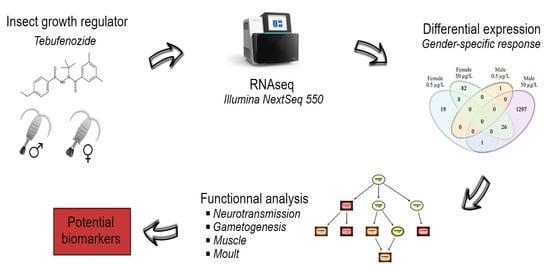
Susceptibility of the Non-Targeted Crustacean Eurytemora affinis to the Endocrine Disruptor Tebufenozide: A Transcriptomic Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Copepod Sampling and Acclimatization
2.2. Chemical Preparation and Experimental Design
2.3. RNA Extraction and Library Preparation and Sequencing
2.4. Differential Gene Expression
2.5. Functional Analysis
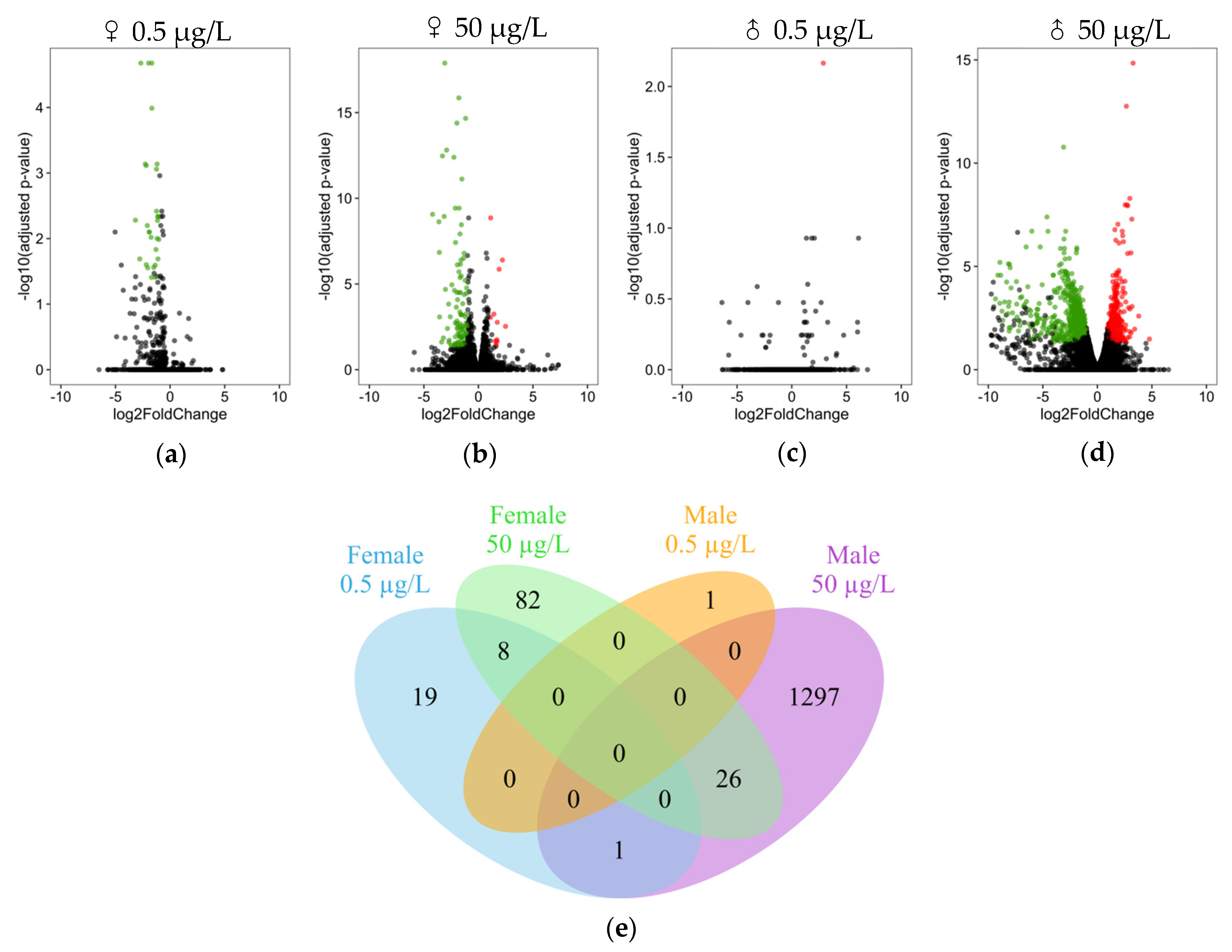
3. Results
3.1. Tebufenozide Concentration in Water
3.2. Differential Expression Analysis
3.3. DEG Functional Analysis
3.4. Mis-Regulated Genes of Interest
4. Discussion
4.1. Sex-Specific Transcriptomic Response
4.2. Defence Systems and Resistance to Insecticide in Males
4.3. Moulting Process and Cuticle Integrity
4.4. Reproductive Capacity and Vitellogenin Status in Males
4.5. Neuromuscular Pathways in Males and Females
4.6. Epigenetic Modifications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martin, J.W.; Davis, G.E. An Updated Classification of the Recent Crustacea; Natural History Museum of Los Angeles County: Los Angeles, CA, USA, 2001; Volume 39, ISBN 1891276271. [Google Scholar]
- Bron, J.E.; Frisch, D.; Goetze, E.; Johnson, S.C.; Lee, C.E.; Wyngaard, G.A. Observing copepods through a genomic lens. Front. Zool. 2011, 8, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Turner, J.T. The importance of small planktonic copepods and their roles in pelagic marine food webs. Zool. Stud. 2004, 43, 255–266. [Google Scholar] [CrossRef]
- Kulkarni, D.; Gergs, A.; Hommen, U.; Ratte, H.T.; Preuss, T.G. A plea for the use of copepods in freshwater ecotoxicology. Environ. Sci. Pollut. Res. 2013, 20, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Kwok, K.W.H.; Souissi, S.; Dur, G.; Won, E.-J.; Lee, J.S. Copepods as References Species in Estuarine and Marine Waters. In Aquatic Ecotoxicology: Advancing Tools for Dealing with Emerging Risks; Academic Press: Cambridge, MA, USA, 2015; pp. 281–308. ISBN 9780128011768. [Google Scholar]
- Arias, A.H.; Souissi, A.; Roussin, M.; Ouddane, B.; Souissi, S. Bioaccumulation of PAHs in marine zooplankton: An experimental study in the copepod Pseudodiaptomus marinus. Environ. Earth Sci. 2016, 75, 1–9. [Google Scholar] [CrossRef]
- Kadiene, E.U.; Ouddane, B.; Hwang, J.-S.; Souissi, S. Bioaccumulation of metals in calanoid copepods by oral intake. Sci. Rep. 2019, 9, 9492. [Google Scholar] [CrossRef] [PubMed]
- Zidour, M.; Boubechiche, Z.; Pan, Y.-J.; Bialais, C.; Cudennec, B.; Grard, T.; Drider, D.; Flahaut, C.; Ouddane, B.; Souissi, S. Population response of the estuarine copepod Eurytemora affinis to its bioaccumulation of trace metals. Chemosphere 2019, 220, 505–513. [Google Scholar] [CrossRef]
- Dalgaard Agersted, M.; Friis Møller, E.; Gustavson, K. Bioaccumulation of oil compounds in the high-Arctic copepod Calanus hyperboreus. Aquat. Toxicol. 2017, 195, 8–14. [Google Scholar] [CrossRef]
- Cailleaud, K.; Forget-Leray, J.; Souissi, S.; Hilde, D.; Lemenach, K.; Budzinski, H. Seasonal variations of hydrophobic organic contaminant concentrations in the water-column of the Seine Estuary and their transfer to a planktonic species Eurytemora affinis (Calanoïda, copepoda). Part 1: PCBs and PAHs. Chemosphere 2007, 70, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Cailleaud, K.; Forget-Leray, J.; Souissi, S.; Lardy, S.; Augagneur, S.; Budzinski, H. Seasonal variation of hydrophobic organic contaminant concentrations in the water-column of the Seine Estuary and their transfer to a planktonic species Eurytemora affinis (CalanoïdCalanoïd, copepoda). Part 2: Alkylphenol-polyethoxylates. Chemosphere 2007, 70, 281–287. [Google Scholar] [CrossRef]
- Hong, H.; Lv, D.; Liu, W.; Huang, L.; Chen, L.; Shen, R.; Shi, D. Toxicity and bioaccumulation of three hexabromocyclododecane diastereoisomers in the marine copepod Tigriopus japonicas. Aquat. Toxicol. Toxicol 2017, 188, 1–9. [Google Scholar] [CrossRef]
- Snape, J.R.; Maund, S.J.; Pickford, D.B.; Hutchinson, T.H. Ecotoxicogenomics: The challenge of integrating genomics into aquatic and terrestrial ecotoxicology. Aquat. Toxicol. 2004, 67, 143–154. [Google Scholar] [CrossRef]
- Prat, O.; Degli-Esposti, D. New Challenges: Omics Technologies in Ecotoxicology. In Ecotoxicology: New Challenges and New Approaches; Gross, E., Garic, J., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 181–208. ISBN 9781785483141. [Google Scholar]
- WHO; UNEP. State of the Science of Endocrine Disrupting Chemicals; World Health Organization (WHO): Geneva, Switzerland; UNEP: Nairobi, Kenya, 2012; ISBN 9780081019832. [Google Scholar]
- Rodríguez, E.M.; Medesani, D.A.; Fingerman, M. Endocrine disruption in crustaceans due to pollutants: A review. Comp. Biochem. Physiol.-A Mol. Integr. Physiol. 2007, 146, 661–671. [Google Scholar] [CrossRef]
- Tunaz, H.; Uygun, N. Insect Growth Regulators for Insect Pest Control. Turk. J. Agric. For. 2004, 28, 377–387. [Google Scholar]
- LeBlanc, G.A. Crustacean endocrine toxicology: A review. Ecotoxicology 2007, 16, 61–81. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.S.; Mykles, D.L. Regulation of crustacean molting: A review and our perspectives. Gen. Comp. Endocrinol. 2011, 172, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Sin, Y.W.; Kenny, N.J.; Qu, Z.; Chan, K.W.; Chan, K.W.S.; Cheong, S.P.S.; Leung, R.W.T.; Chan, T.F.; Bendena, W.G.; Chu, K.H.; et al. Identification of putative ecdysteroid and juvenile hormone pathway genes in the shrimp Neocaridina denticulata. Gen. Comp. Endocrinol. 2015, 214, 167–176. [Google Scholar] [CrossRef]
- Hyde, C.; Elizur, A.; Ventura, T. The crustacean ecdysone cassette: A gatekeeper for molt and metamorphosis. J. Steroid Biochem. Mol. Biol. 2018, 185, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Kusk, K.O.; Wollenberger, L. Towards an internationally harmonized test method for reproductive and developmental effects of endocrine disrupters in marine copepods. Ecotoxicology 2007, 16, 183–195. [Google Scholar] [CrossRef]
- OECD. Detailed Review Paper on Aquatic Arthropods in Life Cycle Toxicity Tests with an Emphasis on Developmental, Reproductive and Endocrine Disruptive Effects; OECD: Paris, France, 2006. [Google Scholar]
- Lee, C.E. Rapid and repeated invasions of fresh water by the copepod Eurytemora affinis. Evolution 1999, 53, 1423–1434. [Google Scholar] [CrossRef] [PubMed]
- Katona, S.K. The Developmental Stages of Eurytemora Affinis (Poppe, 1880) (Copepoda, Calanoida) Raised in Laboratory Cultures, Including a Comparison with the Larvae of Eurytemora Americana Williams, 1906, and Eurytemora Herd Man I Thompson & Scott, 1897. Crustaceana 1971, 21, 5–20. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, B.K.; Buskey, E.; Miller, D.C.; Ritacco, P.J. Effects of copper and cadmium on growth, swimming and predator avoidance in Eurytemora affinis (Copepoda). Mar. Biol. 1983, 77, 299–306. [Google Scholar] [CrossRef]
- Wright, D.; Savitz, J.; Dawson, R.; Magee, J.; Smucker, R. Effect of diflubenzuron on the maturation and reproductive success of the copepod Eurytemora affinis. Ecotoxicology 1996, 5, 47–58. [Google Scholar] [CrossRef]
- Forget-Leray, J.; Landriau, I.; Minier, C.; Leboulenger, F.-O. Impact of endocrine toxicants on survival, development, and reproduction of the estuarine copepod Eurytemora affinis (Poppe). Ecotoxicol. Environ. Saf. 2005, 60, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Cailleaud, K.; Michalec, F.G.; Forget-Leray, J.; Budzinski, H.; Hwang, J.S.; Schmitt, F.G.; Souissi, S. Changes in the swimming behavior of Eurytemora affinis (Copepoda, Calanoida) in response to a sub-lethal exposure to nonylphenols. Aquat. Toxicol. 2011, 102, 228–231. [Google Scholar] [CrossRef]
- Legrand, E.; Boulangé-Lecomte, C.; Restoux, G.; Trémolet, G.; Duflot, A.; Forget-Leray, J. Individual and mixture acute toxicity of model pesticides chlordecone and pyriproxyfen in the estuarine copepod Eurytemora affinis. Environ. Sci. Pollut. Res. 2017, 24, 5976–5984. [Google Scholar] [CrossRef]
- Lesueur, T.; Boulangé-Lecomte, C.; Xuereb, B.; Budzinski, H.; Cachot, J.; Vicquelin, L.; Giusti-Petrucciani, N.; Marie, S.; Petit, F.; Forget-Leray, J. Development of a larval bioassay using the calanoid copepod, Eurytemora affinis to assess the toxicity of sediment-bound pollutants. Ecotoxicol. Environ. Saf. 2013, 94, 60–66. [Google Scholar] [CrossRef]
- Lesueur, T.; Boulangé-Lecomte, C.; Restoux, G.; Deloffre, J.; Xuereb, B.; Le Menach, K.; Budzinski, H.; Petrucciani, N.; Marie, S.; Petit, F.; et al. Toxicity of sediment-bound pollutants in the Seine estuary, France, using a Eurytemora affinis larval bioassay. Ecotoxicol. Environ. Saf. 2015, 113, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Legrand, E.; Forget-Leray, J.; Duflot, A.; Olivier, S.; Thomé, J.-P.; Danger, J.-M.; Boulangé-Lecomte, C. Transcriptome analysis of the copepod Eurytemora affinis upon exposure to endocrine disruptor pesticides: Focus on reproduction and development. Aquat. Toxicol. 2016, 176, 64–75. [Google Scholar] [CrossRef]
- Lee, Y.S.; Lee, S.E.; Son, J.; Kim, Y.; Wee, J.; Cho, K. Toxicity effects and biomarkers of tebufenozide exposure in Yuukianura szeptyckii (Collembola: Neanuridae). Environ. Geochem. Health 2018, 40, 2773–2784. [Google Scholar] [CrossRef] [PubMed]
- Chandler, L.D.; Pair, S.D.; Harrison, W.E. RH-5992, A New Insect Growth Regulator Active Against Corn Earworm and Fall Armyworm (Lepidoptera: Noctuidae). J. Econ. Entomol. 1992, 85, 1099–1103. [Google Scholar] [CrossRef]
- Retnakaran, A.; Gelbic, I.; Sundaram, M.; Tomkins, W.; Ladd, T.; Primavera, M.; Feng, Q.; Arif, B.; Palli, R.; Krell, P. Mode of action of the ecdysone agonist tebufenozide (RH-5992), and an exclusion mechanism to explain resistance to it. Pest Manag. Sci. 2001, 57, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Carlson, G.R. Tebufenozide: A novel caterpillar control agent with unusually high target selectivity. In Green Chemical Syntheses and Processes; American Chemical Society: Washington, DC, USA, 2000; pp. 8–17. [Google Scholar]
- Devreker, D.; Souissi, S.; Winkler, G.; Forget-Leray, J.; Leboulenger, F. Effects of salinity, temperature and individual variability on the reproduction of Eurytemora affinis (Copepoda; Calanoida) from the Seine estuary: A laboratory study. J. Exp. Mar. Biol. Ecol. 2009, 368, 113–123. [Google Scholar] [CrossRef]
- Gouveia, D.; Bonneton, F.; Almunia, C.; Armengaud, J.; Quéau, H.; Degli-Esposti, D.; Geffard, O.; Chaumot, A. Identification, expression, and endocrine-disruption of three ecdysone-responsive genes in the sentinel species Gammarus fossarum. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kreutzweiser, D.P.; Thomas, D.R. Effects of a new molt-inducing insecticide, tebufenozide, on zooplankton communities in lake enclosures. Ecotoxicology 1995, 4, 307–328. [Google Scholar] [CrossRef]
- OECD. Guidance Document on Aqueous-phase. In Aquatic Toxicty Testing of Difficult Test Chemicals; OECD: Paris, France, 2018; Volume 23. [Google Scholar]
- Chhangawala, S.; Rudy, G.; Mason, C.E.; Rosenfeld, J.A. The impact of read length on quantification of differentially expressed genes and splice junction detection. Genome Biol. 2015, 16, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Eyun, S.-I.; Soh, H.Y.; Posavi, M.; Munro, J.B.; Hughes, D.S.T.; Murali, S.C.; Qu, J.; Dugan, S.; Lee, S.L.; Chao, H.; et al. Evolutionary History of Chemosensory Related Gene Families across the Arthropoda. Mol. Biol. Evol. 2017, 34, 1838–1862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Anders, S.; Huber, W. Differential analysis of count data—The DESeq2 package. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Alexa, A.; Rahnenführer, J. Gene Set Enrichment Analysis with topGO; R Package Version 2.36.0; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Tenenbaum, A.D.; Bioconductor, M.; Maintainer, P.; Gentleman, R.; Carlson, M. Package ‘KEGGREST’; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Wickham, H. ggplot2—Elegant Graphics for Data Analysis (2nd Edition). J. Stat. Softw. 2017, 77, 3–5. [Google Scholar] [CrossRef] [Green Version]
- Chen, H. Package ‘VennDiagram’: Generate High-Resolution Venn and Euler Plots; Version 1.6.20; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Li, N.; Arief, N.; Edmands, S. Effects of oxidative stress on sex-specific gene expression in the copepod Tigriopus californicus revealed by single individual RNA-seq. Comp. Biochem. Physiol Part D Genom. Proteom. 2019, 31, 100608. [Google Scholar] [CrossRef] [PubMed]
- Kadiene, E.U.; Ouddane, B.; Gong, H.Y.; Kim, M.S.; Lee, J.S.; Pan, Y.J.; Hwang, J.S.; Souissi, S. Differential gene expression profile of male and female copepods in response to cadmium exposure. Ecotoxicol. Environ. Saf. 2020, 204, 111048. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Flanagan, B.A.; Partridge, M.K.; Huang, E.J.; Edmands, S. Sex differences in early transcriptomic responses to oxidative stress in the copepod Tigriopus californicus. BMC Genom. 2020, 21, 759. [Google Scholar] [CrossRef] [PubMed]
- Medina, M.; Barata, C.; Telfer, T.; Baird, D.J. Age-and Sex-Related Variation in Sensitivity to the Pyrethroid Cypermethrin in the Marine Copepod Acartia tonsa Dana. Arch. Environ. Contam. Toxicol. 2002, 42, 17–22. [Google Scholar] [CrossRef]
- Kadiene, E.; Bialais, C.; Ouddane, B.; Hwang, J.-S.; Souissi, S. Differences in lethal response between male and female calanoid copepods and life cycle traits to cadmium toxicity. Ecotoxicology 2017, 26, 1227–1239. [Google Scholar] [CrossRef]
- Foley, H.B.; Sun, P.Y.; Ramirez, R.; So, B.K.; Venkataraman, Y.R.; Nixon, E.N.; Davies, K.J.A.; Edmands, S. Sex-specific stress tolerance, proteolysis, and lifespan in the invertebrate Tigriopus californicus. Exp. Gerontol. 2019, 119, 146–156. [Google Scholar] [CrossRef]
- Kadiene, E.U.; Meng, P.-J.; Hwang, J.-S.; Souissi, S. Acute and chronic toxicity of cadmium on the copepod Pseudodiaptomus annandalei: A life history traits approach. Chemosphere 2019, 233, 396–404. [Google Scholar] [CrossRef]
- Boulangé-Lecomte, C.; Forget-Leray, J.; Xuereb, B. Sexual dimorphism in Grp78 and Hsp90A heat shock protein expression in the estuarine copepod Eurytemora affinis. Cell Stress Chaperones 2014, 19, 591–597. [Google Scholar] [CrossRef] [Green Version]
- Hemingway, J.; Karunaratne, P. Mosquito carboxylesterases: A review of the molecular biology and biochemistry of a major insecticide resistance mechanism. Artic. Med. Vet. Entomol. 1998, 12, 1–12. [Google Scholar] [CrossRef]
- Scott, J.G. Cytochromes P450 and insecticide resistance. Insect Biochem. Mol. Biol. 1999, 29, 757–777. [Google Scholar] [CrossRef]
- Pavlidi, N.; Vontas, J.; Leeuwen, T. Van The role of glutathione S-transferases (GSTs) in insecticide resistance in crop pests and disease vectors. Curr. Opin. Insect Sci. 2018, 27, 97–102. [Google Scholar] [CrossRef]
- Cerenius, L.; Soderhall, K. The prophenoloxidase-activating system in invertebrates. Immunol. Rev. 2004, 198, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Jiravanichpaisal, P.; Cerenius, L.; Bok, L.L.; Söderhäll, I.; Söderhäll, K. Phenoloxidase is an important component of the defense against Aeromonas hydrophila infection in a crustacean, Pacifastacus leniusculus. J. Biol. Chem. 2007, 282, 33593–33598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terwilliger, N.B. Hemolymph Proteins and Molting in Crustaceans and Insects’. Am. Zool. 1999, 39, 589–599. [Google Scholar] [CrossRef]
- Karim, F.D.; Guild, G.M.; Thummel, C.S. The Drosophila Broad-Complex plays a key role in controlling ecdysoneregulated gene expression at the onset of metamorphosis. Development 1993, 118, 977–988. [Google Scholar] [CrossRef]
- Reynolds, S.E.; Samuels, R.I. Physiology and Biochemistry of Insect Moulting Fluid. Adv. Insect Phys. 1996, 26, 157–232. [Google Scholar] [CrossRef]
- Brookhart, G.L.; Kramer, K.J. Proteinases in molting fluid of the tobacco hornworm, Manduca sexta. Insect Biochem. 1990, 20, 467–477. [Google Scholar] [CrossRef]
- Koga, D.; Jilka, J.; Kramert, K.J. Insect endochitinases: Glycoproteins from moulting fluid, integument and pupal haemolymph of Manduca sexta L. Insect Biochem. 1983, 13, 295–305. [Google Scholar] [CrossRef]
- Van Wormhoudt, A.; Sellos, D.; Donval, A.; Plaire-Goux, S.; Le Moullac, G. Chymotrypsin gene expression during the intermolt cycle in the shrimp Penaeus vannamei (Crustacea; Decapoda). Experientia 1995, 51, 159–163. [Google Scholar] [CrossRef]
- Le Boulay, C.; Van Wormhoudt, A.; Sellos, D. Cloning and expression of cathepsin L-like proteinases in the hepatopancreas of the shrimp Penaeus vannamei during the intermolt cycle. J. Comp. Physiol. 1996, 166, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Kuballa, A.V.; Holton, T.A.; Paterson, B.; Elizur, A. Moult cycle specific differential gene expression profiling of the crab Portunus pelagicus. BMC Genom. 2011, 12, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Zhang, X.; Wei, J.; Sun, X.; Yuan, J.; Li, F.; Xiang, J. Whole transcriptome analysis provides insights into molecular mechanisms for molting in Litopenaeus vannamei. PLoS ONE 2015, 10, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceccaldi, H.J. Anatomy and physiology of digestive tract of Crustaceans Decapods reared in aquaculture. In Actes de Colloques Ifremer, Proceedings of the Advances in Tropical Aquaculture, Workshop, Tahiti, French Polynesia, 20 February–4 March 1989; ScienceOpen, Inc.: Boston, MA, USA, 1989; pp. 243–259. [Google Scholar]
- Hu, K.-J.; Leung, P.-C. Food digestion by cathepsin L and digestion-related rapid cell differentiation in shrimp hepatopancreas. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2007, 146, 69–80. [Google Scholar] [CrossRef]
- Smagghe, G.; Gelman, D.; Tirry, L. In vivo and in vitro effects of tebufenozide and 20-hydroxyecdysone on chitin synthesis. Arch. Insect Biochem. Physiol. 1999, 41, 33–41. [Google Scholar] [CrossRef]
- Doucet, D.; Retnakaran, A. Insect Chitin. Metabolism, Genomics and Pest Management. In Advances in Insect Physiology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 43, pp. 437–511. ISBN 9780123915009. [Google Scholar]
- Perez-Vilar, J.; Hill, R.L. The Structure and Assembly of Secreted Mucins. J. Biol. Chem. 1999, 274, 31751–31754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helvig, C.; Koener, J.F.; Unnithan, G.C.; Feyereisen, R. CYP15A1, the cytochrome P450 that catalyzes epoxidation of methyl farnesoate to juvenile hormone III in cockroach corpora allata. Proc. Natl. Acad. Sci. USA 2004, 101, 4024–4029. [Google Scholar] [CrossRef] [Green Version]
- Riddiford, L.M. How does juvenile hormone control insect metamorphosis and reproduction? Gen. Comp. Endocrinol. 2012, 179, 477–484. [Google Scholar] [CrossRef]
- Laufer, H.; Biggers, W.J. Unifying Concepts Learned from Methyl Farnesoate for Invertebrate Reproduction and Post-Embryonic Development. Am. Zool. 2001, 41, 442–457. [Google Scholar] [CrossRef] [Green Version]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Chalifa-Caspi, V.; Weil, S.; Sharabi, O.; Sagi, A. Post-Embryonic Transcriptomes of the Prawn Macrobrachium rosenbergii: Multigenic Succession through Metamorphosis. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Lin, Y.; Liu, H.; Shen, G.; Luo, J.; Zhang, H.; Peng, Z.; Chen, E.; Xing, R.; Han, C.; et al. The Broad Complex isoform 2 (BrC-Z2) transcriptional factor plays a critical role in vitellogenin transcription in the silkworm Bombyx mori. Biochim. Biophys. Acta 2014, 1840, 2674–2684. [Google Scholar] [CrossRef] [PubMed]
- Buaklin, A.; Sittikankaew, K.; Khamnamtong, B.; Menasveta, P.; Klinbunga, S. Characterization and expression analysis of the Broad-complex (Br-c) gene of the giant tiger shrimp Penaeus monodon. Comp. Biochem. Physiol.-B Biochem. Mol. Biol. 2013, 164, 280–289. [Google Scholar] [CrossRef]
- Piulachs, M.D.; Pagone, V.; Bellés, X. Key roles of the Broad-Complex gene in insect embryogenesis. Insect Biochem. Mol. Biol. 2010, 40, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Chebbo, S.; Josway, S.; Belote, J.M.; Manier, M.K. A putative novel role for Eip74EF in male reproduction in promoting sperm elongation at the cost of male fecundity. J. Exp. Zool. Part B Mol. Dev. Evol. 2020, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sitnik, J.L.; Francis, C.; Hens, K.; Huybrechts, R.; Wolfner, M.F.; Callaerts, P. Neprilysins: An evolutionarily conserved family of metalloproteases that play important roles in reproduction in Drosophila. Genetics 2014, 196, 781–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anaka, M.; MacDonald, C.D.; Barkova, E.; Simon, K.; Rostom, R.; Godoy, R.A.; Haigh, A.J.; Meinertzhagen, I.A.; Lloyd, V. The white gene of Drosophila melanogaster encodes a protein with a role in courtship behavior. J. Neurogenet. 2008, 22, 243–276. [Google Scholar] [CrossRef]
- Hazelrigg, T.; Watkins, W.S.; Marcey, D.; Tu, C.; Karow, M.; Lin, X. The exuperantia gene is required for Drosophila spermatogenesis as well as anteroposterior polarity of the developing oocyte, and encodes overlapping sex-specific transcripts. Genetics 1990, 126, 607–617. [Google Scholar] [CrossRef]
- Parvinen, M. The chromatoid body in spermatogenesis. Int. J. Androl. 2005, 28, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Shao, M.; Qin, Z.; Kyoung, H.K.; Zhang, Z. Cloning, characterization, and expression analysis of the DEAD-box family genes, Fc-vasa and Fc-PL10a, in Chinese shrimp (Fenneropenaeus chinensis). Artic. Chin. J. Oceanol. Limnol. 2010, 28, 37–45. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Han, K.; Zou, Z.; Zhang, Z. A vasa gene from green mud crab Scylla paramamosain and its expression during gonadal development and gametogenesis. Mol. Biol. Rep. 2012, 39, 4327–4335. [Google Scholar] [CrossRef] [PubMed]
- Özhan-Kizil, G.; Havemann, J.; Gerberding, M. Germ cells in the crustacean Parhyale hawaiensis depend on Vasa protein for their maintenance but not for their formation. Dev. Biol. 2009, 327, 230–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, A.; Li, J.; Song, L.; Ji, C.; Böing, M.; Chen, J.; Brand-Saberi, B. GGNBP2 is necessary for testis morphology and sperm development. Sci. Rep. 2017, 7, 2998. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; He, Y.; Guo, K.; Zhou, L.; Li, X.; Tseng, M.; Cai, L.; Lan, Z.J.; Zhou, J.; Wang, H.; et al. Ggnbp2-Null Mutation in Mice Leads to Male Infertility due to a Defect at the Spermiogenesis Stage. Am. J. Pathol. 2017, 187, 2508–2519. [Google Scholar] [CrossRef] [Green Version]
- Tardif, S.; Cormier, N. Role of zonadhesin during sperm-egg interaction: A species-specific acrosomal molecule with multiple functions. Mol. Hum. Reprod. 2011, 17, 661–668. [Google Scholar] [CrossRef] [Green Version]
- Grant, H.J.; Whitfield, P.J. The ultrastructure of the spermatozoon of Lernaeocera branchialis (Copepoda: Pennellidae). Hydrobiologia 1988, 167, 597–605. [Google Scholar] [CrossRef]
- Short, S.; Yang, G.; Kille, P.; Ford, A.T. Vitellogenin is not an appropriate biomarker of feminisation in a Crustacean. Aquat. Toxicol. 2014, 153, 89–97. [Google Scholar] [CrossRef] [Green Version]
- Boulangé-Lecomte, C.; Xuereb, B.; Trémolet, G.; Duflot, A.; Giusti, N.; Olivier, S.; Legrand, E.; Forget-Leray, J. Controversial use of vitellogenin as a biomarker of endocrine disruption in crustaceans: New adverse pieces of evidence in the copepod Eurytemora affinis. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2017, 201, 66–75. [Google Scholar] [CrossRef]
- Hannas, B.R.; Wang, Y.H.; Thomson, S.; Kwon, G.; Li, H.; Leblanc, G.A. Regulation and dysregulation of vitellogenin mRNA accumulation in daphnids (Daphnia magna). Aquat. Toxicol. 2011, 101, 351–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skinner, D.M. Breakdown and reformation of somatic muscle during the molt cycle of the land crab, Gecarcinus lateralis. J. Exp. Zool. 1966, 163, 115–123. [Google Scholar] [CrossRef]
- Song, Y.; Villeneuve, D.L.; Toyota, K.; Iguchi, T.; Tollefsen, K.E. Ecdysone Receptor Agonism Leading to Lethal Molting Disruption in Arthropods: Review and Adverse Outcome Pathway Development. Environ. Sci. Technol. 2017, 51, 4142–4157. [Google Scholar] [CrossRef] [Green Version]
- Güiza, J.; Barría, I.; Sáez, J.C.; Vega, J.L. Innexins: Expression, regulation, and functions. Front. Physiol. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Krishnan, S.N.; Frei, E.; Swain, G.P.; Wyman, R.J. Passover: A gene required for synaptic connectivity in the giant fiber system of Drosophila. Cell 1993, 73, 967–977. [Google Scholar] [CrossRef]
- Phelan, P.; Nakagawa, M.; Wilkin, M.; Moffat, K.G.; O’kane, C.J.; Davies, J.A.; Bacon, J.P. Mutations in shaking-B Prevent Electrical Synapse Formation in the Drosophila Giant Fiber System. J. Neurosci. 1996, 6, 1101–1113. [Google Scholar] [CrossRef] [Green Version]
- Scimemi, A. Structure, function, and plasticity of GABA transporters. Front. Cell Neurosci. 2014, 8. [Google Scholar] [CrossRef] [Green Version]
- Spletter, M.L.; Liu, J.; Liu, J.; Su, H.; Giniger, E.; Komiyama, T.; Quake, S.; Luo, L. Lola regulates Drosophila olfactory projection neuron identity and targeting specificity. Neural. Dev. 2007, 2, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaharbakhshi, E.; Jemc, J.C. Broad-complex, tramtrack, and bric-à-brac (BTB) proteins: Critical regulators of development. Genesis 2016, 54, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Van Vactor, D.; Krantz, D.M.; Reinke, R.; Zipursky, S.L. Analysis of Mutants in Chaoptin, a Photoreceptor Cell-Specific Glycopmtein in Drosophila, Reveals Its Role in Cellular Morphogenesis. Cell 1988, 52, 281–290. [Google Scholar] [CrossRef]
- Zelhof, A.C.; Hardy, R.W.; Becker, A.; Zuker, C.S. Transforming the architecture of compound eyes. Nature 2006, 443, 696–699. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, H.; Hayata, T.; Eisaki, A.; Asashima, M. Cloning a novel developmental regulating gene, Xotx5: Its potential role in anterior formation in Xenopus laevis. Dev. Growth Differ. 2000, 42, 87–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandendries, E.R.; Johnson, D.; Reinke, R. Orthodenticle Is Required for Photoreceptor Cell Development in the Drosophila Eye. Dev. Biol. 1996, 173, 243–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, R.G.P.; Igloi, G.L.; Lichte, B.; Baumann, U.; Maier, D.; Schneider, T.; Brandstatter, J.H.; Frohlich, A.; Fischbach, K.F. The irregular chiasm C-roughest locus of Drosophila, which affects axonal projections and programmed cell death, encodes a novel immunoglobulin-like protein. Genes Dev. 1993, 7, 2533–2547. [Google Scholar] [CrossRef] [PubMed]
- Araujo, H.; Machado, L.C.H.; Octacílio-Silva, S.; Mizutani, C.M.; Silva, M.J.F.; Ramos, R.G.P. Requirement of the roughest gene for differentiation and time of death of interommatidial cells during pupal stages of Drosophila compound eye development. Mech. Dev. 2003, 120, 537–547. [Google Scholar] [CrossRef]
- Martin, G.G.; Speekmann, C.; Beidler, S. Photobehavior of the harpacticoid copepod Tigriopus californicus and the fine structure of its nauplius eye. Invertebr. Biol. 2000, 119, 110–124. [Google Scholar] [CrossRef]
- Fiaz, M.; Martínez, L.C.; Plata-Rueda, A.; Gonçalves, W.G.; Shareef, M.; Zanuncio, J.C.; Serrão, J.E. Toxicological and morphological effects of tebufenozide on Anticarsia gemmatalis (Lepidoptera: Noctuidae) larvae. Chemosphere 2018, 212, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Vandegehuchte, M.B.; Janssen, C.R. Epigeneics and its implications for ecotoxicology. Ecotoxicology 2011, 20, 607–624. [Google Scholar] [CrossRef]
- Head, J.A.; Dolinoy, D.C.; Basu, N. Epigenetics for ecotoxicologists. Environ. Toxicol. Chem. 2012, 31, 221–227. [Google Scholar] [CrossRef]


| Sample | Time | [TEB] (µg/L) |
|---|---|---|
| Control | T0 | <QL 1 |
| TEB 0.5 µg/L | T0 | <QL 1 |
| TEB 50 µg/L | T0 | 13.82 |
| Control | T72h | <QL 1 |
| TEB 0.5 µg/L | T72h | <QL 1 |
| TEB 50 µg/L | T72h | 11.73 |
| Gene Symbol and Description | ♀ 0.5 µg/L | ♀ 50 µg/L | ♂ 50 µg/L | |
|---|---|---|---|---|
| Moulting and metamorphosis | LOC111704351 broad-complex core protein isoforms 1/2/3/4/5-like | - | - | −1.63 * |
| LOC111709681 broad-complex core protein isoforms 1/2/3/4/5-like | - | - | −1.09 * | |
| LOC111694963 broad-complex core protein isoforms 1/2/3/4/5-like | - | - | −1.49 * | |
| LOC111704069 ecdysone-induced protein 74EF-like | - | - | 1.40 ** | |
| LOC111697606 cuticle protein 7-like | −2.68 * | - | - | |
| LOC111698563 cuticle protein 7-like | - | −3.02 * | - | |
| LOC111702984 cuticle protein 16.5-like | −2.18 * | - | - | |
| LOC111704119 cuticle protein 16.5-like | −1.97 * | - | - | |
| LOC111708925 cuticle protein 16.5-like | −2.09 * | - | - | |
| LOC111711095 cuticle protein 16.5-like | - | −1.24 * | - | |
| LOC111717477 methyl farnesoate epoxidase-like | - | 1.53 ** | 2.97 ** | |
| Glycosphingolipids | LOC111699882 ganglioside GM2 activator-like | −1.11 * | - | - |
| LOC111702480 ganglioside GM2 activator-like | - | −1.70 * | - | |
| Mucins | LOC111697205 mucin-5AC-like | −1.90 * | −1.88 * | - |
| LOC111710342 mucin-5AC-like | - | - | −1.23 * | |
| LOC111699105 mucin-5AC-like | - | - | −2.95 * | |
| LOC111716966 mucin-7-like | - | - | −1.26 * | |
| LOC111700935 mucin-17-like | - | - | −7.06 * | |
| LOC111713839 mucin-17-like | - | - | −1.02 * | |
| Protease | LOC111697677 cathepsin L1-like | - | - | 1.11 ** |
| LOC111707394 chymotrypsin-like protease CTRL-1 | - | - | 1.31 ** | |
| LOC111701514 chymotrypsinogen A-like | - | - | 1.31 ** | |
| LOC111697179 endochitinase A-like | - | - | −1.26 * | |
| LOC111716204 trypsin-1-like | - | −1.54 * | - | |
| LOC111703434 trypsin-1-like | - | - | 1.62 ** | |
| Muscle | LOC111696032 actin, clone 403-like | - | - | 1.51 ** |
| LOC111717718 actin, muscle-like | - | - | 1.09 ** | |
| LOC111697494 calpain clp-1-like | - | - | 1.64 ** | |
| LOC111713061 myocyte-specific enhancer factor 2-like | - | - | 1.76 ** | |
| LOC111712575 myosin-1-like | - | - | 2.96 ** | |
| LOC111695443 myosin-9-like | - | - | −1.44 * | |
| LOC111711689 myosin heavy chain, muscle-like | - | −2.25 * | −1.92 * | |
| LOC111711716 myosin heavy chain, muscle-like | - | −3.56 * | −2.86 * | |
| LOC111715464 myosin heavy chain, muscle-like | - | - | 1.61 ** | |
| LOC111708305 myosin regulatory light chain 2-like | - | - | 1.08 ** | |
| LOC111703505 titin homolog | - | - | 1.34 ** | |
| LOC111697095 titin-like | - | - | 1.21 ** | |
| Neuro-transmission | LOC111696990 acetylcholinesterase-like | - | - | 1.62 ** |
| LOC111695263 acetylcholine receptor subunit alpha-like | - | - | 2.45 ** | |
| LOC111697821 acetylcholine receptor subunit alpha-type acr-16-like | - | - | 1.03 ** | |
| LOC111714797 acetylcholine receptor subunit beta-like 2 | - | - | 1.45 ** | |
| LOC111695314 innexin inx2-like | - | - | −3.75 * | |
| LOC111702621 innexin inx2-like | - | - | 1.60 ** | |
| LOC111718086 innexin shaking-B-like | - | - | 1.92 ** | |
| LOC111698781 neurexin-1-like | - | - | 1.45 ** | |
| LOC111698578 neurexin-3-like | - | - | 1.25 ** | |
| LOC111704195 sodium- and chloride-dependent GABA transporter 3-like | - | - | 1.79 ** | |
| LOC111711043 sodium- and chloride-dependent GABA transporter ine-like | - | - | 2.61 ** | |
| LOC111708680 synaptotagmin 1-like | - | - | 1.33 ** | |
| LOC111697845 synaptotagmin 1-like | - | - | 1.75 ** | |
| LOC111703377 synaptotagmin-1-like | - | - | 2.86 ** | |
| LOC111715709 glutamate [NMDA] receptor subunit 1-like | - | - | 1.41 ** | |
| LOC111700830 glutamate receptor ionotropic, kainate 1-like | - | - | 1.28 ** | |
| LOC111715864 glutamate receptor ionotropic, NMDA 2B-like | - | - | 2.48 ** | |
| LOC111712426 longitudinals lacking protein-like | - | - | −1.32 * | |
| Visual perception | LOC111695981 chaoptin-like | - | - | −3.02 * |
| LOC111698143 homeobox protein otx5-A-like | - | - | −1.46 * | |
| LOC111708628 irregular chiasm C-roughest protein-like | - | - | 1.93 ** | |
| LOC111699563 irregular chiasm C-roughest protein-like | - | - | 1.37 ** | |
| Defence response-Insecticide resistance | LOC111716773 phenoloxidase 2-like | - | - | −8.32 * |
| LOC111705105 probable cytochrome P450 6a13 | - | - | 1.38 ** | |
| LOC111715882 esterase FE4-like | - | - | −2.46 * | |
| LOC111698650 ferrochelatase, mitochondrial-like | - | - | −1.56 * | |
| LOC111698532 glutathione S-transferase 1-like | - | - | 1.60 ** | |
| LOC111714366 glutathione S-transferase-like | - | - | 1.76 ** | |
| DNA methylation | LOC111700762 DNA (cytosine-5)-methyltransferase 1-like | - | - | −1.72 * |
| LOC111703921 DNA (cytosine-5)-methyltransferase 1-like | - | - | −1.52 * | |
| LOC111713817 DNA N6-methyl adenine demethylase-like | - | - | −1.43 * | |
| Reproduction | LOC111695973 neprilysin-1-like | - | - | 1.26 ** |
| LOC111714647 neprilysin-2-like | - | - | 1.18 ** | |
| LOC111700675 protein white-like | - | - | 1.63 ** | |
| LOC111704362 vitellogenin-like | - | - | −7.78 * | |
| LOC111702462 ATP-dependent RNA helicase vasa-like | - | - | −3.02 * | |
| LOC111697048 gametogenetin-binding protein 2-like | - | - | −1.77 * | |
| LOC111705270 maternal protein exuperantia-1-like | - | - | −2.15 * | |
| LOC111716440 zonadhesin-like | - | - | −1.58 * | |
| DNA repair | LOC111707470 cell cycle checkpoint protein RAD17-like | - | - | −1.33 * |
| LOC111710432 DNA mismatch repair protein Msh6-like | - | - | −1.32 * | |
| LOC111701439 probable DNA double-strand break repair Rad50 ATPase | - | - | −1.37 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arcanjo, C.; Trémolet, G.; Giusti-Petrucciani, N.; Duflot, A.; Forget-Leray, J.; Boulangé-Lecomte, C. Susceptibility of the Non-Targeted Crustacean Eurytemora affinis to the Endocrine Disruptor Tebufenozide: A Transcriptomic Approach. Genes 2021, 12, 1484. https://doi.org/10.3390/genes12101484
Arcanjo C, Trémolet G, Giusti-Petrucciani N, Duflot A, Forget-Leray J, Boulangé-Lecomte C. Susceptibility of the Non-Targeted Crustacean Eurytemora affinis to the Endocrine Disruptor Tebufenozide: A Transcriptomic Approach. Genes. 2021; 12(10):1484. https://doi.org/10.3390/genes12101484
Chicago/Turabian StyleArcanjo, Caroline, Gauthier Trémolet, Nathalie Giusti-Petrucciani, Aurélie Duflot, Joëlle Forget-Leray, and Céline Boulangé-Lecomte. 2021. "Susceptibility of the Non-Targeted Crustacean Eurytemora affinis to the Endocrine Disruptor Tebufenozide: A Transcriptomic Approach" Genes 12, no. 10: 1484. https://doi.org/10.3390/genes12101484
APA StyleArcanjo, C., Trémolet, G., Giusti-Petrucciani, N., Duflot, A., Forget-Leray, J., & Boulangé-Lecomte, C. (2021). Susceptibility of the Non-Targeted Crustacean Eurytemora affinis to the Endocrine Disruptor Tebufenozide: A Transcriptomic Approach. Genes, 12(10), 1484. https://doi.org/10.3390/genes12101484