosphine (TCEP) [
26]. Kinetic investigation of the S3 reaction using TCEP revealed a pseudo-first order with a half time of 24.8 min (
Figure 2C,D and
Supplementary Figure S15, Table S1, Chart S1).
Construction of a site-specific unnatural amino acid substitution system. We further determined whether the substrates (S1–S4) can be site-specifically incorporated into the
NSUN2 enzyme (
Supplementary Figure S17). Based on the structure of the m
5C modification enzyme,
NSUN6 [
27], cysteine at position 271 plays an important role in RNA m
5C modifications. However, the crystal structure of
NSUN2 has not been established (
Supplementary Figure S16). By screening the
NSUN2 mutant that had been randomly incorporated with our lysine analogs (i.e., our substrates S1–S4), regulation of
NSUN2 enzyme activity was achieved, since they were caged. As for specific substrate S3, terminal 6-amino group was transformed to azido group, compared to lysine. As postulated, the
NSUN2 activity could be restored if the Staudinger reaction was applied to reduce the azido group to the amine functionality [
27]. Moreover, bio-orthogonal reaction of the azido group with various alkynes (attached with fluorescence or biotinylated functionality) was performed, which realized fluorescent labeling or chemical pulldown.
Then, we screened tRNA transferases (MbPylRS) with abilities to specifically recognize lysine derivatives with defined modified groups.
Screening of specific
MbPylRS. First, the mutation library was obtained through random mutations of the
MbPylRS active site. Random mutations were performed in six
MbPylRS sites, including L266, L270, Y271, L274, C313, and W383, with 10
8 mutants being theoretically produced. Then,
MbPylRS mutants with the ability to specifically recognize lysine derivatives (S1–S4) were screened from the mutant library. To facilitate the screening process, we constructed two screening systems [
18,
19,
20]. Through one positive and two negative screening processes, the
MbPylRS-tRNA
CUA system with specific recognition and coding abilities for target lysine derivatives (S1–S4) was identified. Therefore, we achieved efficient replacements of unnatural lysines (S1–S4) at
NSUN2 important sites (
Supplementary Figure S19–S21).
Construction of dual fluorescence reporter system for detection of unnatural lysine substitution. To determine whether screening of unnatural lysine substitution system is feasible, a dual fluorescence reporter system was established. HeLa cells expressing
MbPylRS-tRNA
CUA and dual fluorescent reporter genes were cultured in a DMEM medium with specific lysine derivatives (S1–S4). After 24 h, expressions of fluorescent proteins were detected.
MbPylRS-tRNA
CUA, which specifically recognized the four lysine derivatives (S1–S4), was successfully selected after three rounds of screening. As shown in
Figure 3C, expression of the red fluorescent proteins failed in the absence of S1–S4, while expressions of the two fluorescent proteins were detected in the presence of S1–S4. The results indicated that
MbPylRS-tRNA
CUA could recognize specific unnatural lysine and code the amber codon, implying efficient replacement of lysine with defined S1–S4 on this system.
Substitution with unnatural amino acids at the important site of
NSUN2. The C271 is a critical site of
NSUN4 (
Supplementary Figure S16). It is important for separation of
NSUN from the substrate after catalytic activity completion [
13,
14,
15]. To establish the feasibility of substitution with unnatural lysine on the C271 site, we constructed a C271
NSUN2 mutant in eukaryotic cells. First, the eukaryotic expression system of
NSUN2 was constructed; then, the C271 of
NSUN2 gene sequence was mutated into the TAG amber codon, and the C-terminal of
NSUN2 labeled with the
EGFP fluorescent protein. In
MbPylRS-tRNA
CUA expression cells, the
NSUN2-EGFP eukaryotic expression system was transiently stained. Cells were cultured in DMEM medium supplemented with various lysine derivatives (S1–S4). After 24 h, cells were lysed and total proteins extracted. Protein expressions of
NSUN2 were further analyzed by Western blot assays. We confirmed that expression of the
NSUN2 C271 mutant protein could be detected in the presence of S1–S4 (
Figure 3B). Expression levels of the
NSUN2 protein in the presence of S1 or S3 were relatively high (
Figure 3B), whereas expressions of
NSUN2 protein in the presence of S2 and S4 were much lower level compared to S1 or S3 (
Supplementary Figures S22 and S23). In addition to the desired target expression, a 20 KD band was expressed, it was attributed to the co-transfection process.
Detection of
NSUN2 using S3 (lys-N
3) by a Staudinger reaction with Cy5 in eukaryotic cells. To determine whether
NSUN2 can be located and detected in subcellular regions, click reaction was performed based on azido functionality on
NSUN2 (
Supplementary Schemes S5 and S6 and Figure S18). The unnatural lysine substitution of
NSUN2 mutation C271 was efficiently realized in the presence of S3. Moreover,
NSUN2 with S3 incorporated on the C271 site was labeled with the DBCO-Cy5 fluorescent dye through the click reaction. As such, subcellular location of the
NSUN2 mutant was established (
Figure 3D, red dot). To assess the accuracy of this protein labeling method, the
EGFP fusion protein of
NSUN2 was constructed. Subcellular localization of
NSUN2 was detected by monitoring co-localizations of DBCO-Cy5 (red fluorescence) and
EGFP (green fluorescence) (
Figure 3D). To enhance expression efficacies of
NSUN2, S3 concentrations were optimized. Cells were cultured in DMEM medium with 1.0 mM, 2.0 mM, and 3.0 mM of S3. Respectively. Then, protein localization was detected by fluorescence microscopy after staining with DBCO-Cy5 (
Supplementary Schemes S5 and S6). There was an enhanced substitution efficacy of unnatural lysine during NSUN2 protein expression with elevated concentrations of S3 (lys-N
3). Through the detection of localizations of the green fluorescent protein and the red fluorescence, dual fluorescence were co-located, indicating efficient incorporation of S3 at the NSUN2 C271 site, displaying a potential for real-time tracking of NSUN2. Our findings show that biological orthogonal reactions of unnatural lysine with the azido group together with genetic encoding expansions enable specific NSUN2 protein labeling and tracking (
Figure 3C,D).
Effects of
NSUN2 active site mutations on function. To evaluate the effect of
NSUN2 C271 mutation on the activity of
NSUN2, we investigated the mechanism of its upregulated gene. First, the effect of
NSUN2 C271 mutation to alanine was evaluated. As an RNA methyltransferase, the mutant on
NSUN2 dysregulates RNA methylation, affecting the function and stability of target RNA [
28,
29,
30,
31,
32]. Therefore, we knocked out
NSUN2 in HeLa cells, then replenished the wild-type or C271 mutant of
NSUN2. Proliferative abilities of HeLa cells were then detected using the CCK8 kit, while the effects of
NSUN2 on
CDK1 transcription levels were evaluated by RT-PCR. The
NSUN2 mutant had no significant effect on
CDK1 transcription levels (
Figure 4A). Upon
NSUN2 knock-out, cell proliferation levels were significantly suppressed. When HeLa cells were supplemented with wild-type
NSUN2, cell proliferation levels returned to the wild-type level [
32]. Notably, when
NSUN2 C271 was mutated to alanine, cell proliferation levels were significantly sup-pressed, implying that the C271 mutation significantly inhibited NSUN2 activities (
Figure 4B).
Effects of unnatural lysine substitution on the function of
NSUN2. Regulatory effects of
NSUN2 after replacement of C271 with unnatural lysine (S1–S4) were investigated.
MbPylRS-tRNA
CUA unnatural lysine substitution system and
NSUN2-C271TAG mutant were co-expressed in HeLa cells. Cells were cultured in a DMEM medium containing S1–S4, after which proliferative abilities of tumor cells were detected. Proliferation levels of cells with S1 or S3 substrates were significantly suppressed when compared to wild-type cells, but were significantly higher than for cells with substrates S2 or S4. Therefore, the system with S1 and S3 exhibited higher replacement efficiencies than the one with S2 and S4 at
NSUN2 C271 sites (
Figure 4C).
4. Discussion and Conclusions
First, random mutation of the active site is used to obtain the MbPylRS random mutation library. Here, we mainly performed random mutations at six positions of MbPylRS L266, L270, Y271, L274, C313, W383, and theoretically produced 108 mutants. From this mutant library, MbPylRS mutants that can specifically recognize lysine derivatives were selected. In order to facilitate screening, two screening systems were constructed next. The first screening system is a forward screening system, in which two sites in the tetracycline gene sequence are replaced with TAG amber codons, and in the presence of the MbPylRS-tRNACUA system, if the MbPylRS mutant can specifically recognize the band Lysine derivatives with specific modification groups can convert the amber stop codon into an intentional codon that can encode lysine derivatives. Thus, tetracycline can be expressed smoothly. In the medium containing tetracycline, the strain that can encode TAG can survive, otherwise it cannot survive. The second screening system is a negative screening system. In this system, Barnase ribonuclease is selected. As Barnase has strong cytotoxicity, cells cannot survive in strains expressing Barnase. Here, the two sites in Barnase were also mutated to amber codons. On the basis of positive screening, negative screening was performed here. The culture medium contained no specific lysine derivatives and contained MbPylRS-tRNACUA system and Barnase negative screening system strains, if the strains can survive, it indicates that MbPylRS-tRNACUA has specific recognition for specific lysine derivatives. However, if the strain is unable to survive, it indicates that Barnase is expressed, which means that MbPylRS-tRNACUA has no specific or poor specificity for the recognition of specific lysines, and can also recognize other amino acids. Through the above two screening systems, after two rounds of positive screening and one round of invisible screening, the MbPylRS-tRNACUA system with specific recognition and coding capabilities for target amino acid derivatives can be screened. Through this system, the NSUN2 active site will be further realized.
In order to test whether the screening of the unnatural amino acid replacement system is successful, a dual fluorescent reporter system was further constructed, and two amber codons were inserted between EGFP and DsRed, and the transcription and expression of the two fluorescent genes shared the same set of initiating factors. The HeLa cells expressing MbPylRS-tRNACUA and dual fluorescent reporter gene system were cultured in a DMEM medium containing specific lysine derivatives, and the expression status of fluorescent protein was detected 24 h later. Due to the existence of amber codons, if the unnatural amino acid substitution system can successfully recognize specific lysine derivatives and encode amber codons, the expression of two fluorescent proteins can be detected simultaneously under a fluorescence microscope. To realize the recognition of lysine derivatives, or the encoding of amber codons cannot be realized, the expression of DsRed cannot be further realized after the expression of EGFP. Therefore, only green fluorescence can be detected in this case. The experimental results show that after three rounds of screening, MbPylRS-tRNACUA, which can specifically recognize the above four lysine derivatives, was successfully screened, and the unnatural amino acid substitution of the target protein specific target in eukaryotic cells was successfully achieved. Simultaneous transfection of the dual fluorescence reporter system and unnatural amino acid substitution system in HeLa cells was performed, followed by incubation for 24 h in a DMEM medium without and with S1–S4 (2.0 mM). Then, the expression of fluorescent protein was detected under an inverted fluorescence microscope.
As an m
5C methyltransferase, studies on the function of
NSUN2 are in their infancy stage—specifically, regulation of its RNA substrate [
11]. Regulation of important sites of
NSUN2 may be an efficient approach for modulating the function of these enzymes. There are two active sites of
NSUN2, including C321 and C271. C321 is responsible for methyl transfer while C271 is responsible for completion of cytosine methylation, thereby separating
NSUN2 from the modified cytosine site. We focused on modification of the C271 site of
NSUN2 with varied lysines using genetic encoding expansion and, subsequently, evaluated the effect of
NSUN2 C271 mutations on its activity and gene upregulation. Lysines with four different modifications were successfully installed at the C271 site of
NSUN2. Reports indicate that
CDK1 is an upregulated gene of
NSUN2 [
28,
29,
30].
NSUN2 is highly expressed in various tumor cells, and expression of
NSUN2 promotes the translation process of
CDK1, promoting cell cycle and proliferation [
30,
31,
32]. Furthermore, RT-PCR and cell proliferation assays revealed that the C271 mutation had a minimal influence on mRNA expression levels of upregulated factor
CDK1, but significantly inhibited
CDK1 functions. Moreover, C271 mutations with S1–S4 inhibited HeLa cell proliferation. We postulate that C271 site mutation can inhibit the separation of
NSUN2 from the target RNA sequence after completing the catalysis of m
5C, thereby affecting RNA functions, including the translation process of the target protein,
CDK1 [
28] to inhibit cell proliferation.
First, natural lysines modified with special groups were synthesized by organic synthesis. Second, through two rounds of positive screening and one round of negative screening, we evaluated the MbPylRS-tRNACUA unnatural lysine substitution system, which specifically recognizes lysines with a specific group. Non-natural lysine substitution at C271 active site of NSUN2 and the subsequent fluorescent labeling were realized through the click reaction. Furthermore, we investigated the function of NSUN2 with mutants (S1–S4), its upregulated CDK1 gene, and its effect on cell proliferation. In summary, through genetic encoding expansion, we constructed the NSUN2 model, whose important sites were mutated with unnatural lysine bearing N3, PBin, NO2, and CF3, among others. Moreover, through the click reaction, efficient labeling, and regulation of NSUN2 was verified, laying the basis for further studies on the function and regulatory mechanism of upregulated genes.