Trends in the Application of “Omics” to Ecotoxicology and Stress Ecology
Abstract
1. Introduction
2. Materials and Methods
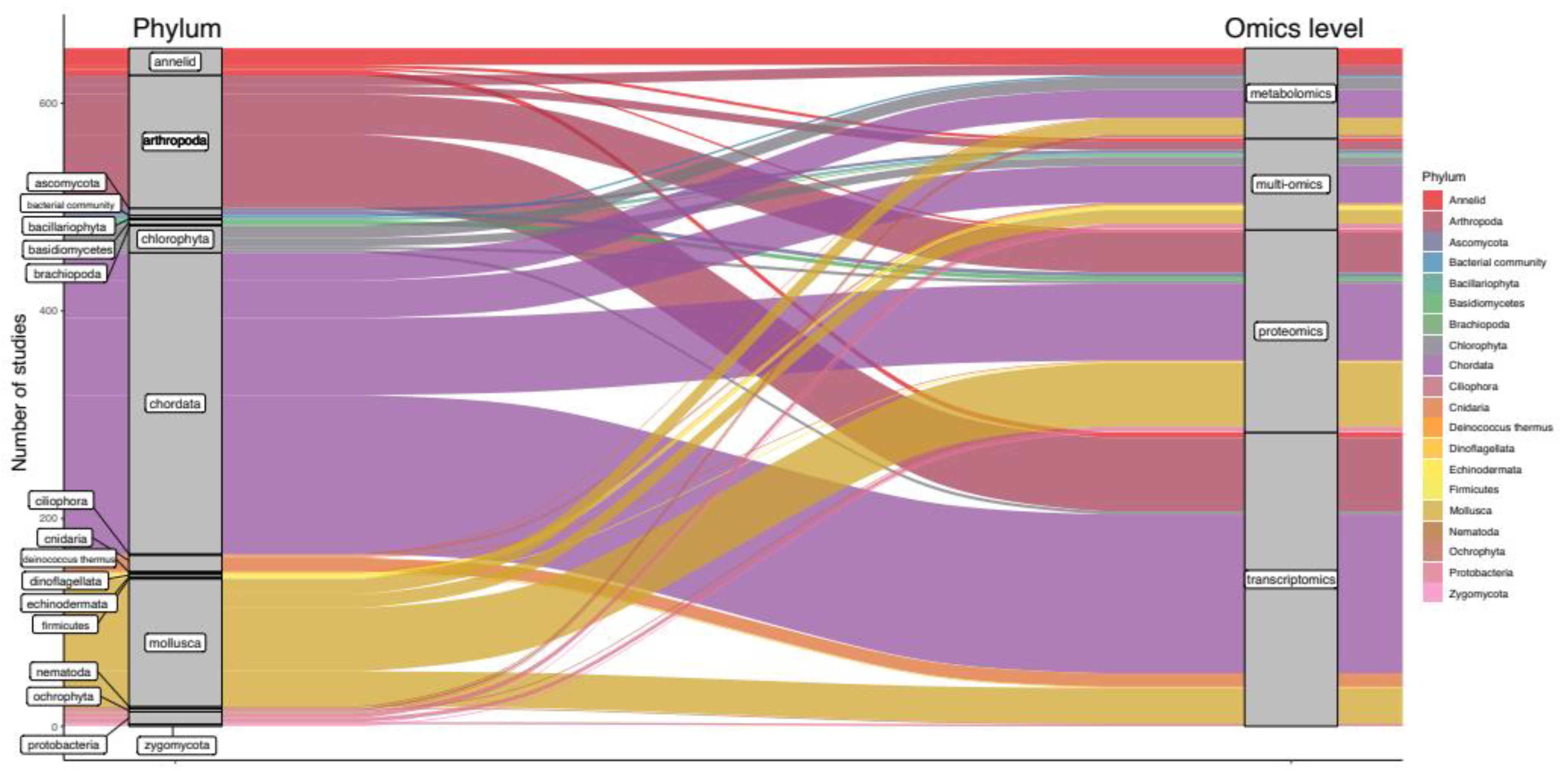
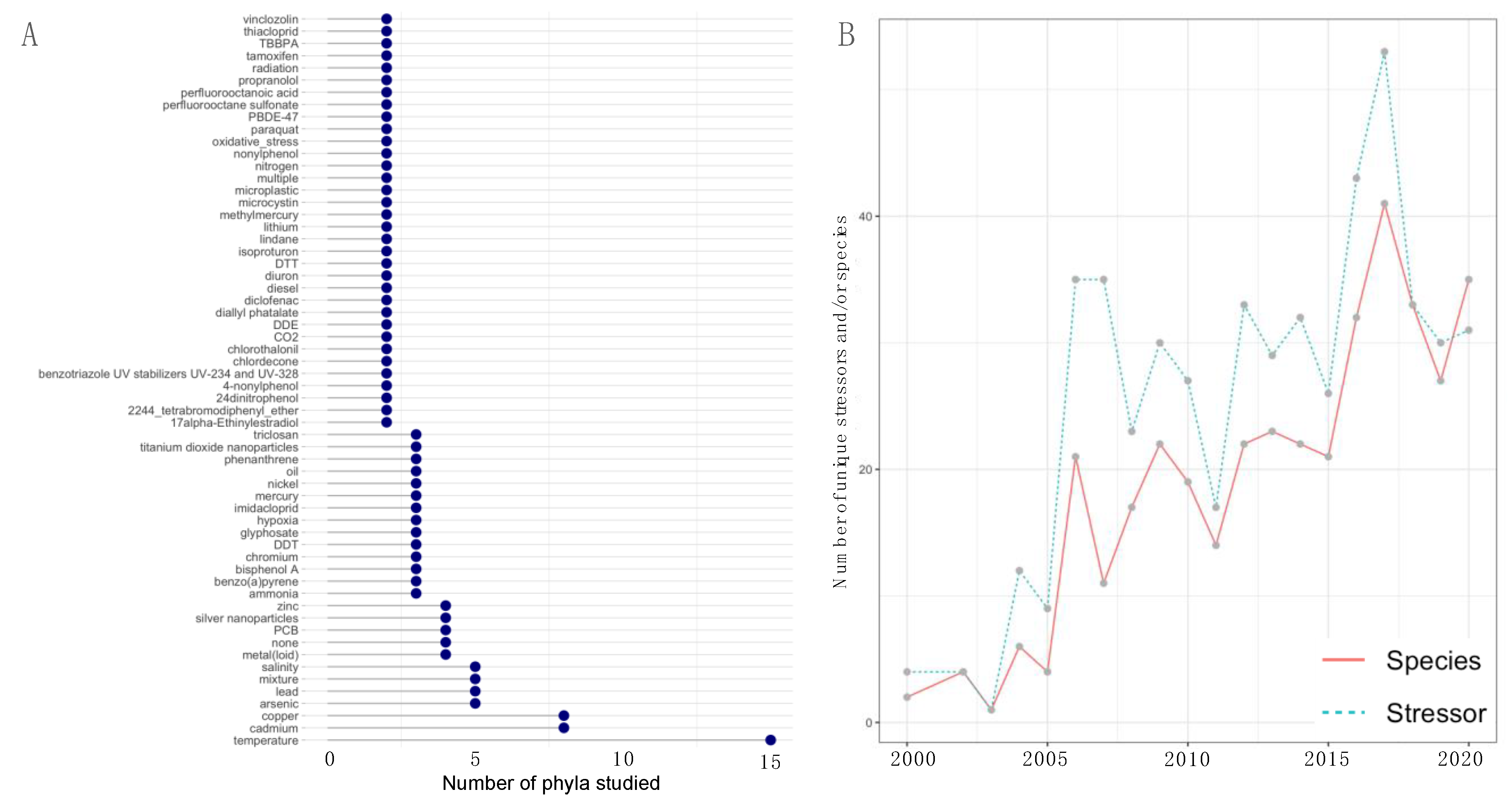
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Walker, C.H.; Sibly, R.M.; Hopkin, S.P.; Peakall, D.B. Principles of Ecotoxicology, 4th ed.; CRC Press: Boca Raton, FL, USA, 2012; ISBN 978-1-4398-6266-7. [Google Scholar]
- Van Straalen, N.M. Ecotoxicology Becomes Stress Ecology. Environ. Sci. Technol. 2003, 37, 324A–330A. [Google Scholar] [CrossRef] [PubMed]
- Heugens, E.H.; Hendriks, A.J.; Dekker, T.; van Straalen, N.M.; Admiraal, W. A Review of the Effects of Multiple Stressors on Aquatic Organisms and Analysis of Uncertainty Factors for Use in Risk Assessment. Crit. Rev. Toxicol. 2001, 31, 247–284. [Google Scholar] [CrossRef]
- Hayward, S.A. Application of Functional ‘Omics’ in Environmental Stress Physiology: Insights, Limitations, and Future Challenges. Curr. Opin. Insect Sci. 2014, 4, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Bludau, I.; Aebersold, R. Proteomic and Interactomic Insights into the Molecular Basis of Cell Functional Diversity. Nat. Rev. Mol. Cell Biol. 2020, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Barabási, A.-L.; Oltvai, Z.N. Network Biology: Understanding the Cell’s Functional Organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef]
- Bruggeman, F.J.; Westerhoff, H.V. The Nature of Systems Biology. Trends Microbiol. 2007, 15, 45–50. [Google Scholar] [CrossRef]
- Côté, I.M.; Darling, E.S.; Brown, C.J. Interactions among Ecosystem Stressors and Their Importance in Conservation. Proc. R. Soc. B Biol. Sci. 2016, 283, 20152592. [Google Scholar] [CrossRef]
- Sturla, S.J.; Boobis, A.R.; FitzGerald, R.E.; Hoeng, J.; Kavlock, R.J.; Schirmer, K.; Whelan, M.; Wilks, M.F.; Peitsch, M.C. Systems Toxicology: From Basic Research to Risk Assessment. Chem. Res. Toxicol. 2014, 27, 314–329. [Google Scholar] [CrossRef]
- Ankley, G.T.; Daston, G.P.; Degitz, S.J.; Denslow, N.D.; Hoke, R.A.; Kennedy, S.W.; Miracle, A.L.; Perkins, E.J.; Snape, J.; Tillitt, D.E.; et al. Toxicogenomics in Regulatory Ecotoxicology. Environ. Sci. Technol. 2006, 40, 4055–4065. [Google Scholar] [CrossRef]
- Leung, K.M. Joining the Dots between Omics and Environmental Management. Integr. Environ. Assess. Manag. 2018, 14, 169–173. [Google Scholar] [CrossRef]
- Kleensang, A.; Maertens, A.; Rosenberg, M.; Fitzpatrick, S.; Lamb, J.; Auerbach, S.; Brennan, R.; Crofton, K.M.; Gordon, B.; Fornace, A.J.; et al. Pathways of Toxicity. ALTEX 2014, 31, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Brockmeier, E.K.; Hodges, G.; Hutchinson, T.H.; Butler, E.; Hecker, M.; Tollefsen, K.E.; Garcia-Reyero, N.; Kille, P.; Becker, D.; Chipman, K.; et al. The Role of Omics in the Application of Adverse Outcome Pathways for Chemical Risk Assessment. Toxicol. Sci. 2017, 158, 252–262. [Google Scholar] [CrossRef]
- Sauer, U.G.; Deferme, L.; Gribaldo, L.; Hackermüller, J.; Tralau, T.; van Ravenzwaay, B.; Yauk, C.; Poole, A.; Tong, W.; Gant, T.W. The Challenge of the Application of ’omics Technologies in Chemicals Risk Assessment: Background and Outlook. Regul. Toxicol. Pharmacol. 2017, 91, S14–S26. [Google Scholar] [CrossRef]
- Feswick, A.; Munkittrick, K.R.; Martyniuk, C.J. Estrogen-Responsive Gene Networks in the Teleost Liver: What Are the Key Molecular Indicators? Environ. Toxicol. Pharmacol. 2017, 56, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Fent, K.; Sumpter, J.P. Progress and Promises in Toxicogenomics in Aquatic Toxicology: Is Technical Innovation Driving Scientific Innovation? Aquat. Toxicol. 2011, 105, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Ankley, G.T.; Bencic, D.C.; Breen, M.S.; Collette, T.W.; Conolly, R.B.; Denslow, N.D.; Edwards, S.W.; Ekman, D.R.; Garcia-Reyero, N.; Jensen, K.M.; et al. Endocrine Disrupting Chemicals in Fish: Developing Exposure Indicators and Predictive Models of Effects Based on Mechanism of Action. Aquat. Toxicol. 2009, 92, 168–178. [Google Scholar] [CrossRef]
- Artigas, J.; Arts, G.; Babut, M.; Caracciolo, A.B.; Charles, S.; Chaumot, A.; Combourieu, B.; Dahllöf, I.; Despréaux, D.; Ferrari, B.; et al. Towards a Renewed Research Agenda in Ecotoxicology. Environ. Pollut. 2012, 160, 201–206. [Google Scholar] [CrossRef]
- Mora, C.; Tittensor, D.P.; Adl, S.; Simpson, A.G.B.; Worm, B. How Many Species Are There on Earth and in the Ocean? PLOS Biology 2011, 9, e1001127. [Google Scholar] [CrossRef]
- Siepel, A.; Bejerano, G.; Pedersen, J.S.; Hinrichs, A.S.; Hou, M.; Rosenbloom, K.; Clawson, H.; Spieth, J.; Hillier, L.W.; Richards, S.; et al. Evolutionarily Conserved Elements in Vertebrate, Insect, Worm, and Yeast Genomes. Genome. Res. 2005, 15, 1034–1050. [Google Scholar] [CrossRef]
- Luo, H.; Gao, F.; Lin, Y. Evolutionary Conservation Analysis between the Essential and Nonessential Genes in Bacterial Genomes. Sci. Rep. 2015, 5, 13210. [Google Scholar] [CrossRef]
- Kunin, V.; Ahren, D.; Goldovsky, L.; Janssen, P.; Ouzounis, C.A. Measuring Genome Conservation across Taxa: Divided Strains and United Kingdoms. Nucleic Acids Res. 2005, 33, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Genomic Approaches for Cross-Species Extrapolation in Toxicology; Benson, W.H., Giulio, R.T.D., Eds.; CRC Press: Boca Raton, FL, USA, 2006; ISBN 978-0-429-14213-0. [Google Scholar]
- Waters, M.D.; Fostel, J.M. Toxicogenomics and Systems Toxicology: Aims and Prospects. Nat. Rev. Genet. 2004, 5, 936–948. [Google Scholar] [CrossRef]
- Martyniuk, C.J. Are We Closer to the Vision? A Proposed Framework for Incorporating Omics into Environmental Assessments. Environ. Toxicol. Pharmacol. 2018, 59, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, S.P.; Lüneburg, J.; Gottschlich, L.; Tams, V.; Cordellier, M. Daphnia s Tressor Database: Taking Advantage of a Decade of Daphnia ‘-Omics’ Data for Gene Annotation. Sci. Rep. 2019, 9, 11135. [Google Scholar] [CrossRef]
- Vandegehuchte, M.B.; Janssen, C.R. Epigenetics in an Ecotoxicological Context. Mutat. Res. Genet. Toxicol. Environ. Mutagenesis 2014, 764–765, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Rey, O.; Eizaguirre, C.; Angers, B.; Baltazar-Soares, M.; Sagonas, K.; Prunier, J.G.; Blanchet, S. Linking Epigenetics and Biological Conservation: Towards a Conservation Epigenetics Perspective. Funct. Ecol. 2020, 34, 414–427. [Google Scholar] [CrossRef]
- Han, X.; Huang, Q. Environmental Pollutants Exposure and Male Reproductive Toxicity: The Role of Epigenetic Modifications. Toxicology 2021, 456, 152780. [Google Scholar] [CrossRef] [PubMed]
- Vandegehuchte, M.B.; Janssen, C.R. Epigenetics and Its Implications for Ecotoxicology. Ecotoxicology 2011, 20, 607–624. [Google Scholar] [CrossRef] [PubMed]
- Šrut, M. Ecotoxicological Epigenetics in Invertebrates: Emerging Tool for the Evaluation of Present and Past Pollution Burden. Chemosphere 2021, 282, 131026. [Google Scholar] [CrossRef]
- Chatterjee, N.; Gim, J.; Choi, J. Epigenetic Profiling to Environmental Stressors in Model and Non-Model Organisms: Ecotoxicology Perspective. Environ. Health Toxicol. 2018, 33, e2018015. [Google Scholar] [CrossRef]
- Jamla, M.; Khare, T.; Joshi, S.; Patil, S.; Penna, S.; Kumar, V. Omics Approaches for Understanding Heavy Metal Responses and Tolerance in Plants. Curr. Plant Biol. 2021, 27, 100213. [Google Scholar] [CrossRef]
- Bickham, J.W. The Four Cornerstones of Evolutionary Toxicology. Ecotoxicology 2011, 20, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Oziolor, E.M.; Matson, C.W. Evolutionary Toxicology: Population Adaptation in Response to Anthropogenic Pollution. In Extremophile Fishes: Ecology, Evolution, and Physiology of Teleosts in Extreme Environments; Riesch, R., Tobler, M., Plath, M., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 247–277. ISBN 978-3-319-13362-1. [Google Scholar]
- Oziolor, E.M.; Bickham, J.W.; Matson, C.W. Evolutionary Toxicology in an Omics World. Evol. Appl. 2017, 10, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Oziolor, E.O.; De Schamphelaere, K.; Matson, C.W. Evolutionary Toxicology: Meta-Analysis of Evolutionary Events in Response to Chemical Stressors. Ecotoxicology 2016, 25, 1858–1866. [Google Scholar] [CrossRef] [PubMed]
- Oziolor, E.M.; DeSchamphelaere, K.; Lyon, D.; Nacci, D.; Poynton, H. Evolutionary Toxicology-An Informational Tool for Chemical Regulation? Environ. Toxicol. Chem. 2020, 39, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.P.; Monosson, E.; Matson, C.W.; Bickham, J.W. Evolutionary Toxicology: Toward a Unified Understanding of Life’s Response to Toxic Chemicals. Evol. Appl. 2017, 10, 745–751. [Google Scholar] [CrossRef]
- Liang, X.; Martyniuk, C.J.; Simmons, D.B.D. Are We Forgetting the “Proteomics” in Multi-Omics Ecotoxicology? Comp. Biochem. Physiol. D-Genom. Proteom. 2020, 36, 100751. [Google Scholar] [CrossRef]
- Ablain, J.; Zon, L.I. Of Fish and Men: Using Zebrafish to Fight Human Diseases. Trends Cell Biol. 2013, 23, 584–586. [Google Scholar] [CrossRef]
- Harris, K.D.M.; Bartlett, N.J.; Lloyd, V.K. Daphnia as an Emerging Epigenetic Model Organism. Genet. Res. Int. 2012, 2012, 147892. [Google Scholar] [CrossRef]
- Seda, J.; Petrusek, A. Daphnia as a Model Organism in Limnology and Aquatic Biology: Introductory Remarks. J. Limnol. 2011, 70, 337–344. [Google Scholar] [CrossRef]
- Caputo, D.R.; Robson, S.C.; Werner, I.; Ford, A.T. Complete Transcriptome Assembly and Annotation of a Critically Important Amphipod Species in Freshwater Ecotoxicological Risk Assessment: Gammarus Fossarum. Environ. Int. 2020, 137, 105319. [Google Scholar] [CrossRef] [PubMed]
- Cogne, Y.; Degli-Esposti, D.; Pible, O.; Gouveia, D.; François, A.; Bouchez, O.; Eché, C.; Ford, A.; Geffard, O.; Armengaud, J.; et al. De Novo Transcriptomes of 14 Gammarid Individuals for Proteogenomic Analysis of Seven Taxonomic Groups. Sci. Data 2019, 6, 184. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-Y.; Choi, B.-S.; Kim, M.-S.; Park, J.C.; Jeong, C.-B.; Han, J.; Lee, J.-S. The Genome of the Freshwater Water Flea Daphnia Magna: A Potential Use for Freshwater Molecular Ecotoxicology. Aquat. Toxicol. 2019, 210, 69–84. [Google Scholar] [CrossRef]
- Colbourne, J.K.; Pfrender, M.E.; Gilbert, D.; Thomas, W.K.; Tucker, A.; Oakley, T.H.; Tokishita, S.; Aerts, A.; Arnold, G.J.; Basu, M.K.; et al. The Ecoresponsive Genome of Daphnia Pulex. Science 2011, 331, 555–561. [Google Scholar] [CrossRef]
- May, P.; Wienkoop, S.; Kempa, S.; Usadel, B.; Christian, N.; Rupprecht, J.; Weiss, J.; Recuenco-Munoz, L.; Ebenhöh, O.; Weckwerth, W.; et al. Metabolomics- and Proteomics-Assisted Genome Annotation and Analysis of the Draft Metabolic Network of Chlamydomonas Reinhardtii. Genetics 2008, 179, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Shrager, J.; Hauser, C.; Chang, C.-W.; Harris, E.H.; Davies, J.; McDermott, J.; Tamse, R.; Zhang, Z.; Grossman, A.R. Chlamydomonas Reinhardtii Genome Project. A Guide to the Generation and Use of the CDNA Information. Plant Physiol. 2003, 131, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Dziedzic, K.E.; Elder, H.; Tavalire, H.; Meyer, E. Heritable Variation in Bleaching Responses and Its Functional Genomic Basis in Reef-Building Corals (Orbicella Faveolata). Mol. Ecol. 2019, 28, 2238–2253. [Google Scholar] [CrossRef]
- Snelling, J.; Dziedzic, K.; Guermond, S.; Meyer, E. Development of an Integrated Genomic Map for a Threatened Caribbean Coral (Orbicella Faveolata). bioRxiv 2017, 183467. [Google Scholar] [CrossRef]
- Anderson, D.A.; Walz, M.E.; Weil, E.; Tonellato, P.; Smith, M.C. RNA-Seq of the Caribbean Reef-Building Coral Orbicella Faveolata (Scleractinia-Merulinidae) under Bleaching and Disease Stress Expands Models of Coral Innate Immunity. PeerJ 2016, 4, e1616. [Google Scholar] [CrossRef]
- Brandl, J.; Andersen, M.R. Aspergilli: Models for Systems Biology in Filamentous Fungi. Curr. Opin. Syst. Biol. 2017, 6, 67–73. [Google Scholar] [CrossRef]
- Boontawon, T.; Nakazawa, T.; Inoue, C.; Osakabe, K.; Kawauchi, M.; Sakamoto, M.; Honda, Y. Efficient Genome Editing with CRISPR/Cas9 in Pleurotus Ostreatus. AMB Express 2021, 11, 30. [Google Scholar] [CrossRef] [PubMed]
- Canzler, S.; Schor, J.; Busch, W.; Schubert, K.; Rolle-Kampczyk, U.E.; Seitz, H.; Kamp, H.; von Bergen, M.; Buesen, R.; Hackermüller, J. Prospects and Challenges of Multi-Omics Data Integration in Toxicology. Arch. Toxicol. 2020, 94, 371–388. [Google Scholar] [CrossRef]
- Schwartz, T.S. The Promises and the Challenges of Integrating Multi-Omics and Systems Biology in Comparative Stress Biology. Integr. Comp. Biol. 2020, 60, 89–97. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, S.T.; Dzurisin, J.D.K.; Williams, C.M.; Lobo, N.F.; Higgins, J.K.; Deines, J.M.; Carmichael, R.D.; Zeng, E.; Tan, J.C.; Wu, G.C.; et al. Gene Expression in Closely Related Species Mirrors Local Adaptation: Consequences for Responses to a Warming World. Mol. Ecol. 2014, 23, 2686–2698. [Google Scholar] [CrossRef] [PubMed]
- Ebner, J.N.; Ritz, D.; von Fumetti, S. Abiotic and Past Climatic Conditions Drive Protein Abundance Variation among Natural Populations of the Caddisfly Crunoecia Irrorata. Sci. Rep. 2020, 10, 15538. [Google Scholar] [CrossRef] [PubMed]
- Simmons, D.B.D.; Benskin, J.P.; Cosgrove, J.R.; Duncker, B.P.; Ekman, D.R.; Martyniuk, C.J.; Sherry, J.P. Omics for Aquatic Ecotoxicology: Control of Extraneous Variability to Enhance the Analysis of Environmental Effects. Environ. Toxicol. Chem. 2015, 34, 1693–1704. [Google Scholar] [CrossRef]
- Feder, M.E.; Walser, J.C. The Biological Limitations of Transcriptomics in Elucidating Stress and Stress Responses. J. Evol. Biol. 2005, 18, 901–910. [Google Scholar]
- Madeira, C.; Costa, P.M. Proteomics in systems toxicology. In Advances in Protein Chemistry and Structural Biology; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar]
- Liang, X.; Martyniuk, C.J.; Simmons, D.B.D. Current Topics in Omics, Ecotoxicology, and Environmental Science. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 38, 100782. [Google Scholar] [CrossRef]
- Hall, D.R.; Peng, H. Characterizing Physical Protein Targets of Chemical Contaminants with Chemical Proteomics: Is It Time to Fill a Crucial Environmental Toxicology Knowledge Gap? Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 34, 100655. [Google Scholar] [CrossRef]
- Evans, T.G. Considerations for the Use of Transcriptomics in Identifying the ‘Genes That Matter’ for Environmental Adaptation. J. Exp. Biol. 2015, 218, 1925–1935. [Google Scholar] [CrossRef]
- Garcia-Reyero, N.; Habib, T.; Pirooznia, M.; Gust, K.A.; Gong, P.; Warner, C.; Wilbanks, M.; Perkins, E. Conserved Toxic Responses across Divergent Phylogenetic Lineages: A Meta-Analysis of the Neurotoxic Effects of RDX among Multiple Species Using Toxicogenomics. Ecotoxicology 2011, 20, 580. [Google Scholar] [CrossRef]
- Lu, Y.; Huggins, P.; Bar-Joseph, Z. Cross Species Analysis of Microarray Expression Data. Bioinformatics 2009, 25, 1476–1483. [Google Scholar] [CrossRef] [PubMed]
- Sharan, R.; Suthram, S.; Kelley, R.M.; Kuhn, T.; McCuine, S.; Uetz, P.; Sittler, T.; Karp, R.M.; Ideker, T. Conserved Patterns of Protein Interaction in Multiple Species. Proc. Natl. Acad. Sci. USA 2005, 102, 1974–1979. [Google Scholar] [CrossRef]
- Hoeng, J.; Talikka, M.; Martin, F.; Sewer, A.; Yang, X.; Iskandar, A.; Schlage, W.K.; Peitsch, M.C. Case Study: The Role of Mechanistic Network Models in Systems Toxicology. Drug Discov. Today 2014, 19, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Rugard, M.; Coumoul, X.; Carvaillo, J.-C.; Barouki, R.; Audouze, K. Deciphering Adverse Outcome Pathway Network Linked to Bisphenol F Using Text Mining and Systems Toxicology Approaches. Toxicol. Sci. 2020, 173, 32–40. [Google Scholar] [CrossRef]
- Hartung, T.; FitzGerald, R.E.; Jennings, P.; Mirams, G.R.; Peitsch, M.C.; Rostami-Hodjegan, A.; Shah, I.; Wilks, M.F.; Sturla, S.J. Systems Toxicology: Real World Applications and Opportunities. Chem. Res. Toxicol. 2017, 30, 870–882. [Google Scholar] [CrossRef] [PubMed]
- Ives, C.; Campia, I.; Wang, R.-L.; Wittwehr, C.; Edwards, S. Creating a Structured Adverse Outcome Pathway Knowledgebase via Ontology-Based Annotations. Appl. Vitr. Toxicol. 2017, 3, 298–311. [Google Scholar] [CrossRef]
- Shi, J.; Walker, M.G. Gene Set Enrichment Analysis (GSEA) for Interpreting Gene Expression Profiles. Curr. Bioinform. 2007, 2, 133–137. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Narad, P.; Chaurasia, A.; Wadhwab, G.; Upadhyayaa, K.C. Net2Align: An Algorithm For Pairwise Global Alignment of Biological Networks. Bioinformation 2016, 12, 408–411. [Google Scholar] [CrossRef][Green Version]
- Koutrouli, M.; Karatzas, E.; Paez-Espino, D.; Pavlopoulos, G.A. A Guide to Conquer the Biological Network Era Using Graph Theory. Front. Bioeng. Biotechnol. 2020, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.-L.; Edwards, S.; Ives, C. Ontology-Based Semantic Mapping of Chemical Toxicities. Toxicology 2019, 412, 89–100. [Google Scholar] [CrossRef]
- Wang, R.-L. Semantic Characterization of Adverse Outcome Pathways. Aquat. Toxicol. 2020, 222, 105478. [Google Scholar] [CrossRef] [PubMed]
- Kelder, T.; van Iersel, M.P.; Hanspers, K.; Kutmon, M.; Conklin, B.R.; Evelo, C.T.; Pico, A.R. WikiPathways: Building Research Communities on Biological Pathways. Nucleic Acids Res. 2012, 40, D1301–1307. [Google Scholar] [CrossRef]
- Ruepp, A.; Zollner, A.; Maier, D.; Albermann, K.; Hani, J.; Mokrejs, M.; Tetko, I.; Güldener, U.; Mannhaupt, G.; Münsterkötter, M.; et al. The FunCat, a Functional Annotation Scheme for Systematic Classification of Proteins from Whole Genomes. Nucleic Acids Res. 2004, 32, 5539–5545. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam Protein Families Database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Finn, R.D.; Mistry, J.; Tate, J.; Coggill, P.; Heger, A.; Pollington, J.E.; Gavin, O.L.; Gunasekaran, P.; Ceric, G.; Forslund, K.; et al. The Pfam Protein Families Database. Nucleic Acids Res. 2010, 38, D211–222. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kitano, H. Systems Biology: A Brief Overview. Science 2002, 295, 1662–1664. [Google Scholar] [CrossRef] [PubMed]
- Chuang, H.-Y.; Hofree, M.; Ideker, T. A Decade of Systems Biology. Annu. Rev. Cell Dev. Biol. 2010, 26, 721–744. [Google Scholar] [CrossRef] [PubMed]
- Ideker, T.; Galitski, T.; Hood, L. A New Approach to Decoding Life: Systems Biology. Annu. Rev. Genom. Hum. Genet. 2001, 2, 343–372. [Google Scholar] [CrossRef]
- Wolkenhauer, O.; Mesarović, M. Feedback Dynamics and Cell Function: Why Systems Biology Is Called Systems Biology. Mol. BioSystems 2005, 1, 14–16. [Google Scholar] [CrossRef] [PubMed]
- Benfey, P.N.; Mitchell-Olds, T. From Genotype to Phenotype: Systems Biology Meets Natural Variation. Science 2008, 320, 495–497. [Google Scholar] [CrossRef] [PubMed]
- Pain, G.; Hickey, G.; Mondou, M.; Crump, D.; Hecker, M.; Basu, N.; Maguire, S. Drivers of and Obstacles to the Adoption of Toxicogenomics for Chemical Risk Assessment: Insights from Social Science Perspectives. Environ. Health Perspect. 2020, 128, 105002. [Google Scholar] [CrossRef]
- Buesen, R.; Chorley, B.N.; da Silva Lima, B.; Daston, G.; Deferme, L.; Ebbels, T.; Gant, T.W.; Goetz, A.; Greally, J.; Gribaldo, L.; et al. Applying ’omics Technologies in Chemicals Risk Assessment: Report of an ECETOC Workshop. Regul. Toxicol. Pharmacol. 2017, 91, S3–S13. [Google Scholar] [CrossRef]
- Hendrickx, D.M.; Aerts, H.J.W.L.; Caiment, F.; Clark, D.; Ebbels, T.M.D.; Evelo, C.T.; Gmuender, H.; Hebels, D.G.A.J.; Herwig, R.; Hescheler, J.; et al. DiXa: A Data Infrastructure for Chemical Safety Assessment. Bioinformatics 2015, 31, 1505–1507. [Google Scholar] [CrossRef] [PubMed]
- Canzler, S.; Hackermüller, J.; Schor, J. MOD-Finder: Identify Multi-Omics Data Sets Related to Defined Chemical Exposure. arXiv 2019, arXiv:1907.06346. [Google Scholar]
- Darde, T.A.; Gaudriault, P.; Beranger, R.; Lancien, C.; Caillarec-Joly, A.; Sallou, O.; Bonvallot, N.; Chevrier, C.; Mazaud-Guittot, S.; Jégou, B.; et al. TOXsIgN: A Cross-Species Repository for Toxicogenomic Signatures. Bioinformatics 2018, 34, 2116–2122. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.; Stasiewicz, S.; Merrick, B.A.; Tomer, K.; Bushel, P.; Paules, R.; Stegman, N.; Nehls, G.; Yost, K.J.; Johnson, C.H.; et al. CEBS--Chemical Effects in Biological Systems: A Public Data Repository Integrating Study Design and Toxicity Data with Microarray and Proteomics Data. Nucleic Acids Res. 2008, 36, D892–900. [Google Scholar] [CrossRef]
- Lea, I.A.; Gong, H.; Paleja, A.; Rashid, A.; Fostel, J. CEBS: A Comprehensive Annotated Database of Toxicological Data. Nucleic Acids Res. 2017, 45, D964–D971. [Google Scholar] [CrossRef]
- Davis, A.P.; Murphy, C.G.; Johnson, R.; Lay, J.M.; Lennon-Hopkins, K.; Saraceni-Richards, C.; Sciaky, D.; King, B.L.; Rosenstein, M.C.; Wiegers, T.C.; et al. The Comparative Toxicogenomics Database: Update 2013. Nucleic Acids Res. 2013, 41, D1104–D1114. [Google Scholar] [CrossRef]
- Fan, G.; Song, Y.; Yang, L.; Huang, X.; Zhang, S.; Zhang, M.; Yang, X.; Chang, Y.; Zhang, H.; Li, Y.; et al. Initial Data Release and Announcement of the 10,000 Fish Genomes Project (Fish10K). GigaScience 2020, 9. [Google Scholar] [CrossRef]
- i5K Consortium The I5K Initiative: Advancing Arthropod Genomics for Knowledge, Human Health, Agriculture, and the Environment. J. Hered. 2013, 104, 595–600. [CrossRef] [PubMed]
- Lewin, H.A.; Robinson, G.E.; Kress, W.J.; Baker, W.J.; Coddington, J.; Crandall, K.A.; Durbin, R.; Edwards, S.V.; Forest, F.; Gilbert, M.T.P.; et al. Earth BioGenome Project: Sequencing Life for the Future of Life. Proc. Natl. Acad. Sci. USA 2018, 115, 4325–4333. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.E.; Sadler, K.C. Casting a Wide Net: Use of Diverse Model Organisms to Advance Toxicology. Dis. Models Mech. 2020, 13. [Google Scholar] [CrossRef]
- Robinson, G.E.; Hackett, K.J.; Purcell-Miramontes, M.; Brown, S.J.; Evans, J.D.; Goldsmith, M.R.; Lawson, D.; Okamuro, J.; Robertson, H.M.; Schneider, D.J. Creating a Buzz About Insect Genomes. Science 2011, 331, 1386. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.D.; Turan, N.; Diab, A.M.; Wu, H.; Mackenzie, C.; Bartie, K.L.; Hrydziuszko, O.; Lyons, B.P.; Stentiford, G.D.; Herbert, J.M.; et al. Towards a System Level Understanding of Non-Model Organisms Sampled from the Environment: A Network Biology Approach. PLOS Comput. Biol. 2011, 7, e1002126. [Google Scholar] [CrossRef] [PubMed]
- Ekblom, R.; Galindo, J. Applications of next Generation Sequencing in Molecular Ecology of Non-Model Organisms. Heredity 2011, 107, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Armengaud, J.; Trapp, J.; Pible, O.; Geffard, O.; Chaumot, A.; Hartmann, E.M. Non-Model Organisms, a Species Endangered by Proteogenomics. J. Proteom. 2014, 105, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Forbes, V.E.; Palmqvist, A.; Bach, L. The Use and Misuse of Biomarkers in Ecotoxicology. Environ. Toxicol. Chem. 2006, 25, 272–280. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ebner, J.N. Trends in the Application of “Omics” to Ecotoxicology and Stress Ecology. Genes 2021, 12, 1481. https://doi.org/10.3390/genes12101481
Ebner JN. Trends in the Application of “Omics” to Ecotoxicology and Stress Ecology. Genes. 2021; 12(10):1481. https://doi.org/10.3390/genes12101481
Chicago/Turabian StyleEbner, Joshua Niklas. 2021. "Trends in the Application of “Omics” to Ecotoxicology and Stress Ecology" Genes 12, no. 10: 1481. https://doi.org/10.3390/genes12101481
APA StyleEbner, J. N. (2021). Trends in the Application of “Omics” to Ecotoxicology and Stress Ecology. Genes, 12(10), 1481. https://doi.org/10.3390/genes12101481





