Structural Variability, Expression Profile, and Pharmacogenetic Properties of TMPRSS2 Gene as a Potential Target for COVID-19 Therapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Structural Variability Data
2.2. Bioinformatics Analysis of Gene Expression, Mirna Interactions, and Pharmacogenomics
3. Results and Discussion
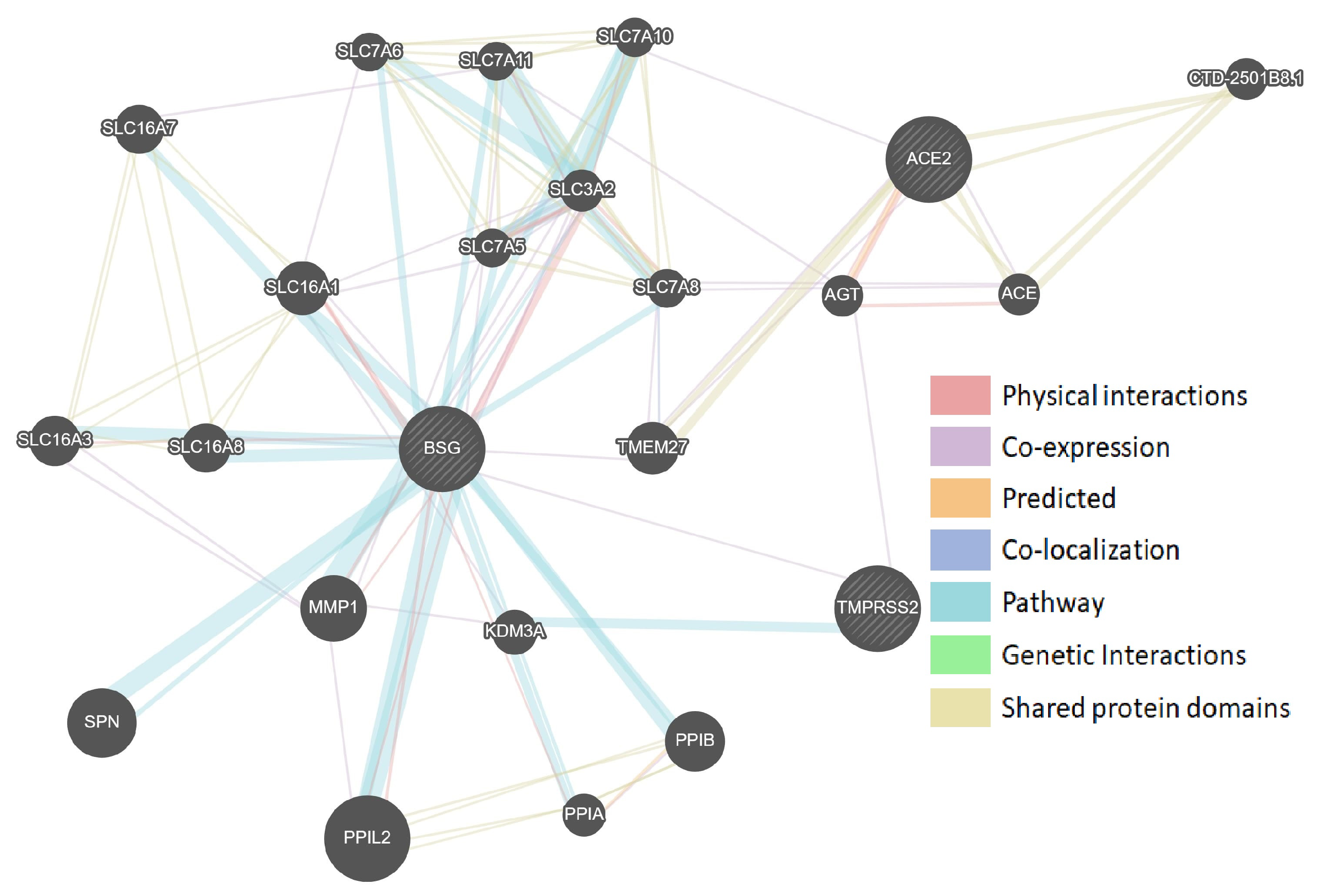
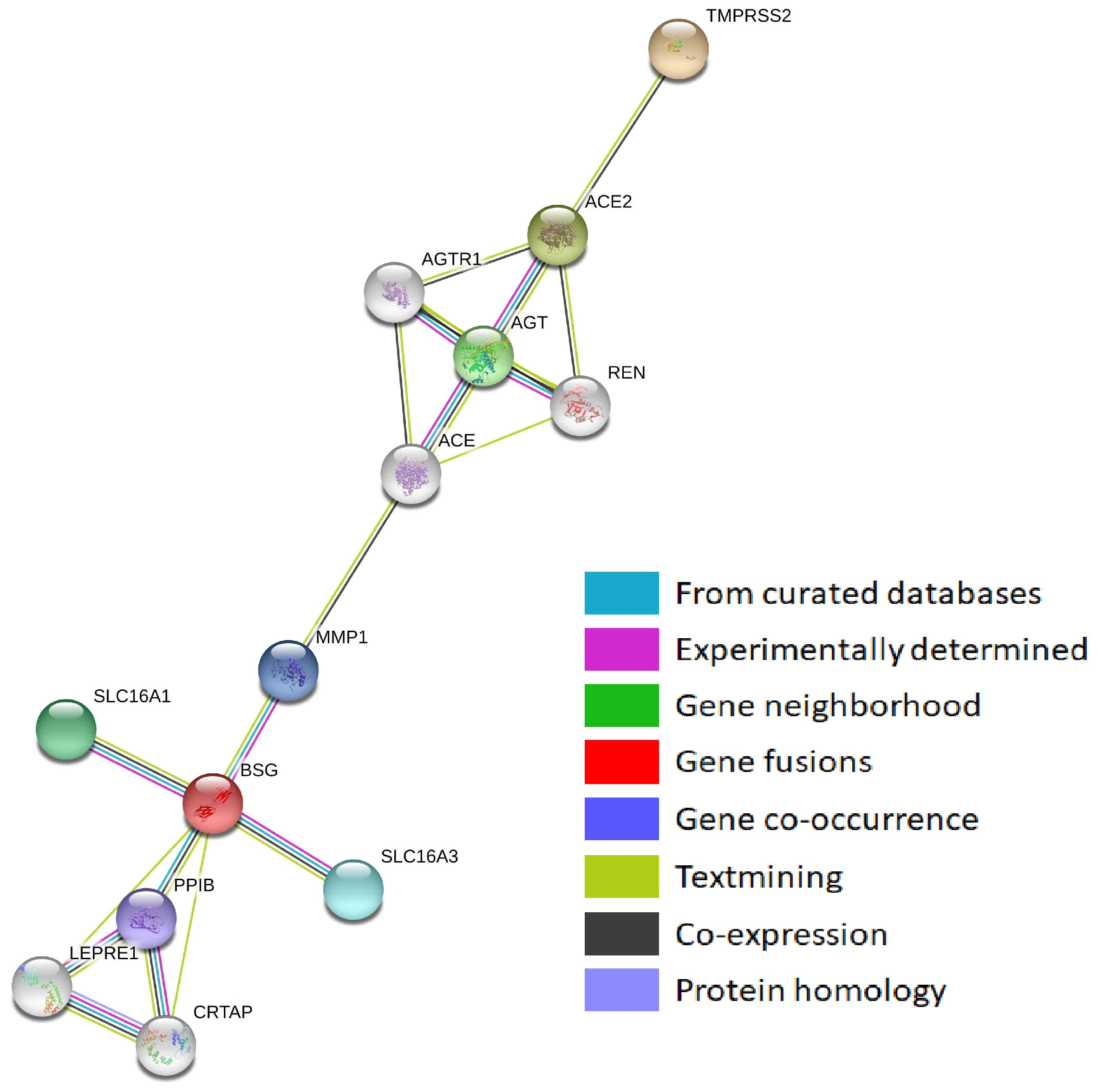
3.1. Protein–Protein Interaction Networks of Sars-Cov-2-Interacting Genes
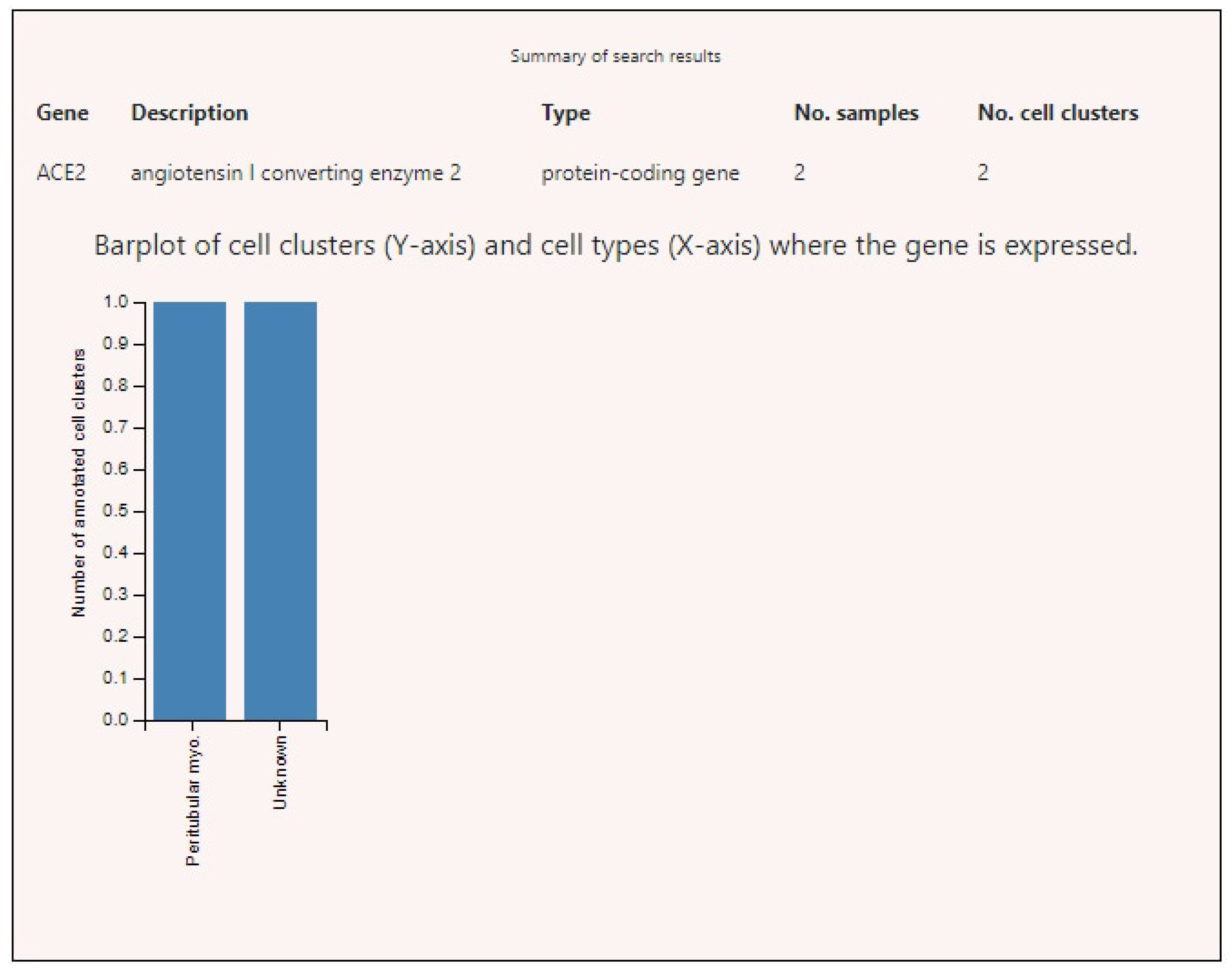
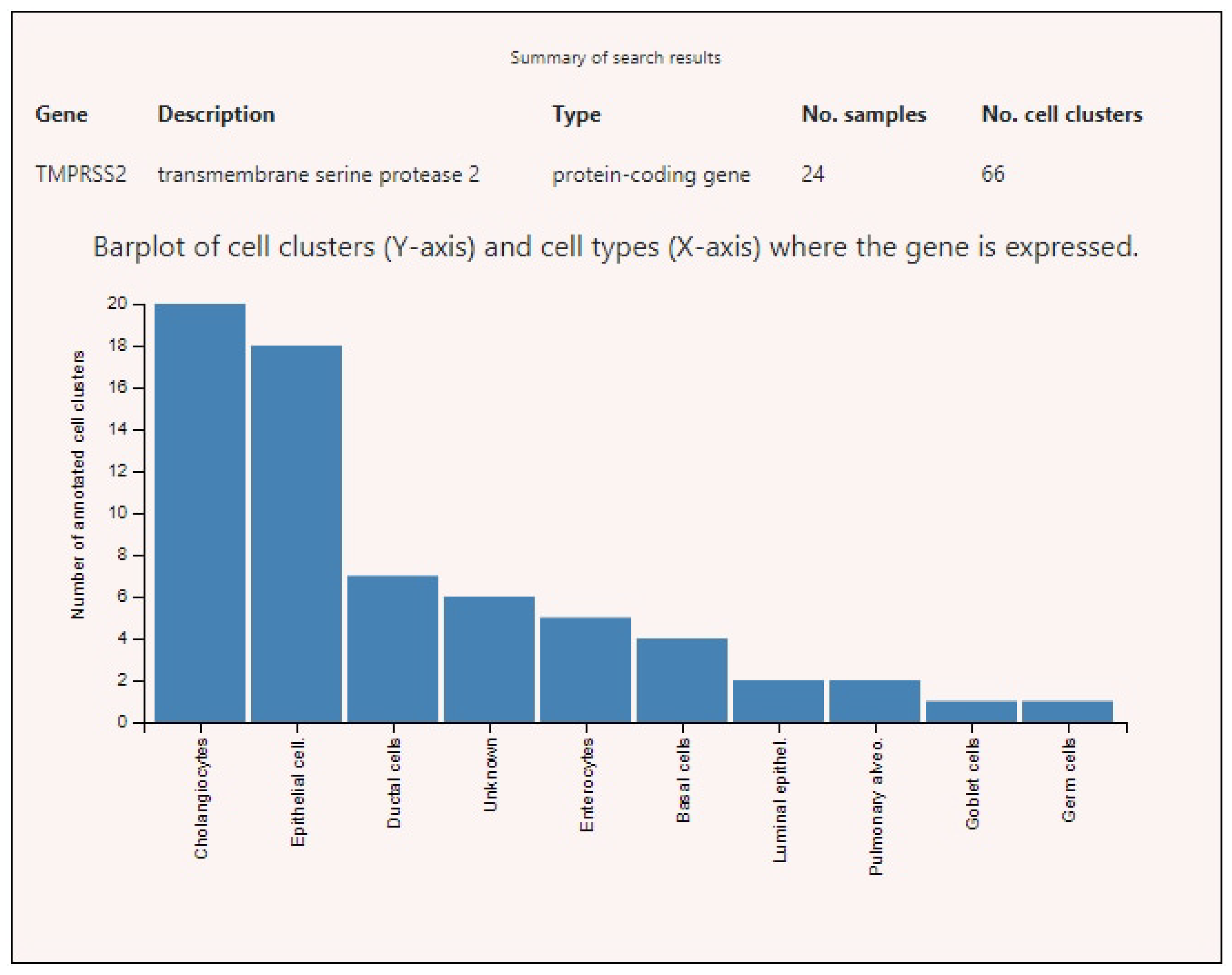
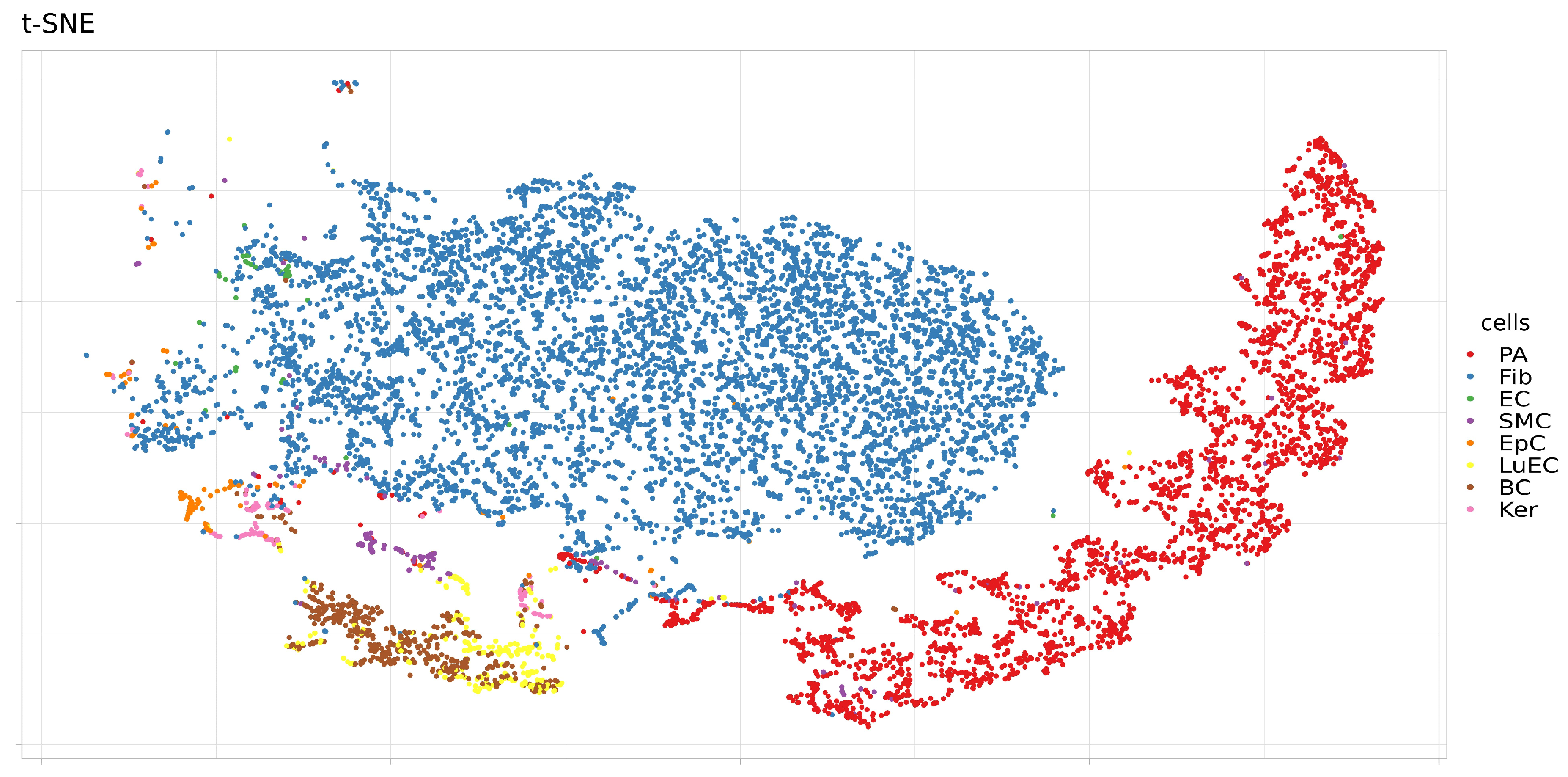
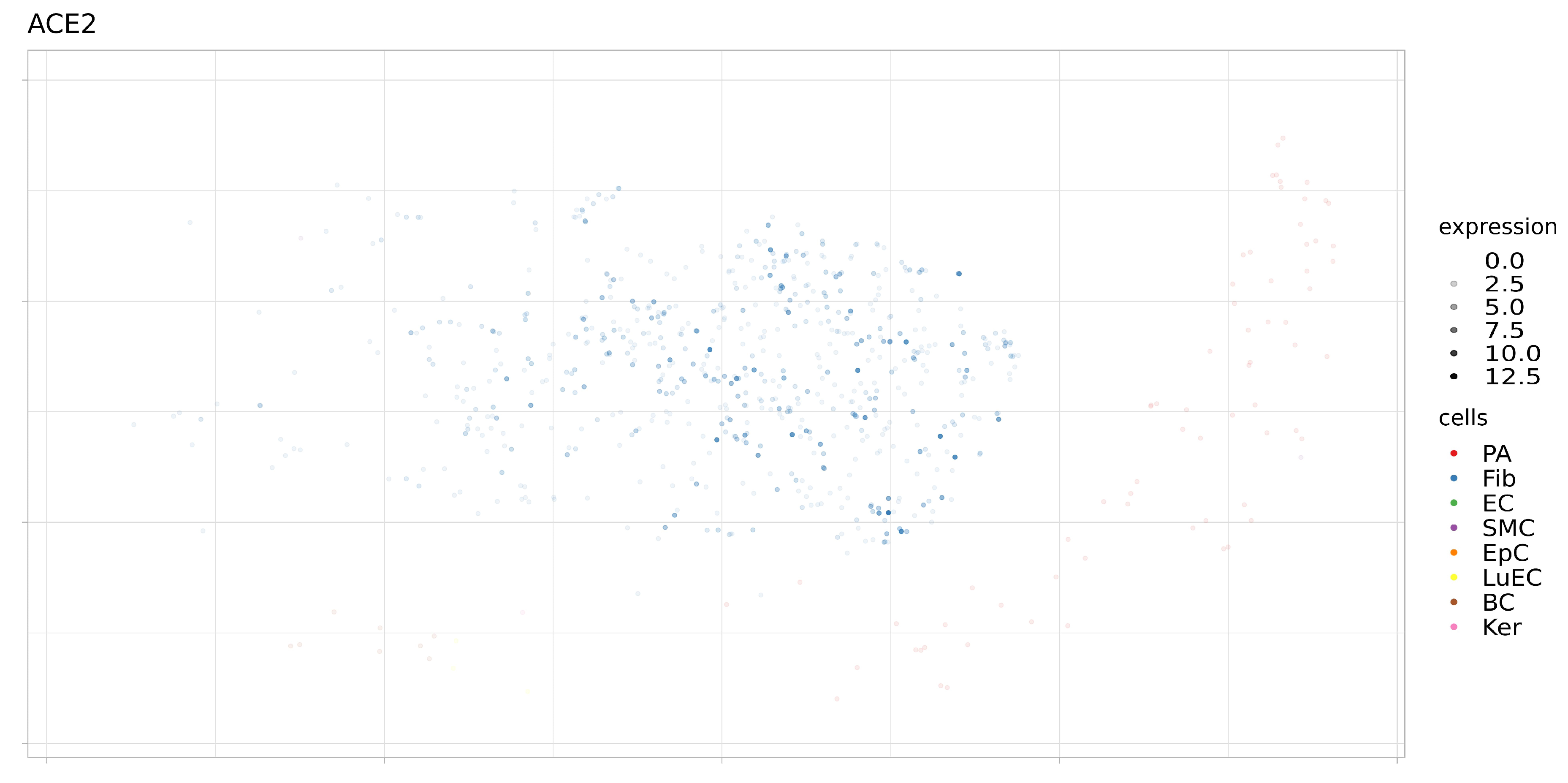
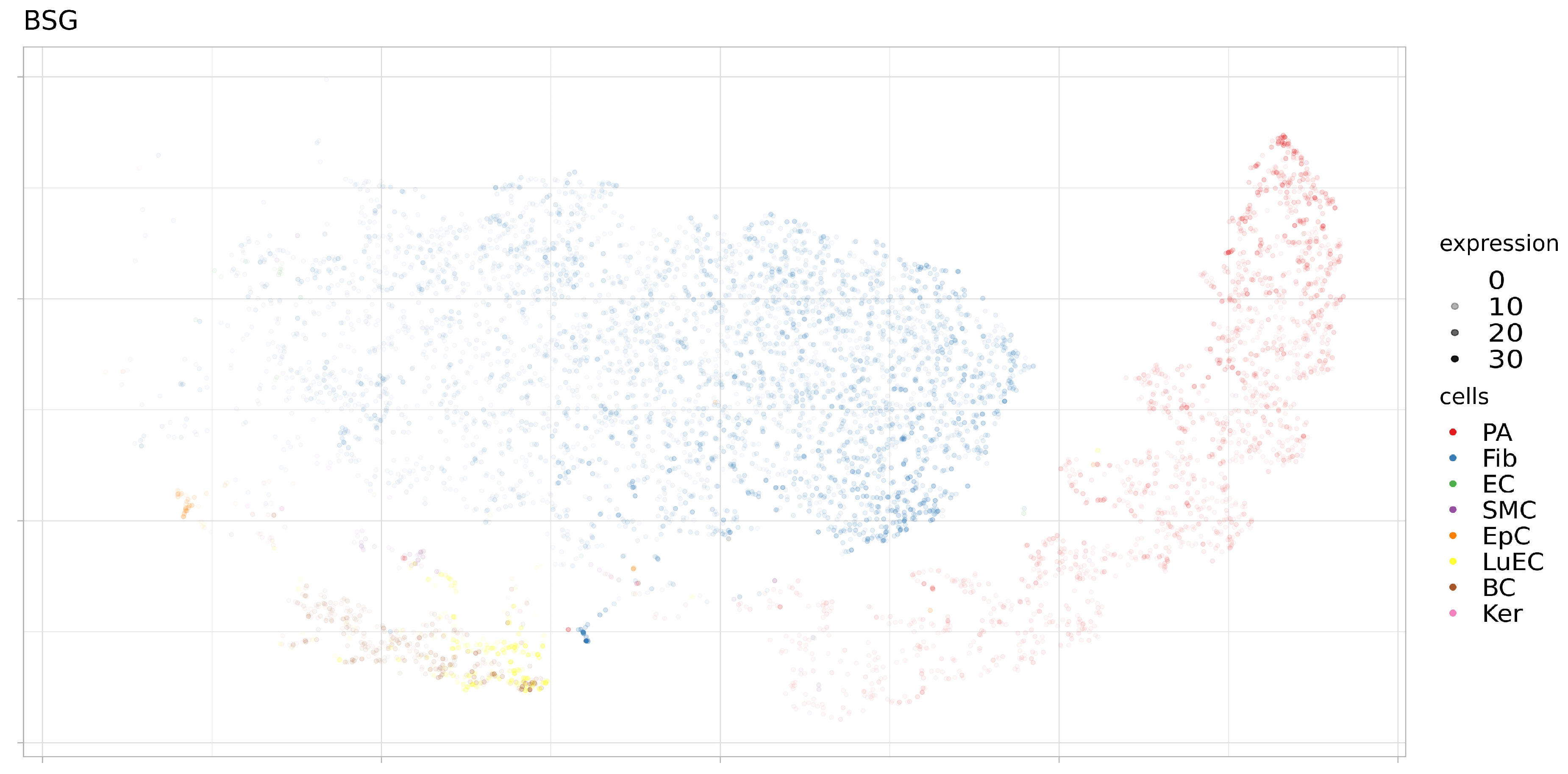
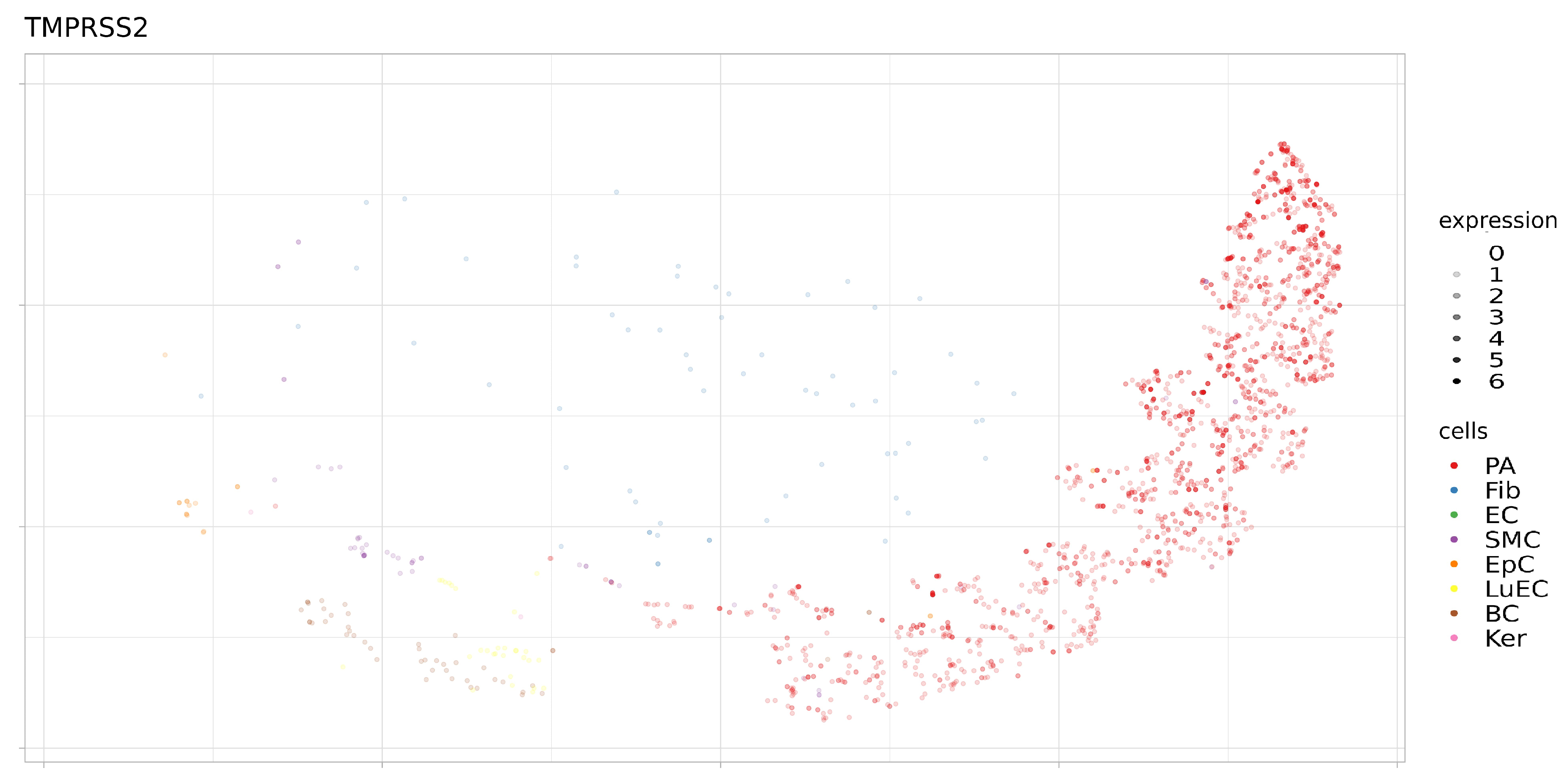
3.2. Expression of ACE2, BSG, and TMPRSS2 in Single Cells
3.3. Snv and Indel Variants of the Tmprss2 Gene
3.4. Frequency of Protein-Changing Allelic Variants of the Tmprss2 Gene in Populations of North Eurasia
3.5. Regulation of Expression of Tmprss2
3.5.1. Eqtls
3.5.2. Mirnas
3.5.3. Pharmacotranscriptomics of Tmprss2
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, K.; Chen, W.; Zhou, Y.S.; Lian, J.Q.; Zhang, Z.; Du, P.; Gong, L.; Zhang, Y.; Cui, H.Y.; Geng, J.J.; et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 16, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, A.; Graff, R.E.; Bauer, S.R.; Pitt, M.J.; Lis, R.T.; Stack, E.C.; Martin, N.E.; Kunz, L.; Penney, K.L.; Ligon, A.H.; et al. The TMPRSS2: ERG rearrangement, ERG expression, and prostate cancer outcomes: A cohort study and meta-analysis. Cancer Epidemiol. Biomarkers Prev. 2012, 21, 1497–1509. [Google Scholar] [CrossRef]
- Saramäki, O.R.; Harjula, A.E.; Martikainen, P.M.; Vessella, R.L.; Tammela, T.L.; Visakorpi, T. TMPRSS2: ERG fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin. Cancer Res. 2008, 14, 3395–3400. [Google Scholar] [CrossRef] [PubMed]
- Shulla, A.; Heald-Sargent, T.; Subramanya, G.; Zhao, J.; Perlman, S.; Gallagher, T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011, 85, 873–882. [Google Scholar] [CrossRef]
- Glowacka, I.; Bertram, S.; Müller, M.A.; Allen, P.; Soilleux, E.; Pfefferle, S.; Steffen, I.; Tsegaye, T.S.; He, Y.; Gnirss, K.; et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol 2011, 85, 4122–4134. [Google Scholar] [CrossRef]
- Bertram, S.; Dijkman, R.; Habjan, M.; Heurich, A.; Gierer, S.; Glowacka, I.; Welsch, K.; Winkler, M.; Schneider, H.; Hofmann-Winkler, H.; et al. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 2013, 87, 6150–6160. [Google Scholar] [CrossRef]
- Abe, M.; Tahara, M.; Sakai, K.; Yamaguchi, H.; Kanou, K.; Shirato, K.; Kawase, M.; Noda, M.; Kimura, H.; Matsuyama, S.; et al. TMPRSS2 is an activating protease for respiratory parainfluenza viruses. J. Virol. 2013, 87, 11930–11935. [Google Scholar] [CrossRef] [PubMed]
- Shirato, K.; Kawase, M.; Matsuyama, S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 2013, 87, 12552–12561. [Google Scholar] [CrossRef] [PubMed]
- Heurich, A.; Hofmann-Winkler, H.; Gierer, S.; Liepold, T.; Jahn, O.; Pöhlmann, S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014, 88, 1293–1307. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 2019, 531210. [Google Scholar] [CrossRef]
- Koike, A.; Nishida, N.; Yamashita, D.; Tokunaga, K. Comparative analysis of copy number variation detection methods and database construction. BMC Genet. 2011, 12, 29. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hadley, D.; Liu, R.; Glessner, J.; Grant, S.F.; Hakonarson, H.; Bucan, M. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007, 17, 1665–1674. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Human Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef]
- Zuberi, K.; Franz, M.; Rodriguez, H.; Montojo, J.; Lopes, C.T.; Bader, G.D.; Morris, Q. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 2013, 41, W115–W122. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Franzén, O.; Gan, L.M.; Björkegren, J.L. PanglaoDB: A web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, 2019. [Google Scholar] [CrossRef]
- Leinonen, R.; Sugawara, H.; Shumway, M.; International Nucleotide Sequence Database Collaboration. The sequence read archive. Nucleic Acids Res. 2010, 39, D19–D21. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., III; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive integration of single-cell data. Cell 2019, 177, 1888–1902. [Google Scholar] [CrossRef]
- Huang, H.Y.; Lin, Y.C.D.; Li, J.; Huang, K.Y.; Shrestha, S.; Hong, H.C.; Tang, Y.; Chen, Y.G.; Jin, C.N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef]
- Kehl, T.; Kern, F.; Backes, C.; Fehlmann, T.; Stöckel, D.; Meese, E.; Lenhof, H.P.; Keller, A. miRPathDB 2.0: A novel release of the miRNA Pathway Dictionary Database. Nucleic Acids Res. 2020, 48, D142–D147. [Google Scholar] [CrossRef] [PubMed]
- De Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Åström, G.; Babina, M.; Bertin, N.; Burroughs, A.M.; et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017, 35, 872. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, C.; Manoharan, C.; Wilson, M.C.; Manoharan, C.; Wilson, M.C.; Sessions, R.B.; Halestrap, A.P. The role of charged residues in the transmembrane helices of monocarboxylate transporter 1 and its ancillary protein basigin in determining plasma membrane expression and catalytic activity. Mol. Membr. Biol. 2006, 23, 486–498. [Google Scholar] [CrossRef]
- Castorino, J.J.; Gallagher-Colombo, S.M.; Levin, A.V.; FitzGerald, P.G.; Polishook, J.; Kloeckener-Gruissem, B.; Ostertag, E.; Philp, N.J. Juvenile cataract-associated mutation of solute carrier SLC16A12 impairs trafficking of the protein to the plasma membrane. Invest. Ophthalmol. Vis. Sci. 2011, 52, 6774–6784. [Google Scholar] [CrossRef]
- Rusu, V.; Hoch, E.; Mercader, J.M.; Tenen, D.E.; Gymrek, M.; Hartigan, C.R.; DeRan, M.; von Grotthuss, M.; Fontanillas, P.; Spooner, A.; et al. Type 2 diabetes variants disrupt function of SLC16A11 through two distinct mechanisms. Cell 2017, 170, 199–212. [Google Scholar] [CrossRef]
- Huang, Q.; Li, J.; Xing, J.; Li, W.; Li, H.; Ke, X.; Zhang, J.; Ren, T.; Shang, Y.; Yang, H.; et al. CD147 promotes reprogramming of glucose metabolism and cell proliferation in HCC cells by inhibiting the p53-dependent signaling pathway. J. Hepatol. 2014, 61, 859–866. [Google Scholar] [CrossRef]
- Zhao, P.; Zhang, W.; Wang, S.J.; Yu, X.L.; Tang, J.; Huang, W.; Li, Y.; Cui, H.Y.; Guo, Y.S.; Tavernier, J.; et al. HAb18G/CD147 promotes cell motility by regulating annexin II-activated RhoA and Rac1 signaling pathways in hepatocellular carcinoma cells. Hepatology 2011, 54, 2012–2024. [Google Scholar] [CrossRef]
- Lu, M.; Wu, J.; Hao, Z.W.; Shang, Y.K.; Xu, J.; Nan, G.; Li, X.; Chen, Z.N.; Bian, H. Basolateral CD147 induces hepatocyte polarity loss by E-cadherin ubiquitination and degradation in hepatocellular carcinoma progress. Hepatology 2018, 68, 317–332. [Google Scholar] [CrossRef]
- Zhang, M.Y.; Zhang, Y.; Wu, X.D.; Zhang, K.; Lin, P.; Bian, H.J.; Qin, M.M.; Huang, W.; Wei, D.; Zhang, Z.; et al. Disrupting CD147-RAP2 interaction abrogates erythrocyte invasion by Plasmodium falciparum. Blood 2018, 131, 1111–1121. [Google Scholar] [CrossRef]
- Castro, A.P.V.; Carvalho, T.M.; Moussatché, N.; Damaso, C.R. Redistribution of cyclophilin A to viral factories during vaccinia virus infection and its incorporation into mature particles. J. Virol. 2003, 77, 9052–9068. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Mi, L.; Xu, J.; Yu, J.; Wang, X.; Jiang, J.; Xing, J.; Shang, P.; Qian, A.; Li, Y.; et al. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2005, 191, 755–760. [Google Scholar] [CrossRef] [PubMed]
- Bian, H.; Zheng, Z.H.; Wei, D.; Zhang, Z.; Kang, W.Z.; Hao, C.Q.; Dong, K.; Kang, W.; Xia, J.L.; Miao, J.L.; et al. Meplazumab treats COVID-19 pneumonia: An open-labelled, concurrent controlled add-on clinical trial. MedRxiv 2020. [Google Scholar] [CrossRef]
- Bhanushali, A.; Rao, P.; Raman, V.; Kokate, P.; Ambekar, A.; Mandva, S.; Bhatia, S.; Das, B. Status of TMPRSS2–ERG fusion in prostate cancer patients from India: Correlation with clinico-pathological details and TMPRSS2 Met160Val polymorphism. Prostate Int. 2018, 6, 145–150. [Google Scholar] [CrossRef]
- FitzGerald, L.M.; Agalliu, I.; Johnson, K.; Miller, M.A.; Kwon, E.M.; Hurtado-Coll, A.; Fazli, L.; Rajput, A.B.; Gleave, M.E.; Cox, M.E.; et al. Association of TMPRSS2-ERG gene fusion with clinical characteristics and outcomes: Results from a population-based study of prostate cancer. BMC Cancer 2008, 8, 230. [Google Scholar] [CrossRef]
- Paniri, A.; Hosseini, M.M.; Akhavan-Niaki, H. First comprehensive computational analysis of functional consequences of TMPRSS2 SNPs in susceptibility to SARS-CoV-2 among different populations. J. Biomol. Struct. Dyn. 2020, 1–18. [Google Scholar] [CrossRef]
- Alhazzani, W.; Møller, M.H.; Arabi, Y.M.; Loeb, M.; Gong, M.N.; Fan, E.; Oczkowski, S.; Levy, M.M.; Derde, L.; Dzierba, A.; et al. Surviving Sepsis Campaign: Guidelines on the management of critically ill adults with Coronavirus Disease 2019 (COVID-19). Intensive Care Med. 2020, 1–34. [Google Scholar] [CrossRef]
- Khaerunnisa, S.; Kurniawan, H.; Awaluddin, R.; Suhartati, S.; Soetjipto, S. Potential inhibitor of COVID-19 main protease (Mpro) from several medicinal plant compounds by molecular docking study. Preprints 2020, 20944, 1–14. Available online: https://www.preprints.org/manuscript/202003.0226/v1 (accessed on 10 June 2020).

| Variant Type | Maf > 0.01 | Maf < 0.01 |
|---|---|---|
| 3_prime_UTR_variant | 0 | 22 |
| 5_prime_UTR_variant | 0 | 5 |
| frameshift_variant | 0 | 17 |
| inframe_deletion | 0 | 1 |
| inframe_insertion | 0 | 2 |
| intron_variant | 13 | 409 |
| missense_variant | 2 | 332 |
| splice_acceptor_variant | 0 | 4 |
| splice_donor_variant | 0 | 5 |
| splice_region_variant | 0 | 54 |
| stop_gained | 0 | 13 |
| stop_lost | 0 | 1 |
| synonymous_variant | 5 | 140 |
| Region | N | Frequency % | Copy Number | Gene Name |
|---|---|---|---|---|
| Chr21:42857241-42863723 | 164 | 1.2195122 | 1 | TMPRSS2 |
| Population | N | rs148125094 | rs143597099 | rs12329760 | rs201093031 |
|---|---|---|---|---|---|
| Easten Europe | 419 | 0.0012 | 0.0012 | 0.2983 | 0.0000 |
| Bashkirs Burzyan | 29 | 0.0000 | 0.0000 | 0.1552 | 0.0000 |
| Bashkirs Perm | 15 | 0.0000 | 0.0000 | 0.3000 | 0.0000 |
| Bashkirs Salavat | 15 | 0.0000 | 0.0000 | 0.3667 | 0.0000 |
| Besermians | 16 | 0.0000 | 0.0000 | 0.1563 | 0.0000 |
| Chuvash | 26 | 0.0000 | 0.0000 | 0.3077 | 0.0000 |
| Karelians | 29 | 0.0172 | 0.0000 | 0.3966 | 0.0000 |
| Komi | 30 | 0.0000 | 0.0000 | 0.3333 | 0.0000 |
| Mari | 30 | 0.0000 | 0.0000 | 0.2500 | 0.0000 |
| Mordvins Erzya | 16 | 0.0000 | 0.0000 | 0.3125 | 0.0000 |
| Mordvins Moksha | 30 | 0.0000 | 0.0000 | 0.3000 | 0.0000 |
| Mordvins Shoksha | 14 | 0.0000 | 0.0000 | 0.3214 | 0.0000 |
| Russians | 33 | 0.0000 | 0.0000 | 0.3333 | 0.0000 |
| Tatars Kazan | 30 | 0.0000 | 0.0000 | 0.3000 | 0.0000 |
| Udmurts | 30 | 0.0000 | 0.0000 | 0.3000 | 0.0000 |
| Udmurts Balezino | 28 | 0.0000 | 0.0000 | 0.3214 | 0.0000 |
| Udmurts Sharkan | 18 | 0.0000 | 0.0000 | 0.2500 | 0.0000 |
| Veps | 30 | 0.0000 | 0.0167 | 0.3333 | 0.0000 |
| North Caucasus (excl. Dagestan) | 274 | 0.0018 | 0.0000 | 0.1989 | 0.0000 |
| Abkhaz | 30 | 0.0167 | 0.0000 | 0.3000 | 0.0000 |
| Adyghe | 10 | 0.0000 | 0.0000 | 0.1500 | 0.0000 |
| Balkars | 50 | 0.0000 | 0.0000 | 0.1800 | 0.0000 |
| Chechens | 27 | 0.0000 | 0.0000 | 0.2222 | 0.0000 |
| Cherkess | 30 | 0.0000 | 0.0000 | 0.2167 | 0.0000 |
| Ingush | 30 | 0.0000 | 0.0000 | 0.1500 | 0.0000 |
| Karachays | 22 | 0.0000 | 0.0000 | 0.2045 | 0.0000 |
| Mingrelians | 28 | 0.0000 | 0.0000 | 0.1607 | 0.0000 |
| North Ossetians | 30 | 0.0000 | 0.0000 | 0.1833 | 0.0000 |
| South Ossetians | 17 | 0.0000 | 0.0000 | 0.2059 | 0.0000 |
| Dagestan | 538 | 0.0000 | 0.0000 | 0.2309 | 0.0000 |
| Aghuls | 24 | 0.0000 | 0.0000 | 0.2292 | 0.0000 |
| Akhvakhs | 24 | 0.0000 | 0.0000 | 0.3125 | 0.0000 |
| Andis | 17 | 0.0000 | 0.0000 | 0.2353 | 0.0000 |
| Archins | 24 | 0.0000 | 0.0000 | 0.3333 | 0.0000 |
| Avars | 24 | 0.0000 | 0.0000 | 0.1875 | 0.0000 |
| Bagulals | 23 | 0.0000 | 0.0000 | 0.3261 | 0.0000 |
| Bezhtins | 22 | 0.0000 | 0.0000 | 0.2273 | 0.0000 |
| Botlikhs | 16 | 0.0000 | 0.0000 | 0.1250 | 0.0000 |
| Chamalals | 24 | 0.0000 | 0.0000 | 0.2083 | 0.0000 |
| Dargins | 28 | 0.0000 | 0.0000 | 0.2321 | 0.0000 |
| Ginukhs | 19 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| Gunzibians | 17 | 0.0000 | 0.0000 | 0.0294 | 0.0000 |
| Karanogais | 19 | 0.0000 | 0.0000 | 0.2368 | 0.0000 |
| Karatins | 24 | 0.0000 | 0.0000 | 0.3333 | 0.0000 |
| Khvarshins | 15 | 0.0000 | 0.0000 | 0.1000 | 0.0000 |
| Kumyks | 37 | 0.0000 | 0.0000 | 0.2703 | 0.0000 |
| Laks | 24 | 0.0000 | 0.0000 | 0.3125 | 0.0000 |
| Lezgins | 28 | 0.0000 | 0.0000 | 0.2037 | 0.0000 |
| Nogais | 20 | 0.0000 | 0.0000 | 0.2750 | 0.0000 |
| Rutuls | 22 | 0.0000 | 0.0000 | 0.1818 | 0.0000 |
| Tabasarans | 21 | 0.0000 | 0.0000 | 0.2619 | 0.0000 |
| Tindins | 18 | 0.0000 | 0.0000 | 0.2222 | 0.0000 |
| Tsakhurs | 24 | 0.0000 | 0.0000 | 0.2292 | 0.0000 |
| Tsez | 24 | 0.0000 | 0.0000 | 0.2708 | 0.0000 |
| Central Asia | 128 | 0.0000 | 0.0000 | 0.3565 | 0.0000 |
| Dungans | 23 | 0.0000 | 0.0000 | 0.4130 | 0.0000 |
| Kazakh Junior Horde | 29 | 0.0000 | 0.0000 | 0.2931 | 0.0000 |
| Kazakh Great Horde | 26 | 0.0000 | 0.0000 | 0.4423 | 0.0000 |
| Kyrgyz | 28 | 0.0000 | 0.0000 | 0.3704 | 0.0000 |
| Uzbeks | 22 | 0.0000 | 0.0000 | 0.2619 | 0.0000 |
| Siberia | 404 | 0.0000 | 0.0000 | 0.3540 | 0.0013 |
| Altaians Maymalar | 24 | 0.0000 | 0.0000 | 0.3958 | 0.0000 |
| Altaians Kizhi | 25 | 0.0000 | 0.0000 | 0.3600 | 0.0000 |
| Buryats Aginskoe | 23 | 0.0000 | 0.0000 | 0.4130 | 0.0000 |
| Buryats Kurumkan | 28 | 0.0000 | 0.0000 | 0.3929 | 0.0000 |
| Chulyms | 22 | 0.0000 | 0.0000 | 0.3636 | 0.0000 |
| Evenks Yakutia | 28 | 0.0000 | 0.0000 | 0.2857 | 0.0000 |
| Evenks Zabaykalsky Krai | 25 | 0.0000 | 0.0000 | 0.3200 | 0.0000 |
| Kalmyks | 29 | 0.0000 | 0.0000 | 0.3103 | 0.0000 |
| Kets | 15 | 0.0000 | 0.0000 | 0.3333 | 0.0000 |
| Khakas Kachins | 26 | 0.0000 | 0.0000 | 0.4423 | 0.0000 |
| Khakas Sagays | 29 | 0.0000 | 0.0000 | 0.6379 | 0.0000 |
| Khanty Kazym | 30 | 0.0000 | 0.0000 | 0.1333 | 0.0000 |
| Khanty Russkinskie | 26 | 0.0000 | 0.0000 | 0.2500 | 0.0000 |
| Tomsk Tatas | 20 | 0.0000 | 0.0000 | 0.3250 | 0.0000 |
| Tuvans | 28 | 0.0000 | 0.0000 | 0.3036 | 0.0185 |
| Yakuts | 26 | 0.0000 | 0.0000 | 0.4038 | 0.0000 |
| North East Asia | 73 | 0.0000 | 0.0000 | 0.2671 | 0.0284 |
| Chukchi | 25 | 0.0000 | 0.0000 | 0.3000 | 0.0000 |
| Koryaks | 20 | 0.0000 | 0.0000 | 0.3500 | 0.0000 |
| Nivkhs | 13 | 0.0000 | 0.0000 | 0.1538 | 0.0769 |
| Udege | 15 | 0.0000 | 0.0000 | 0.2000 | 0.0714 |
| rs148125094 | rs12329760 | rs201093031 | ||||
|---|---|---|---|---|---|---|
| Population | N | Frequency | N | Frequency | N | Frequency |
| European | 77147 | 0.0014 | 76846 | 0.2549 | 77117 | 0.0000 |
| Finnish | 12560 | 0.0016 | 12544 | 0.3725 | 12561 | 0.0000 |
| Estonian | 2416 | 0.0060 | 2394 | 0.3074 | 2406 | 0.0000 |
| Southern European | 5805 | 0.0013 | 5778 | 0.1748 | 5802 | 0.0000 |
| North-western European | 25402 | 0.0012 | 25348 | 0.2212 | 25391 | 0.0000 |
| Other non-Finnish European | 16562 | 0.0012 | 16439 | 0.2286 | 16557 | 0.0000 |
| Swedish | 13067 | 0.0010 | 13013 | 0.2722 | 13066 | 0.0000 |
| Bulgarian | 1335 | 0.0007 | 1330 | 0.1970 | 1334 | 0.0000 |
| South Asian | 15308 | 0.0007 | 15298 | 0.2477 | 15303 | 0.0000 |
| Latino | 17718 | 0.0003 | 17705 | 0.1533 | 17697 | 0.0000 |
| African | 12480 | 0.0002 | 12448 | 0.2918 | 12480 | 0.0000 |
| Ashkenazi Jewish | 5185 | 0.0000 | 5163 | 0.1352 | 5179 | 0.0000 |
| East Asian | 9196 | 0.0000 | 9188 | 0.3810 | 9193 | 0.0024 |
| Japanese | 76 | 0.0000 | 76 | 0.4013 | 76 | 0.0000 |
| Korean | 1909 | 0.0000 | 1909 | 0.3675 | 1909 | 0.0018 |
| Other East Asian | 7211 | 0.0000 | 7203 | 0.3844 | 7208 | 0.0026 |
| N SNP | Average Maf | Average Slope | |
|---|---|---|---|
| down | 60 | 0.3722896 | −0.09795966 |
| up | 76 | 0.4537386 | 0.09709619 |
| miRNA | Cell Ontology |
|---|---|
| hsa-miR-4476 | B cell |
| hsa-miR-5187-3p | myeloid leukocyte |
| hsa-miR-5187-3p | hematopoietic cell |
| hsa-miR-7849-3p | endothelial cell |
| hsa-miR-7849-3p | blood vessel endothelial cell |
| hsa-miR-7849-3p | endothelial cell of vascular tree |
| hsa-miR-7849-3p | neutrophil |
| Drug | Drug Groups | Change | References |
|---|---|---|---|
| Acetaminophen | Approved | downregulated | 21420995 |
| Acyline | Investigational | downregulated | 17510436 |
| Stanolone | Illicit Investigational | downregulated | 12711008 |
| Stanolone | Illicit Investigational | upregulated | 20601956, 23708653 |
| Estradiol | Approved Investigational Vet Approved | downregulated | 24758408 |
| Estradiol | Approved Investigational Vet Approved | upregulated | 19619570 |
| Curcumin | Approved Experimental Investigational | downregulated | 18719366, 22258452 |
| Cyclosporine | Approved Investigational Vet Approved | downregulated | 20106945 |
| Calcitriol | Approved Nutraceutical | upregulated | 21592394, 26485663 |
| Entinostat | Investigational | upregulated | 26272509 |
| Ethinylestradiol | Approved | downregulated | 18936297 |
| Genistein | Investigational | downregulated | 15378649, 26865667 |
| Metribolone | Experimental | downregulated | 12711008 |
| Metribolone | Experimental | upregulated | 17010675, 21440447 |
| Resveratrol | Investigational | downregulated | 18586690 |
| Selenium | Approved Investigational Vet Approved | upregulated | 19244175 |
| Testosterone | Approved Investigational | upregulated | 21592394 |
| Tretinoin | Approved Investigational Nutraceutical | upregulated | 23830798 |
| Valproic acid | Approved Investigational | upregulated | 23179753, 24383497, 26272509 |
| Zoledronic acid | Approved | upregulated | 24714768 |
| Drug | Drug Groups | Change | References |
|---|---|---|---|
| Amiodarone | Approved Investigational | upregulated | 19774075 |
| Arsenic trioxide | Approved Investigational | downregulated | 23232515 |
| Estradiol | Approved Investigational Vet Approved | upregulated | 19167446 |
| Methotrexate | Approved | downregulated | 25339124 |
| Quercetin | Experimental Investigational | upregulated | 21632981 |
| Isotretinoin | Approved | downregulated | 20436886 |
| Silicon dioxide | Approved | downregulated | 25895662 |
| Valproic acid | Approved Investigational | downregulated | 23179753 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zarubin, A.; Stepanov, V.; Markov, A.; Kolesnikov, N.; Marusin, A.; Khitrinskaya, I.; Swarovskaya, M.; Litvinov, S.; Ekomasova, N.; Dzhaubermezov, M.; et al. Structural Variability, Expression Profile, and Pharmacogenetic Properties of TMPRSS2 Gene as a Potential Target for COVID-19 Therapy. Genes 2021, 12, 19. https://doi.org/10.3390/genes12010019
Zarubin A, Stepanov V, Markov A, Kolesnikov N, Marusin A, Khitrinskaya I, Swarovskaya M, Litvinov S, Ekomasova N, Dzhaubermezov M, et al. Structural Variability, Expression Profile, and Pharmacogenetic Properties of TMPRSS2 Gene as a Potential Target for COVID-19 Therapy. Genes. 2021; 12(1):19. https://doi.org/10.3390/genes12010019
Chicago/Turabian StyleZarubin, Aleksei, Vadim Stepanov, Anton Markov, Nikita Kolesnikov, Andrey Marusin, Irina Khitrinskaya, Maria Swarovskaya, Sergey Litvinov, Natalia Ekomasova, Murat Dzhaubermezov, and et al. 2021. "Structural Variability, Expression Profile, and Pharmacogenetic Properties of TMPRSS2 Gene as a Potential Target for COVID-19 Therapy" Genes 12, no. 1: 19. https://doi.org/10.3390/genes12010019
APA StyleZarubin, A., Stepanov, V., Markov, A., Kolesnikov, N., Marusin, A., Khitrinskaya, I., Swarovskaya, M., Litvinov, S., Ekomasova, N., Dzhaubermezov, M., Maksimova, N., Sukhomyasova, A., Shtygasheva, O., Khusnutdinova, E., Radzhabov, M., & Kharkov, V. (2021). Structural Variability, Expression Profile, and Pharmacogenetic Properties of TMPRSS2 Gene as a Potential Target for COVID-19 Therapy. Genes, 12(1), 19. https://doi.org/10.3390/genes12010019






