Abstract
The taxonomy and phylogenetics of Neotropical deer have been mostly based on morphological criteria and needs a critical revision on the basis of new molecular and cytogenetic markers. In this study, we used the variation in the sequence, copy number, and chromosome localization of satellite I-IV DNA to evaluate evolutionary relationships among eight Neotropical deer species. Using FISH with satI-IV probes derived from Mazama gouazoubira, we proved the presence of satellite DNA blocks in peri/centromeric regions of all analyzed deer. Satellite DNA was also detected in the interstitial chromosome regions of species of the genus Mazama with highly reduced chromosome numbers. In contrast to Blastocerus dichotomus, Ozotoceros bezoarticus, and Odocoileus virginianus, Mazama species showed high abundance of satIV DNA by FISH. The phylogenetic analysis of the satellite DNA showed close relationships between O. bezoarticus and B. dichotomus. Furthermore, the Neotropical and Nearctic populations of O. virginianus formed a single clade. However, the satellite DNA phylogeny did not allow resolving the relationships within the genus Mazama. The high abundance of the satellite DNA in centromeres probably contributes to the formation of chromosomal rearrangements, thus leading to a fast and ongoing speciation in this genus, which has not yet been reflected in the satellite DNA sequence diversification.
1. Introduction
Among large mammals, Neotropical deer (Cervidae, Pecora, Ruminantia, Artiodactyla) [1,2] represent an interesting group of species still lacking comprehensive scientific data. Their taxonomy has been established mostly on the basis of morphology indicating a need for its critical revision and a future systematic research [3,4]. Neotropical deer involve genera Pudu, Mazama, Hippocamelus, Blastocerus, Ozotoceros, and Odocoileus grouped in the tribe Rangiferini, subfamily Capreolinae [4,5]. As in other Cervidae, a variety of karyotypes has been observed in Neotropical deer, ranging from 2n = 70 in Mazama gouazoubira or Odocoileus virginianus, to 2n = 32–34+ Bs in Mazama bororo [6,7]. The 2n = 70 karyotype considered to reflect cervid ancestral karyotype has derived from the hypothetical ancestral karyotype of Pecora (2n = 58) by six chromosome fissions [6,8,9]. However, a series of evolutionary chromosome rearrangements occurred in many deer taxa, which led to a significant diversification of their karyotypes [9]. As with the other Neotropical deer species, a rapid karyotype evolution has been observed in Mazama americana, a taxon grouping several cryptic species currently classified as cytotypes on the basis of their karyotype differences and geographical distribution and reported as M. americana species complex [4,10,11,12]. There is no doubt that the taxonomy and phylogenetics of the Neotropical deer would benefit from new approaches and utilization of new molecular markers.
A useful source of information can be found in satellite DNA, which consists of rapidly evolving, tandemly organized repetitive sequences, and might serve, to some extent, as molecular cytogenetic marker to trace individual and species origin and phylogeny. Satellite DNA located in centromeres and pericentromeric chromosome regions probably represents a structure linked to centromeric functions and chromosome segregation [13,14,15,16]. However, the functional roles of satellite DNA have not been fully elucidated yet. It is known that despite the relative uniformity of monomer lengths within satellite DNA families, they often show variations in sequence, copy numbers, and chromosome distribution even among related species [13,17], which can be used in phylogenetic studies [16,18,19,20,21,22,23,24,25,26,27].
Six satellite DNA families were described in Cervidae so far, of which satI-satIV were characterized in terms of sequence and chromosomal distribution in a number of Eurasian and North American cervid species [24,28,29,30,31,32,33,34]. However, there is a complete lack of data on satellite DNA sequences and their chromosome distribution in deer inhabiting South America. The sole exception is O. virginianus, a species spread throughout the American continent, in which a representative of its northern population has been under study recently [24].
In this study, we isolated four main groups of cervid satellite DNA sequences (satI-IV) in eight Neotropical deer species: Mazama gouazoubira, Mazama nemorivaga, Mazama nana, Mazama bororo, M. americana, Blastocerus dichotomus, Ozotoceros bezoarticus, and O. virginianus of South American origin. We performed intra- and inter-species comparisons of the obtained satellite DNA sequences and their physical localization on metaphase chromosomes using fluorescence in situ hybridization (FISH). We also searched the obtained sequences for a presence of the 17-bp binding motif for the CENP-B centromeric protein. Finally, we reconstructed phylogenetic trees of the satellite DNA sequences and compared them with the mt-cyb gene phylogeny to infer the evolutionary relationships among Neotropical deer species and their position within Cervidae.
2. Material and Methods
2.1. Species and Samples
Fibroblast tissue cultures prepared according to standard techniques from skin samples of eight Neotropical deer species and available at NUPECCE (Jaboticabal, Brazil) were used in this study for DNA isolation and FISH. No animals were euthanized in this study. The samples are listed in Table 1. To expand our knowledge on satellite DNAs in Cervidae, we also performed an analysis of partial satIII DNA sequence in eight Old world deer species (see Table 1) still missing data on satIII DNA sequence variability. Genomic DNA obtained previously [24] from peripheral lymphocytes was used for the analysis. Taxonomic nomenclature published by Groves and Grubb (2011) was used [5].

Table 1.
Species analyzed in this study.
2.2. Satellite DNA Isolation
Genomic DNA was obtained from fixed suspensions of cultured fibroblasts of the available Neotropical deer using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) after washing in PBS. Satellite DNA was isolated from the genomic DNA by PCR amplification using previously published primer sets [24] (Table S1). SatI, satII, and satIV DNA sequences were isolated from all Neotropical deer species. SatIII DNA was obtained only from M. gouazoubira, and the satIII DNA internal fragment (satIII-part) was isolated from all remaining Neotropical and Old-World deer samples available for this study. All PCR reactions were performed using Hot Start Combi PPP Master Mix (Top-Bio, Prague, Czech Republic) according to the manufacturer’s instructions. The obtained PCR products were cloned into the pDrive Cloning Vector (Qiagen, Hilden, Germany). Four different clones of each of satI, satII, satIII-part, and satIV DNA were selected in each species on the basis of their Hae III RFLP patterns (recognition site GG*CC) and subjected to sequencing.
2.3. Sequence Analysis
All satellite DNA sequences obtained in this study were screened for interspersed repeats using RepeatMasker (http://www.repeatmasker.org). The GC content was calculated using DNA/RNA GC Content Calculator (http://www.endmemo.com). All satellite sequences were also screened for a presence of the 17 bp CENP-B binding motif (NTTCGNNNNANNCGGGN) and the satI for the 31-bp subrepeat unit motif [35,36] using FIMO (version 5.1.0) software (http://meme-suite.org) [37]. The satellite DNA sequences obtained in this study were compared to cervid satellite sequences available in the NCBI database using BLASTN (https://blast.ncbi.nlm.nih.gov) and BLAST2 software was used to assess the sequence homology.
2.4. FISH
Cloned satI, satII, satIII, and satIV DNA of M. gouazoubira were labelled with Orange- or Green-dUTP (Abbott, Abbott Park, IL, USA) using Nick Translation Reagent Kit (Abbott) to serve as probes for comparative FISH. FISH was performed using standard protocols [19]. Hybridization signals were examined using Zeiss Axio Imager.Z2 fluorescence microscope (Carl Zeiss Microimaging GmbH, Jena, Germany) equipped with appropriate fluorescent filters and the Metafer Slide Scanning System (MetaSystems, Altlussheim, Germany). Images of well-spread metaphase cells were captured and analyzed using ISIS3 software (MetaSystems).
2.5. Phylogenetic Analysis
Multiple sequence alignments were constructed in MAFFT 7.474 [38] using the L-INS-i algorithm [39] for each satellite sequence separately. Alignments of cervid satellite DNA contain 1–8% of gaps [24], and gaps can influence phylogenetic reconstruction [40]. To capture phylogenetic information in gaps, the indels in the satI-IV alignments were re-coded to presence/absence data and used as a partition in phylogenetic reconstruction [22]. For the indel partition, gaps in a sequence were coded as 1, and all nucleotides were coded as 0. Optimal substitution models for DNA sequences were selected with the smart model selection algorithm 1.8.4 based on the Bayesian Information Criterion that utilized likelihood estimation implemented in PhyML 3.3 [41,42]. The phylogenetic trees were reconstructed in MrBayes 3.2 [43] in the partitioned analysis, capturing the DNA sequence variation and the phylogenetic information in the indels. The Markov Chains Monte Carlo (MCMC) were run for 2 million generations, sampled every thousandth generation. Two runs of four MCMC were run to ascertain efficient treespace search and check for convergence, following discarding 30% of initial samples as burn-in. The analyses were considered converged when the average standard deviation of split frequencies was <0.01 at the end of the run, potential scale reduction factors were ≈1.000 for each model parameter and frequency of swaps between neighboring chains was between 0.3 and 0.7. The trees were visualized in R [44] with help from packages ape [45], treeio [46], phytools [47], and RColorBrewer [48], where nodes with posterior probability ≥0.95 were considered supported. The trees were rooted at midpoint.
We downloaded cervid reference sequences of the mt-cyb gene from the NCBI database to reconstruct a phylogenetic tree from a mitochondrial marker and to compare the satDNA and mtDNA phylogenies. The analysis was performed analogically to the analysis of satDNA sequences, with the difference that the mtDNA marker did not contain gaps and the phylogeny was reconstructed from a single partition containing the DNA sequences.
3. Results
3.1. Sequence Analysis
In this study, newly obtained satI, satII, satIII-part, and satIV DNA sequences were analyzed in the Neotropical deer species including different M. americana cytotypes. The PCR product lengths, GC content, and sequence similarities among the individual satellite DNA clones are displayed in Table 2. Moreover, the satIII-part sequence was isolated and analyzed in C. elaphus, D. dama, R. eldii, M. reevesi, C. capreolus, R. tarandus, and A. alces. All satellite DNA sequences obtained in this study were deposited in the NCBI database (accession numbers MW273496–MW273692).

Table 2.
Characteristics of the satI-IV sequences based on four clones of each satellite DNA analyzed in each sample of Neotropical deer (see text for abbreviations of species names).
Using RepeatMasker, we did not find any SINE, LINE, or LTR elements in the analyzed satI, satII, and satIV DNA sequences. In all analyzed species, the predicted CENP-B binding motif was detected in the satII DNA starting at position 145–148 bp (Table S2). Eighteen to 21 copies of the 31-subrepeat unit motif were revealed in the satI DNA sequences (Table 2). The sequences of the 31-bp subrepeat unit showed a substantial intra- and interspecies variability. A higher similarity was observed between the subrepeat sequences in a particular satI monomer position in different species than among the subrepeat sequences in the same satI monomer. The 31-bp subrepeat sequence variance and its positions in the satI sequence in the analysed Neotropical deer are shown in Table S3.
Sequence similarity among satellite DNAs of Neotropical deer, Capreolinae, and Cervinae was compared using both sequences obtained in this study, and those available in the NCBI database (Table 3). SatI DNA sequences showed the highest intra- and interspecies variability. A high intra- and interspecies satellite DNA sequence similarity was observed in satII-satIV, even when the Neotropical deer satellite DNA was compared with sequences previously published in Capreolinae and Cervinae (Table 3).

Table 3.
Satellite DNA sequence similarity among Neotropical deer, Capreolinae, and Cervinae (Cervini and Muntjacini).
3.2. FISH
Probes for satI, satII, satIII, and satIV DNA obtained from M. gouazoubira were used for comparative FISH in the 11 Neotropical deer samples. The FISH results are summarized in Table 4 and displayed in Figure 1, Figure 2, Figure 3 and Figure 4.

Table 4.
Fluorescence in situ hybridization (FISH) patterns of the MGO satI-IV probes in Neotropical deer.
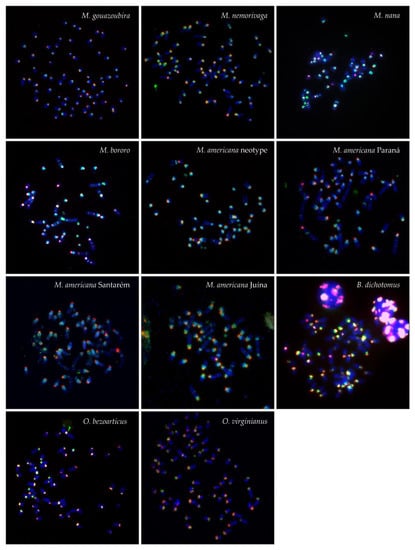
Figure 1.
FISH patterns of the satI (green) and satII (red) DNA probe in the analyzed species.
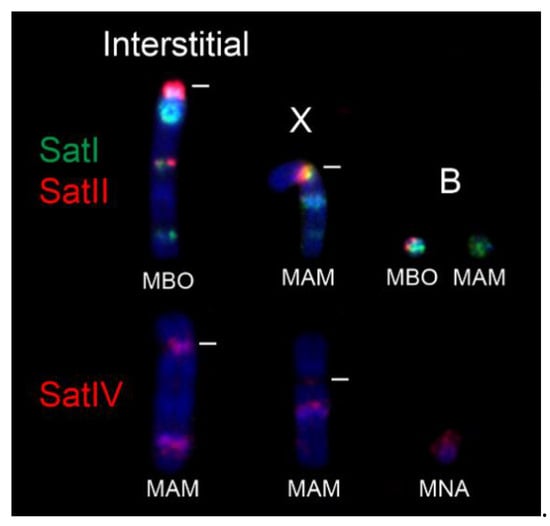
Figure 2.
Examples of centromeric and interstitial satI, satII, and satIV signals on autosomes, X chromosomes, and B chromosomes. MBO—M. bororo, MNA—M. nana, MAM—M. americana. Centromeres are indicated by white lines.
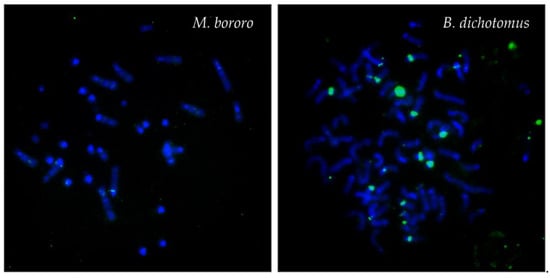
Figure 3.
Examples of FISH patterns of the satIII (green) DNA probe in selected species. FISH pattern similar to the results in M. bororo was observed in all species except B. dichotomus.
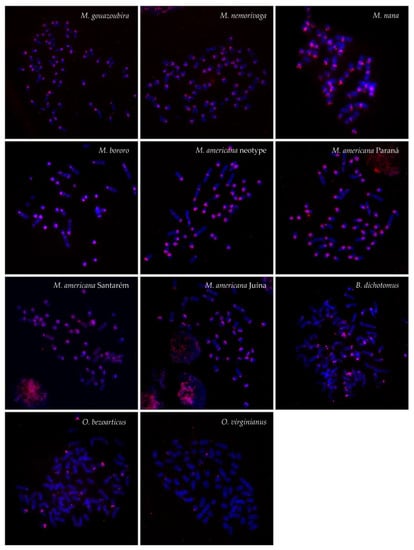
Figure 4.
FISH patterns of the satIV (red) DNA probe in the analyzed species.
In general, the MGO satI and satII probe produced centromeric signals on all autosomes (both acrocentric and bi-armed) and on the X chromosome in all analysed Neotropical deer (Figure 1 and Figure 2). In all animals with B chromosomes, the satI probe also marked Bs. Moreover, the satI probe produced interstitial signals on one or more chromosomes in M. nana, M. bororo and in all four analyzed cytotypes of M. americana. Signals of the satII probe were also observed interstitially in M. nana and M. bororo and on B chromosomes in M. bororo. Examples of the interstitial signals and signals on B chromosomes are displayed in detail in Figure 2.
Using the MGO satIII probe, none or only weak subcentromeric signals were detected in the Neotropical Cervidae. The only exception was B. dichotomus showing also large signals on several autosomes (Figure 3).
The MGO satIV probe hybridized to large regions of centromeric heterochromatin of all chromosomes in most Neotropical deer species and produced also interstitial signals in M. bororo and in all M. americana cytotypes. In addition, B chromosomes were marked in M. nana (Figure 2 and Figure 4). A different pattern was observed in B. dichotomus, O. bezoarticus, and the Brazilian O. virginianus, only showing very weak centromeric signals of the satIV probe on a few autosomes together with several intense signals in B. dichotomus and O. bezoarticus.
3.3. Phylogenetic Analysis
Together with previously published sequences, multiple sequence alignments consisted of 64 to 76 satI-IV sequences and 18 mt-cyb sequences of species from the family Cervidae (Table S4). The smart model selection algorithm suggested the GTR substitution model for the satI and satIII alignments, K80 model for the satII, and HKY model for the satIV and mt-cyb alignments. In all satDNA alignments, rate heterogeneity between sites was modelled according to the Γ distribution, and in the mt-cyb according to the proportion of invariable sites (Table S4). The satII and satIV phylogenies differentiated currently recognized tribes, with satII sequencing supporting monophyletic groups of Cervini, Muntiacini, Alceini, Capreolini, and Rangiferini (Figure 5B and Figure 6B).
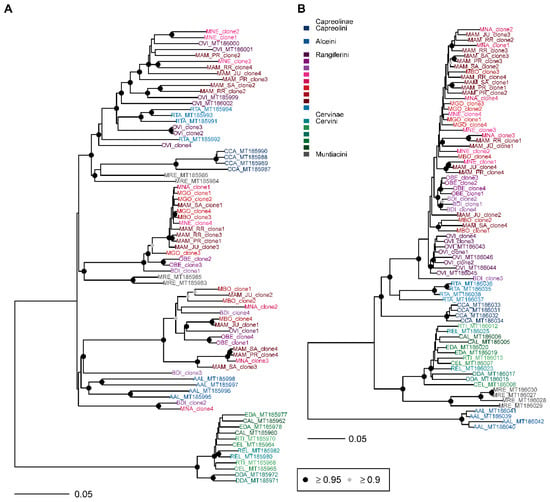
Figure 5.
Bayesian phylogenetic trees constructed from cervid satellite sequences. (A) SatI, (B) satII. AAL—Alces alces, BDI —Blastocerus dichotomus, CAL—Cervus albirostris, CCA—Capreolus capreolus, CEL—Cervus elaphus, DDA—Dama dama, EDA—Elaphurus davidianus, MAM—Mazama americana, MBO—Mazama bororo, MGO—Mazama gouazoubira, MNA—Mazama nana, MNE—Mazama nemorivaga, MRE—Muntiacus reevesi, OBE—Ozotoceros bezoarticus, OVI—Odocoileus virginianus, REL—Rucervus eldii, RTA—Rangifer tarandus, RTI—Rusa timorensis. Circles at nodes signify nodes with posterior probability ≥0.95 (black) and ≥0.90 (grey). Unmarked nodes were not supported.
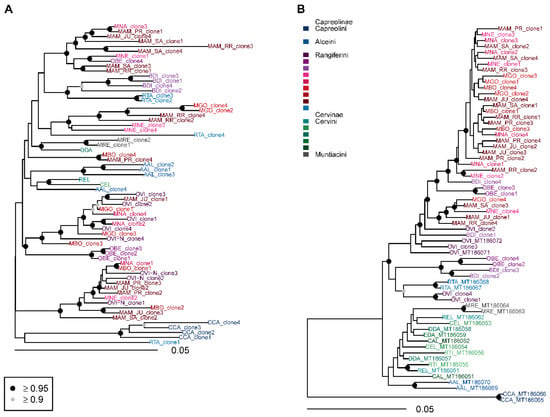
Figure 6.
Bayesian phylogenetic trees constructed from cervid satellite sequences. (A) SatIII, (B) satIV. AAL—Alces alces, BDI—Blastocerus dichotomus, CAL—Cervus albirostris, CCA—Capreolus capreolus, CEL—Cervus elaphus, DDA—Dama dama, EDA—Elaphurus davidianus, MAM—Mazama americana, MBO—Mazama bororo, MGO—Mazama gouazoubira, MNA—Mazama nana, MNE—Mazama nemorivaga, MRE—Muntiacus reevesi, OBE—Ozotoceros bezoarticus, OVI—Odocoileus virginianus, REL—Rucervus eldii, RTA—Rangifer tarandus, RTI—Rusa timorensis. Circles at nodes signify nodes with posterior probability ≥0.95 (black) and ≥0.90 (grey). Unmarked nodes were not supported.
In satI phylogeny, a monophyletic relationship of satellite sequences was supported for Cervini, Alceini, and Capreolini, however Rangiferini and Muntiacini were polyphyletic (Figure 5A). The Neotropical deer satI sequences were diverged and formed three deeply differentiated lineages (Figure 6A). Clones from multiple species (M. americana, M. nemorivaga, M. bororo, M. nana, O. bezoarticus, B. dichotomus) were represented in more than one lineage. In M. americana, clones from all sampled regions were present in all three lineages. The first lineage included Mazama, Odocoileus (both populations), and Holarctic Rangifer in incomplete sorting of sequences at the species level, and the lineage formed a sister relationship to Capreolus and Muntiacus. The second Neotropical deer satI lineage consisted of Mazama, Ozotoceros, and Blastocerus as a sister group to two Muntiacus satI sequences. SatI clones from M. gouazoubira all belonged to the second lineage. The genera of Neotropical deer grouping in the third satI lineage were also Mazama, Ozotoceros, and Blastocerus, but no clear sister relationship was identified in the satI phylogeny (Figure 6A).
Neotropical deer formed a single supported lineage in the satII sequences with supported monophyly of Odocoileus, Ozotoceros, and Blastocerus (Figure 6B). The latter two taxa formed a paraphyletic sister relationship with respect to Mazama. Notably, BDI_clone3 formed a long branch at the base of Neotropical Rangiferini. Similar relationships were retrieved in the satIV phylogeny, but additional clones from all Neotropical genera grouped within the Mazama lineage (Figure 6B).
Low divergence in satIII-part (Table 3) resulted in incomplete lineage sorting in Neotropical deer and a polytomy at the deep divergence of Cervidae (Figure 6A).
The mt-cyb phylogeny showed monophyletic groups representing the tribes Rangiferini and Cervini (Figure S1). Muntiacini were sister to Cervini, and Capreolini and Alceini diverged rapidly close to the root of the tree. In Neotropical deer, mtDNA phylogeny did not support monophyly of Mazama similarly as was shown in the satDNA trees. Instead, Mazama were paraphyletic, with M. nana, M. bororo, and M. americana forming a single lineage sister to O. virginianus, and M. gouazoubira and M. nemorivaga forming an unresolved group with B. dichotomus and O. bezoarticus (Figure S1).
4. Discussion
Sequence Comparisons
Cervidae is a diverse group of species distributed in Eurasia and North and South America. Among them, Neotropical deer species have still been understudied in terms of current taxonomy and phylogenetic relationships [49]. In this study, we performed a comparative sequence and FISH analysis of the four main cervid satellite DNA families (satI-IV) isolated from a variety of Neotropical deer species, including several specific cytotypes.
Our sequence comparisons revealed close relationships among the studied Neotropical deer in satII, satIII and satIV DNA that also showed a high similarity to satellite DNA sequences available in the NCBI database for other species of Capreolinae. Regarding satI DNA, it showed the highest intra- and interspecific variability indicating a fast satI sequence evolution at the time of early divergence of the Capreolinae subfamily. The satI and satII monomer lengths were comparable to previously studied Capreolinae [24]. However, the number of the 31-bp satI internal subrepeat units was lower in the studied Neotropical deer than in other Capreolinae, and closer to that published in Cervini [24]. The general occurrence of this subrepeat throughout many bovid and cervid genomes [21,24,28,50] indicates its possible biological function. This suggestion is also supported by the interspecies similarity in the 31-bp subrepaeat sequences at the individual positions of the satI DNA monomers. This sequence might form a part of a 3D structure involved in protein or siRNA binding during heterochromatin formation, cell division, or transcription regulation. However, further studies are needed to elucidate its functions.
The highest GC content and the presence of the CENP-B binding motif, known to be associated with the centromeric function [51,52], were detected in the satII DNA in all studied Neotropical deer. These findings support the previously published hypothesis that satII DNA might represent the most important satellite DNA family in Cervidae [24] but the satII DNA significance has yet to be confirmed by functional studies. The sequence of the 17-bp CENP-B motif was identical throughout most analyzed Neotropical deer samples indicating a high level of conservation. The most common CENP-B motif sequence detected in this study (TTTGGAGGCAGGCGGGG) contained the published human core recognition sequence (NTTCGNNNNANNCGGGN) [53] with one nucleotide difference (C-G substitution). However, three different one-nucleotide substitutions from the deer core sequence were revealed in M. bororo, indicating a surprising CENP-B motif variance in this species that probably originated during its separate evolution.
In Cervidae, satI and satII DNA sequences are highly abundant, occupying 2–35% of the genomic DNA [33]. On the other hand, low copy numbers of satIII and satIV DNA not detectable by FISH were previously reported in many deer species [24]. In this study, we also observed significant differences in the abundance of the satellite DNA families by FISH. The high copy numbers of satI and satII DNA, reflected by large hybridization signals, were in contrast to very weak satIII signals only present on one or a few chromosomes in all studied species except B. dichotomus. Regarding satIV DNA, there were significant differences in its abundance between O. virginianus, B. dichotomus and O. bezoarticus on one side, and species of the genus Mazama on the other, the latter showing large satIV DNA blocks. The observed interspecies differences in the satellite DNA abundance at the relatively high sequence similarity of the repeat units can be related to the satellite DNA evolution. It is generally accepted that satellite DNAs are formed by a fast amplification of monomers existing in ancestral genomes [54]. As suggested by a library model, related species share a common collection of satellite DNAs, which vary in their abundance, with specific sequences being differentially amplified in individual species [55,56,57]. Satellite DNA arrays are probably formed by a mechanism of rolling circle replication with subsequent further amplification by unequal crossing over [58,59,60,61]. The existing sequences are then diversified by mutations, which spread through the genome by mechanisms of concerted evolution, leading to the formation of species- and chromosome-specific satellite sequences [13,62,63,64]. Accordingly, interspecies differences in satellite DNA abundance and sequences were found to be highly consistent with species phylogeny [16,22,24,56,63,65].
In this study, the satII DNA sequence phylogeny well corresponds to the mtDNA (mt-cyb gene) phylogeny (Figure S1) and the published taxonomic divergences in Cervidae [24]. The satII phylogeny differentiated the currently recognized tribes, suggesting monophyletic origin of Cervini, Muntiacini, Alceini, Capreolini, and Rangiferini. In Neotropical deer, O. bezoarticus was closely related to B. dichotomus, and both Neotropical and Nearctic populations of O. virginianus formed one lineage without defined intraspecific relationships. SatII and satIV sequences of Mazama showed unresolved relationships both at the level of species differentiation as well as at the genus level. Incomplete lineage sorting of Mazama species with respect to other Neotropical deer consistently occurs in satellite DNA (this study), mtDNA phylogenies (Figure S1 and [66,67]), and even with the molecular and morphological data combined [68]. Neotropical deer, as descendants of Nearctic ancestors that arrived to South America during the Great America Biotic Interchange between late Miocene and late Pleistocene [69], diverged in an explosive radiation, forming morphologically well-defined but genetically unresolved genera. As in other Neotropical mammals, morphological convergence in genetically diverged taxa could be attributed to ecological adaptations to specific niche partitioning in newly colonized regions [70,71]). In this study, the explosive radiation in Neotropical deer can be best observed in the satI sequence phylogeny. At least four distinct lineages first began to diverge at the time of the split of Capreolinae. Genera Rangifer, Capreolus, and Alces each retained one lineage of satI sequences, but in Neotropical deer, satI sequences diversified three times independently.
The presence of genomic satellite DNA arrays can facilitate the formation of chromosome rearrangements and thus karyotype and species evolution [29,72,73]. The deer chromosome evolution from the ancestral karyotype (2n = 70) was driven by centric and tandem fusions at the simultaneous reduction in the chromosome number [9]. Moreover, the existence of centric fusion polymorphisms previously described in the genus Mazama indicate that chromosome fusions represent an important source of the recent and ongoing karyotype evolution of this taxon [74,75,76]. Despite the predominantly peri/centromeric location of the satellite DNA, we also observed interstitial satI, satII, and satIV FISH signals in Mazama species with highly reduced chromosome numbers (M. nana, M. bororo, M. americana cytotypes). Similar finding of interstitial satellite DNA signals at the tandem fusion sites, and even their co-localization with telomeric sequences was previously reported in muntjacs [30,32,77,78,79].
In M. americana, a multiple sexual system resulting from evolutionary X-autosomal fusions was previously described [12,80]. Among deer, the XY1Y2 system was found also in the genus Muntiacus [81,82]. It is known that sex-autosomal translocations are usually associated with a disturbed process of X chromosome inactivation and with a meiotic disruption [83,84,85]. However, heterochromatin blocks intercalated between the gonosomal and autosomal parts of the rearranged sex-autosomes can serve as effective barriers for spreading of somatic X-chromosome inactivation in females and for regulation of meiotic processes in males [86,87,88]. In our study, the presence of intercalated heterochromatin in the X-autosomal fusion region suggested by a distinct DAPI band was proved by detection of satI, less frequently satII and satIV hybridisation signals in all M. americana cytotypes. As discussed in the previous paragraph, also these interstitial satellite DNA signals probably map to a historical fusion site and represent former centromeric heterochromatin of the autosome fused to the ancestral X chromosome.
Another interesting karyotype feature of Mazama species analyzed in this study is the presence of B chromosomes [7,12,75,80]. B chromosomes are supernumerary, mitotically unstable chromosomes that were described in some animal, plant, and fungal species [89]. Their numbers vary in different individuals and even individual cells of the organism, and, despite a presence of duplicated coding genes detected in Bs in some species [90,91], their biological function is unknown. A recent study based on comparative FISH and next-generation sequencing of B chromosomes in two deer species, Capreolus pygargus (Capreolini) and M. gouazoubira (Rangiferini) demonstrated an independent origin of B chromosomes in these species, and their different evolutionary history [92]. In all animals carrying B chromosomes in our study, the Bs showed FISH signals of the satI DNA probe. The independent divergent lineages observed in Neotropical deer in the satI phylogeny might be attributed to satI sequences on the B chromosomes. Moreover, the satII DNA signals were detected on Bs in M. bororo. This might indicate either an independent origin or a more primitive stage of the B chromosomes in M. bororo, which have not yet led to an evolutionary satII DNA sequence degeneration in this species.
5. Conclusions
The Neotropical deer species show high intra- and interspecies satellite DNA sequence similarities indicating close evolutionary relationships. The high satellite DNA abundance probably stands behind the cervid karyotype differentiation driven by centric and tandem fusions at the simultaneous reduction of the chromosome number.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/12/1/123/s1: Table S1. Primers and annealing temperatures for satellite DNA isolation. Table S2. Positions and sequences of the CENP-B binding motif in individual satII DNA clones. Table S3. The 31-bp subrepeat sequence variance and its positions in the satI DNA sequence. Table S4. Multiple sequence alignment composition and selected substitution models of cervid satellite and mitochondrial sequences. Figure S1. Bayesian phylogenetic tree constructed from cervid mt-cyb sequences. AAL—Alces alces, BDI—Blastocerus dichotomus, CAL—Cervus albirostris, CCA—Capreolus capreolus, CEL—Cervus elaphus, DDA—Dama dama, EDA—Elaphurus davidianus, MAM—Mazama americana, MBO—Mazama bororo, MGO—Mazama gouazoubira, MNA—Mazama nana, MNE—Mazama nemorivaga, MRE—Muntiacus reevesi, OBE—Ozotoceros bezoarticus, OVI—Odocoileus virginianus, REL—Rucervus eldii, RTA—Rangifer tarandus, RTI—Rusa timorensis. Circles at nodes signify nodes with posterior probability ≥0.95 (black) and ≥0.90 (grey). Unmarked nodes were not supported.
Author Contributions
Conceptualization, S.K., M.V. and J.R.; investigation, M.V., S.K., N.M., A.M.B., D.J.G., H.C., D.K. and P.M.; methodology, M.V. and S.K. and N.M.; supervision, J.R. and J.M.D.; writing—original draft, M.V., S.K. and N.M.; writing—review and editing, D.J.G., J.M.D. and J.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Czech Science Foundation grant number 20-22517J, the São Paulo Research Foundation FAPESP 2019/06940-1-GACR, FONDECYT grant No. 116-2017, Ministry of Agriculture of the Czech Republic, grant No. RO 0520, and Ministry of Education, Youth and Sports of the Czech Republic under the project CEITEC 2020, grant No. LQ1601.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee on Animal Use of the School of Agricultural and Veterinarian Sciences, São Paulo State University (protocol code 005433/19).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the article and supplementary material.
Acknowledgments
The authors wish to thank to the whole NUPECCE team for their collaborative support.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Ethical Statement
The biological material for tissue culture was obtained by a veterinarian during medical examination of the animals. All procedures performed in this study were in accordance with the ethical standards of the Veterinary Research Institute (Brno, Czech Republic), which complies with the Czech and European Union Legislation for the protection of animals used for scientific purposes.
References
- Wilson, D.E.; Reeder, D.M. Mammal Species of the World: A Taxonomic and Geographic Reference; Johns Hopkins University Press: Baltimore, MD, USA, 2005; ISBN 978-0-8018-8221-0. [Google Scholar]
- Chen, L.; Qiu, Q.; Jiang, Y.; Wang, K.; Lin, Z.; Li, Z.; Bibi, F.; Yang, Y.; Wang, J.; Nie, W.; et al. Large-Scale Ruminant Genome Sequencing Provides Insights into Their Evolution and Distinct Traits. Science 2019, 364. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, E.E.; Helgen, K.M.; McDonough, M.M.; Bauer, F.; Hawkins, M.T.R.; Escobedo-Morales, L.A.; Patterson, B.D.; Maldonado, J.E. A Gene-Tree Test of the Traditional Taxonomy of American Deer: The Importance of Voucher Specimens, Geographic Data, and Dense Sampling. ZooKeys 2017, 697, 87–131. [Google Scholar] [CrossRef]
- Duarte, J.M.B.; González, S.; Maldonado, J.E. The Surprising Evolutionary History of South American Deer. Mol. Phylogenet. Evol. 2008, 49, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Groves, C.; Grubb, P. Ungulate Taxonomy, 1st ed.; Johns Hopkins University Press: Baltimore, MD, USA, 2011; ISBN 978-1-4214-0093-8. [Google Scholar]
- Fontana, F.; Rubini, M. Chromosomal Evolution in Cervidae. BioSystems 1990, 24, 157–174. [Google Scholar] [CrossRef]
- Duarte, J.M.B.; Jorge, W. Morphologic and Cytogenetic Description of the Small Red Brocket (Mazama Bororo Duarte,1996) in Brazil. Mammalia 2009, 67, 403–410. [Google Scholar] [CrossRef]
- Huang, L.; Chi, J.; Nie, W.; Wang, J.; Yang, F. Phylogenomics of Several Deer Species Revealed by Comparative Chromosome Painting with Chinese Muntjac Paints. Genetica 2006, 127, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Nietzel, H. Chromosome Evolution of Cervidae: Karyotypic and Molecular Aspects. In Cytogenetics: Basic and Applied Aspects; Obe, G., Basler, A., Eds.; Springer: Berlin/Heidelberg, Germany, 1987; ISBN 978-3-642-72804-4. [Google Scholar]
- Cifuentes-Rincón, A.; Morales-Donoso, J.A.; Sandoval, E.D.P.; Tomazella, I.M.; Mantellatto, A.M.B.; de Thoisy, B.; Duarte, J.M.B. Designation of a Neotype for Mazama Americana (Artiodactyla, Cervidae) Reveals a Cryptic New Complex of Brocket Deer Species. ZooKeys 2020, 958, 143–164. [Google Scholar] [CrossRef]
- Cursino, M.S.; Salviano, M.B.; Abril, V.V.; Zanetti, E.; dos, S.; Duarte, J.M. The Role of Chromosome Variation in the Speciation of the Red Brocket Deer Complex: The Study of Reproductive Isolation in Females. BMC Evol. Biol. 2014, 14, 40. [Google Scholar] [CrossRef]
- Abril, V.V.; Carnelossi, E.A.G.; González, S.; Duarte, J.M.B. Elucidating the Evolution of the Red Brocket Deer Mazama Americana Complex (Artiodactyla; Cervidae). Cytogenet. Genome Res. 2010, 128, 177–187. [Google Scholar] [CrossRef]
- Plohl, M.; Luchetti, A.; Mestrović, N.; Mantovani, B. Satellite DNAs between Selfishness and Functionality: Structure, Genomics and Evolution of Tandem Repeats in Centromeric (Hetero)Chromatin. Gene 2008, 409, 72–82. [Google Scholar] [CrossRef]
- Ugarković, D.; Plohl, M. Variation in Satellite DNA Profiles—Causes and Effects. EMBO J. 2002, 21, 5955–5959. [Google Scholar] [CrossRef] [PubMed]
- Schalch, T.; Steiner, F.A. Structure of Centromere Chromatin: From Nucleosome to Chromosomal Architecture. Chromosoma 2017, 126, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Kunze, B.; Traut, W.; Garagna, S.; Weichenhan, D.; Redi, C.A.; Winking, H. Pericentric Satellite DNA and Molecular Phylogeny in Acomys (Rodentia). Chromosome Res. 1999, 7, 131. [Google Scholar] [CrossRef] [PubMed]
- Louzada, S.; Vieira-da-Silva, A.; Mendes-da-Silva, A.; Kubickova, S.; Rubes, J.; Adega, F.; Chaves, R. A Novel Satellite DNA Sequence in the Peromyscus Genome (PMSat): Evolution via Copy Number Fluctuation. Mol. Phylogenet. Evol. 2015, 92, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Baicharoen, S.; Miyabe-Nishiwaki, T.; Arsaithamkul, V.; Hirai, Y.; Duangsa-ard, K.; Siriaroonrat, B.; Domae, H.; Srikulnath, K.; Koga, A.; Hirai, H. Locational Diversity of Alpha Satellite DNA and Intergeneric Hybridization Aspects in the Nomascus and Hylobates Genera of Small Apes. PLoS ONE 2014, 9, e109151. [Google Scholar] [CrossRef]
- Vozdova, M.; Kubickova, S.; Cernohorska, H.; Fröhlich, J.; Vodicka, R.; Rubes, J. Comparative Study of the Bush Dog (Speothos venaticus) Karyotype and Analysis of Satellite DNA Sequences and Their Chromosome Distribution in Six Species of Canidae. Cytogenet. Genome Res. 2019, 159, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Vozdova, M.; Kubickova, S.; Cernohorska, H.; Fröhlich, J.; Rubes, J. Satellite DNA Sequences in Canidae and Their Chromosome Distribution in Dog and Red Fox. Cytogenet. Genome Res. 2016, 150, 118–127. [Google Scholar] [CrossRef]
- Jobse, C.; Buntjer, J.B.; Haagsma, N.; Breukelman, H.J.; Beintema, J.J.; Lenstra, J.A. Evolution and Recombination of Bovine DNA Repeats. J. Mol. Evol. 1995, 41, 277–283. [Google Scholar] [CrossRef]
- Kopecna, O.; Kubickova, S.; Cernohorska, H.; Cabelova, K.; Vahala, J.; Martinkova, N.; Rubes, J. Tribe-Specific Satellite DNA in Non-Domestic Bovidae. Chromosome Res. 2014, 22, 277–291. [Google Scholar] [CrossRef]
- Chaves, R.; Guedes-Pinto, H.; Heslop-Harrison, J.S. Phylogenetic Relationships and the Primitive X Chromosome Inferred from Chromosomal and Satellite DNA Analysis in Bovidae. Proc. Biol. Sci. 2005, 272, 2009–2016. [Google Scholar] [CrossRef]
- Vozdova, M.; Kubickova, S.; Cernohorska, H.; Fröhlich, J.; Martínková, N.; Rubes, J. Sequence Analysis and FISH Mapping of Four Satellite DNA Families among Cervidae. Genes 2020, 11, 584. [Google Scholar] [CrossRef] [PubMed]
- Slamovits, H.C.; Cook, J.A.; Lessa, E.P.; Rossi, M.S. Recurrent Amplifications and Deletions of Satellite DNA Accompanied Chromosomal Diversification in South American Tuco-Tucos (Genus Ctenomys, Rodentia: Octodontidae): A Phylogenetic Approach. Mol. Biol. Evol. 2001, 18, 1708–1719. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, L.; Sperlich, D. Gradual Evolution of a Specific Satellite DNA Family in Drosophila Ambigua, D. Tristis, and D. Obscura. Mol. Biol. Evol. 1993, 10, 647–659. [Google Scholar] [PubMed]
- Garrido-Ramos, M.A.; de la Herrán, R.; Jamilena, M.; Lozano, R.; Ruiz Rejón, C.; Ruiz Rejón, M. Evolution of Centromeric Satellite DNA and Its Use in Phylogenetic Studies of the Sparidae Family (Pisces, Perciformes). Mol. Phylogenet. Evol. 1999, 12, 200–204. [Google Scholar] [CrossRef]
- Lee, C.; Court, D.R.; Cho, C.; Haslett, J.L.; Lin, C.C. Higher-Order Organization of Subrepeats and the Evolution of Cervid Satellite I DNA. J. Mol. Evol. 1997, 44, 327–335. [Google Scholar] [CrossRef]
- Li, Y.-C.; Lin, C.-C. Cervid Satellite DNA and Karyotypic Evolution of Indian Muntjac. Genes Genom. 2012, 34, 7–11. [Google Scholar] [CrossRef]
- Liu, Y.; Nie, W.; Huang, L.; Wang, J.; Su, W.; Lin, C.; Yang, F. Cloning, Characterization, and FISH Mapping of Four Satellite DNAs from Black Muntjac (Muntiacus crinifrons) and Fea’s Muntjac (M. feae). Zoolog. Res. 2008, 2008, 225–235. [Google Scholar] [CrossRef][Green Version]
- Lin, C.C.; Li, Y.C. Chromosomal Distribution and Organization of Three Cervid Satellite DNAs in Chinese Water Deer (Hydropotes inermis). Cytogenet. Genome Res. 2006, 114, 147–154. [Google Scholar] [CrossRef]
- Li, Y.C.; Lee, C.; Chang, W.S.; Li, S.-Y.; Lin, C.C. Isolation and Identification of a Novel Satellite DNA Family Highly Conserved in Several Cervidae Species. Chromosoma 2002, 111, 176–183. [Google Scholar] [CrossRef]
- Blake, R.D.; Wang, J.Z.; Beauregard, L. Repetitive Sequence Families in Alces Alces Americana. J. Mol. Evol. 1997, 44, 509–520. [Google Scholar] [CrossRef]
- Li, Y.C.; Lee, C.; Hseu, T.H.; Li, S.Y.; Lin, C.C.; Hsu, T.H. Direct Visualization of the Genomic Distribution and Organization of Two Cervid Centromeric Satellite DNA Families. Cytogenet. Cell Genet. 2000, 89, 192–198. [Google Scholar] [CrossRef]
- Płucienniczak, A.; Skowroński, J.; Jaworski, J. Nucleotide Sequence of Bovine 1.715 Satellite DNA and Its Relation to Other Bovine Satellite Sequences. J. Mol. Biol. 1982, 158, 293–304. [Google Scholar] [CrossRef]
- Masumoto, H.; Masukata, H.; Muro, Y.; Nozaki, N.; Okazaki, T. A Human Centromere Antigen (CENP-B) Interacts with a Short Specific Sequence in Alphoid DNA, a Human Centromeric Satellite. J. Cell Biol. 1989, 109, 1963–1973. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for Occurrences of a given Motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, D.K. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT Version 5: Improvement in Accuracy of Multiple Sequence Alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Machado, D.J.; Castroviejo-Fisher, S.; Grant, T. Evidence of Absence Treated as Absence of Evidence: The Effects of Variation in the Number and Distribution of Gaps Treated as Missing Data on the Results of Standard Maximum Likelihood Analysis. Mol. Phylogenet. Evol. 2021, 154, 106966. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Paradis, E.; Schliep, K. Ape 5.0: An Environment for Modern Phylogenetics and Evolutionary Analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Wang, L.-G.; Lam, T.T.-Y.; Xu, S.; Dai, Z.; Zhou, L.; Feng, T.; Guo, P.; Dunn, C.W.; Jones, B.R.; Bradley, T.; et al. Treeio: An R Package for Phylogenetic Tree Input and Output with Richly Annotated and Associated Data. Mol. Biol. Evol. 2020, 37, 599–603. [Google Scholar] [CrossRef]
- Revell, L.J. Phytools: An R Package for Phylogenetic Comparative Biology (and Other Things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Neuwirth, E. RColorBrewer: ColorBrewer Palettes. 2014. Available online: https://CRAN.R-project.org/package=RColorBrewer (accessed on 7 December 2014).
- Barbanti Duarte, J.M.; González, S. Neotropical Cervidology, Biology and Medicine of Latin American Deer. Available online: https://www.iucn.org/content/neotropical-cervidology (accessed on 23 November 2020).
- Bogenberger, J.M.; Neumaier, P.S.; Fittler, F. The Muntjak Satellite IA Sequence Is Composed of 31-Base-Pair Internal Repeats That Are Highly Homologous to the 31-Base-Pair Subrepeats of the Bovine Satellite 1. Eur. J. Biochem. 1985, 148, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Vafa, O.; Shelby, R.D.; Sullivan, K.F. CENP-A Associated Complex Satellite DNA in the Kinetochore of the Indian Muntjac. Chromosoma 1999, 108, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Carroll, C.W.; Silva, M.C.C.; Godek, K.M.; Jansen, L.E.T.; Straight, A.F. Centromere Assembly Requires the Direct Recognition of CENP-A Nucleosomes by CENP-N. Nat. Cell Biol. 2009, 11, 896–902. [Google Scholar] [CrossRef] [PubMed]
- Suntronpong, A.; Kugou, K.; Masumoto, H.; Srikulnath, K.; Ohshima, K.; Hirai, H.; Koga, A. CENP-B Box, a Nucleotide Motif Involved in Centromere Formation, Occurs in a New World Monkey. Biol. Lett. 2016, 12. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-Throughput Analysis of the Satellitome Illuminates Satellite DNA Evolution. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Mestrović, N.; Plohl, M.; Mravinac, B.; Ugarković, D. Evolution of Satellite DNAs from the Genus Palorus--Experimental Evidence for the “Library” Hypothesis. Mol. Biol. Evol. 1998, 15, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Gimenez, O.M.; Milani, D.; Song, H.; Marti, D.A.; López-León, M.D.; Ruiz-Ruano, F.J.; Camacho, J.P.M.; Cabral-de-Mello, D.C. Eight Million Years of Satellite DNA Evolution in Grasshoppers of the Genus Schistocerca Illuminate the Ins and Outs of the Library Hypothesis. Genome Biol. Evol. 2020, 12, 88–102. [Google Scholar] [CrossRef]
- Fry, K.; Salser, W. Nucleotide Sequences of HS-α Satellite DNA from Kangaroo Rat Dipodomys Ordii and Characterization of Similar Sequences in Other Rodents. Cell 1977, 12, 1069–1084. [Google Scholar] [CrossRef]
- Walsh, J.B. Persistence of Tandem Arrays: Implications for Satellite and Simple-Sequence DNAs. Genetics 1987, 115, 553–567. [Google Scholar] [CrossRef] [PubMed]
- Okumura, K.; Kiyama, R.; Oishi, M. Sequence Analyses of Extrachromosomal Sau3A and Related Family DNA: Analysis of Recombination in the Excision Event. Nucleic Acids Res. 1987, 15, 7477–7489. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alkan, C.; Eichler, E.E.; Bailey, J.A.; Sahinalp, S.C.; Tüzün, E. The Role of Unequal Crossover in Alpha-Satellite DNA Evolution: A Computational Analysis. J. Comput. Biol. 2004, 11, 933–944. [Google Scholar] [CrossRef] [PubMed]
- Stephan, W. Recombination and the Evolution of Satellite DNA. Genet. Res. 1986, 47, 167–174. [Google Scholar] [CrossRef]
- Dover, G.A. Molecular Drive in Multigene Families: How Biological Novelties Arise, Spread and Are Assimilated. Trends Genet. 1986, 2, 159–165. [Google Scholar] [CrossRef]
- Lorite, P.; Muñoz-López, M.; Carrillo, J.A.; Sanllorente, O.; Vela, J.; Mora, P.; Tinaut, A.; Torres, M.I.; Palomeque, T. Concerted Evolution, a Slow Process for Ant Satellite DNA: Study of the Satellite DNA in the Aphaenogaster Genus (Hymenoptera, Formicidae). Org. Divers. Evol. 2017, 17, 595–606. [Google Scholar] [CrossRef]
- Durfy, S.J.; Willard, H.F. Concerted Evolution of Primate Alpha Satellite DNA: Evidence for an Ancestral Sequence Shared by Gorilla and Human X Chromosome Alpha Satellite. J. Mol. Biol. 1990, 216, 555–566. [Google Scholar] [CrossRef]
- Mravinac, B.; Plohl, M. Parallelism in Evolution of Highly Repetitive DNAs in Sibling Species. Mol. Biol. Evol. 2010, 27, 1857–1867. [Google Scholar] [CrossRef]
- Mantellatto, A.M.B.; González, S.; Duarte, J.M.B. Molecular Identification of Mazama Species (Cervidae: Artiodactyla) from Natural History Collections. Genet. Mol. Biol. 2020, 43. [Google Scholar] [CrossRef]
- Escobedo-Morales, L.A.; Mandujano, S.; Eguiarte, L.E.; Rodríguez-Rodríguez, M.A.; Maldonado, J.E. First Phylogenetic Analysis of Mesoamerican Brocket Deer Mazama Pandora and Mazama Temama (Cetartiodactyla: Cervidae) Based on Mitochondrial Sequences: Implications for Neotropical Deer Evolution. Mammal. Biol. 2016, 81, 303–313. [Google Scholar] [CrossRef]
- Heckeberg, N.S. The Systematics of the Cervidae: A Total Evidence Approach. PeerJ 2020, 8, e8114. [Google Scholar] [CrossRef]
- Carrillo, J.D.; Faurby, S.; Silvestro, D.; Zizka, A.; Jaramillo, C.; Bacon, C.D.; Antonelli, A. Disproportionate Extinction of South American Mammals Drove the Asymmetry of the Great American Biotic Interchange. Proc. Natl. Acad. Sci. USA 2020, 117, 26281–26287. [Google Scholar] [CrossRef]
- Carrizo, L.V.; Tulli, M.J.; Santos, D.A.D.; Abdala, V. Interplay between Postcranial Morphology and Locomotor Types in Neotropical Sigmodontine Rodents. J. Anat. 2014, 224, 469–481. [Google Scholar] [CrossRef]
- Pečnerová, P.; Moravec, J.C.; Martínková, N. A Skull Might Lie: Modeling Ancestral Ranges and Diet from Genes and Shape of Tree Squirrels. Syst. Biol. 2015, 64, 1074–1088. [Google Scholar] [CrossRef]
- Barra, V.; Fachinetti, D. The Dark Side of Centromeres: Types, Causes and Consequences of Structural Abnormalities Implicating Centromeric DNA. Nat. Commun. 2018, 9, 4340. [Google Scholar] [CrossRef]
- Puppo, I.L.; Saifitdinova, A.F.; Tonyan, Z.N. The Role of Satellite DNA in Causing Structural Rearrangements in Human Karyotype. Russ. J. Genet. 2020, 56, 41–47. [Google Scholar] [CrossRef]
- Aquino, C.I.; Abril, V.V.; Duarte, J.M.B. Meiotic Pairing of B Chromosomes, Multiple Sexual System, and Robertsonian Fusion in the Red Brocket Deer Mazama Americana (Mammalia, Cervidae). Genet. Mol. Res. GMR 2013, 12, 3566–3574. [Google Scholar] [CrossRef]
- Abril, V.V.; Duarte, J.M.B. Chromosome Polymorphism in the Brazilian Dwarf Brocket Deer, Mazama Nana (Mammalia, Cervidae). Genet. Mol. Biol. 2008, 31, 53–57. [Google Scholar] [CrossRef]
- Valeri, P.M.; Tomazella, I.M.; Duarte, J.M.B. Intrapopulation Chromosomal Polymorphism in Mazama Gouazoubira (Cetartiodactyla; Cervidae): The Emergence of a New Species? Cytogenet. Genome Res. 2018, 154, 147–152. [Google Scholar] [CrossRef]
- Li, Y.C.; Lee, C.; Sanoudou, D.; Hseu, T.H.; Li, S.Y.; Lin, C.C.; Hsu, T.H. Interstitial Colocalization of Two Cervid Satellite DNAs Involved in the Genesis of the Indian Muntjac Karyotype. Chrom. Res. 2000, 8, 363–373. [Google Scholar] [CrossRef]
- Yang, F.; O’Brien, P.C.; Wienberg, J.; Neitzel, H.; Lin, C.C.; Ferguson-Smith, M.A. Chromosomal Evolution of the Chinese Muntjac (Muntiacus Reevesi). Chromosoma 1997, 106, 37–43. [Google Scholar] [CrossRef]
- Hartmann, N.; Scherthan, H. Characterization of Ancestral Chromosome Fusion Points in the Indian Muntjac Deer. Chromosoma 2004, 112, 213–220. [Google Scholar] [CrossRef]
- Fiorillo, B.F.; Sarria-Perea, J.A.; Abril, V.V.; Duarte, J.M.B. Cytogenetic Description of the Amazonian Brown Brocket Mazama Nemorivaga (Artiodactyla, Cervidae). Comp. cytogenet. 2013, 7, 25–31. [Google Scholar] [CrossRef]
- Yang, F.; Carter, N.P.; Shi, L.; Ferguson-Smith, M.A. A Comparative Study of Karyotypes of Muntjacs by Chromosome Painting. Chromosoma 1995, 103, 642–652. [Google Scholar] [CrossRef]
- Yang, F.; O’Brien, P.C.; Wienberg, J.; Ferguson-Smith, M.A. A Reappraisal of the Tandem Fusion Theory of Karyotype Evolution in Indian Muntjac Using Chromosome Painting. Chrom. Res. 1997, 5, 109–117. [Google Scholar] [CrossRef]
- Ashley, T. X-Autosome Translocations, Meiotic Synapsis, Chromosome Evolution and Speciation. Cytogenet. Genome Res. 2002, 96, 33–39. [Google Scholar] [CrossRef]
- Cotton, A.M.; Chen, C.-Y.; Lam, L.L.; Wasserman, W.W.; Kobor, M.S.; Brown, C.J. Spread of X-Chromosome Inactivation into Autosomal Sequences: Role for DNA Elements, Chromatin Features and Chromosomal Domains. Hum. Mol. Genet. 2014, 23, 1211–1223. [Google Scholar] [CrossRef][Green Version]
- Kalz-Füller, B.; Sleegers, E.; Schwanitz, G.; Schubert, R. Characterisation, Phenotypic Manifestations and X-Inactivation Pattern in 14 Patients with X-Autosome Translocations. Clin. Genet. 1999, 55, 362–366. [Google Scholar] [CrossRef]
- Dobigny, G.; Ozouf-Costaz, C.; Bonillo, C.; Volobouev, V. Viability of X-Autosome Translocations in Mammals: An Epigenomic Hypothesis from a Rodent Case-Study. Chromosoma 2004, 113, 34–41. [Google Scholar] [CrossRef]
- Ratomponirina, C.; Viegas-Péquignot, E.; Dutrillaux, B.; Petter, F.; Rumpler, Y. Synaptonemal Complexes in Gerbillidae: Probable Role of Intercalated Heterochromatin in Gonosome-Autosome Translocations. Cytogenet. Cell Genet. 1986, 43, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Vozdova, M.; Ruiz-Herrera, A.; Fernandez, J.; Cernohorska, H.; Frohlich, J.; Sebestova, H.; Kubickova, S.; Rubes, J. Meiotic Behaviour of Evolutionary Sex-Autosome Translocations in Bovidae. Chrom. Res. 2016, 24, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Camacho, J.P.; Sharbel, T.F.; Beukeboom, L.W. B-Chromosome Evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Trifonov, V.A.; Dementyeva, P.V.; Larkin, D.M.; O’Brien, P.C.M.; Perelman, P.L.; Yang, F.; Ferguson-Smith, M.A.; Graphodatsky, A.S. Transcription of a Protein-Coding Gene on B Chromosomes of the Siberian Roe Deer (Capreolus pygargus). BMC Biol. 2013, 11, 90. [Google Scholar] [CrossRef] [PubMed]
- Duke Becker, S.E.; Thomas, R.; Trifonov, V.A.; Wayne, R.K.; Graphodatsky, A.S.; Breen, M. Anchoring the Dog to Its Relatives Reveals New Evolutionary Breakpoints across 11 Species of the Canidae and Provides New Clues for the Role of B Chromosomes. Chrom. Res. 2011, 19, 685–708. [Google Scholar] [CrossRef]
- Makunin, A.I.; Kichigin, I.G.; Larkin, D.M.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Yang, F.; Proskuryakova, A.A.; Vorobieva, N.V.; Chernyaeva, E.N.; O’Brien, S.J.; et al. Contrasting Origin of B Chromosomes in Two Cervids (Siberian Roe Deer and Grey Brocket Deer) Unravelled by Chromosome-Specific DNA Sequencing. BMC Genom. 2016, 17, 618. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).