The Investigation of Perennial Sunflower Species (Helianthus L.) Mitochondrial Genomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Mitochondrial DNA Extraction
2.2. Next-Generation Sequencing
2.3. Mitochondrial Genome Assembly and Annotation
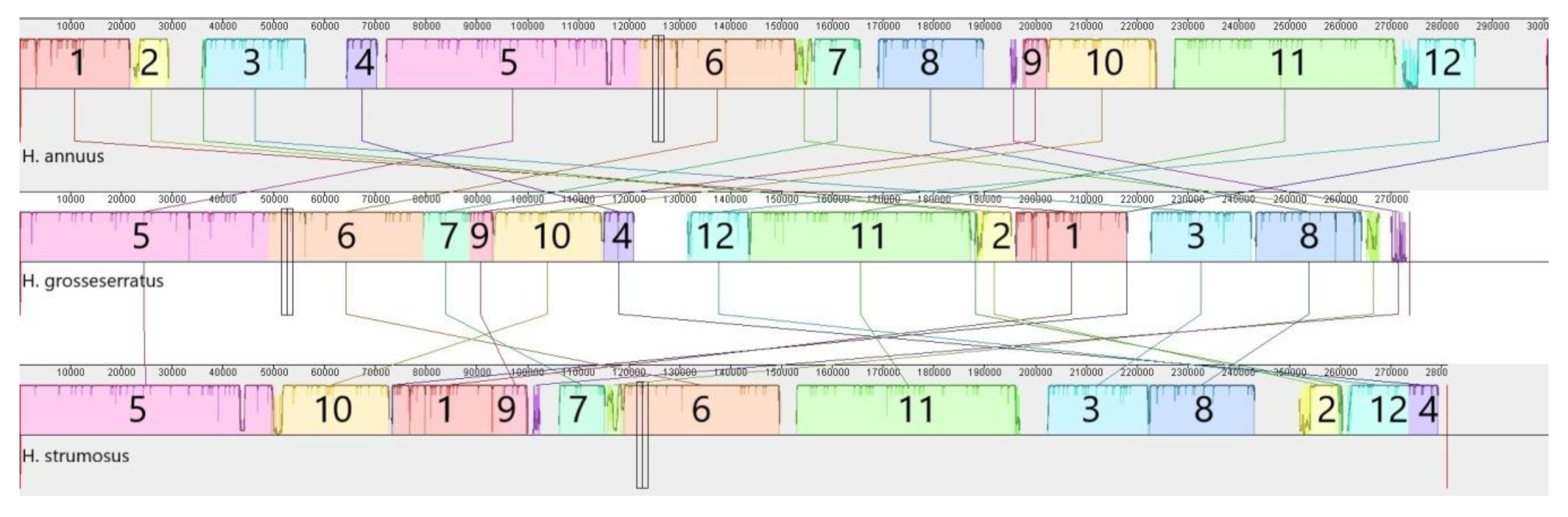
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Flood, P.J.; Theeuwen, T.P.J.M.; Schneeberger, K.; Keizer, P.; Kruijer, W.; Severing, E.; Kouklas, E.; Hageman, J.A.; Wijfjes, R.; Calvo-Baltanas, V.; et al. Reciprocal cybrids reveal how organellar genomes affect plant phenotypes. Nat. Plants 2020, 6, 13–21. [Google Scholar] [CrossRef]
- Liu, H.; Cui, P.; Zhan, K.; Lin, Q.; Zhuo, G.; Guo, X.; Ding, F.; Yang, W.; Liu, D.; Hu, S.; et al. Comparative analysis of mitochondrial genomes between a wheat K-type cytoplasmic male sterility (CMS) line and its maintainer line. BMC Genom. 2011, 12, 163. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Newton, K.J. Plant mitochondrial genomes: Dynamics and mechanisms of mutation. Annu. Rev. Plant Biol. 2017, 68, 225–252. [Google Scholar] [CrossRef] [PubMed]
- Logacheva, M.D.; Schelkunov, M.I.; Fesenko, A.N.; Kasianov, A.S.; Penin, A.A. Mitochondrial genome of Fagopyrum esculentum and the genetic diversity of extranuclear genomes in buckwheat. Plants 2020, 9, 618. [Google Scholar] [CrossRef] [PubMed]
- Seiler, G.J.; Rieseberg, L.H. Systematics, origin, and germplasm resources of the wild and domesticated sunflower. In Sunflower Technology and Production; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 21–65. ISBN 978-0-89118-227-6. [Google Scholar]
- Timme, R.E.; Simpson, B.B.; Linder, C.R. High-resolution phylogeny for Helianthus (Asteraceae) using the 18S–26S ribosomal DNA external transcribed spacer. Am. J. Bot. 2007, 94, 1837–1852. [Google Scholar] [CrossRef] [PubMed]
- Bock, D.G.; Kane, N.C.; Ebert, D.P.; Rieseberg, L.H. Genome skimming reveals the origin of the Jerusalem Artichoke tuber crop species: Neither from Jerusalem nor an artichoke. New Phytol. 2014, 201, 1021–1030. [Google Scholar] [CrossRef]
- Makarenko, M.S.; Usatov, A.V.; Tatarinova, T.V.; Azarin, K.V.; Logacheva, M.D.; Gavrilova, V.A.; Horn, R. Characterization of the mitochondrial genome of the MAX1 type of cytoplasmic male-sterile sunflower. BMC Plant Biol. 2019, 19, 51. [Google Scholar] [CrossRef]
- Garcia, L.; Edera, A.; Marfil, C.; Sanchez-Puerta, M. Male sterility and somatic hybridization in plant breeding. Preprints 2019. [Google Scholar] [CrossRef]
- Tyagi, V.; Dhillon, S.K.; Kaur, G.; Kaushik, P. Heterotic effect of different cytoplasmic combinations in sunflower hybrids cultivated under diverse irrigation regimes. Plants 2020, 9, 465. [Google Scholar] [CrossRef]
- Seiler, G.J.; Qi, L.L.; Marek, L.F. Utilization of sunflower crop wild relatives for cultivated sunflower improvement. Crop Sci. 2017, 57, 1083–1101. [Google Scholar] [CrossRef]
- Jan, C.-C.; Seiler, G.J.; Hammond, J.J. Effect of wild Helianthus cytoplasms on agronomic and oil characteristics of cultivated sunflower (Helianthus annuus L.). Plant Breed. 2014, 133, 262–267. [Google Scholar] [CrossRef]
- Jocković, J.; Rajčević, N.; Terzić, S.; Zorić, L.; Jocković, M.; Miladinović, D.; Luković, J. Pericarp features of wild perennial Helianthus L. species as a potential source for improvement of technical and technological properties of cultivated sunflower. Ind. Crop. Prod. 2020, 144, 112030. [Google Scholar] [CrossRef]
- Makarenko, M.S.; Usatov, A.V.; Markin, N.V.; Azarin, K.V.; Gorbachenko, O.F.; Usatov, N.A. Comparative genomics of domesticated and wild sunflower: Complete chloroplast and mitochondrial genomes. Online J. Biol. Sci. 2016, 16, 71–75. [Google Scholar] [CrossRef][Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.; Korobeynikov, A.; Lapidus, A.; Prjibelsky, A.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling genomes and mini-metagenomes from highly chimeric reads. In Lecture Notes in Computer Science, Proceedings of the Research in Computational Molecular Biology, Beijing, China, 7–10 April 2013; Deng, M., Jiang, R., Sun, F., Zhang, X., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 158–170. [Google Scholar]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Milne, I.; Stephen, G.; Bayer, M.; Cock, P.J.A.; Pritchard, L.; Cardle, L.; Shaw, P.D.; Marshall, D. Using tablet for visual exploration of second-generation sequencing data. Brief. Bioinform. 2013, 14, 193–202. [Google Scholar] [CrossRef]
- Alverson, A.J.; Wei, X.; Rice, D.W.; Stern, D.B.; Barry, K.; Palmer, J.D. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 2010, 27, 1436–1448. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Varré, J.-S.; D′Agostino, N.; Touzet, P.; Gallina, S.; Tamburino, R.; Cantarella, C.; Ubrig, E.; Cardi, T.; Drouard, L.; Gualberto, J.M.; et al. Complete sequence, multichromosomal architecture and transcriptome analysis of the Solanum tuberosum mitochondrial genome. Int. J. Mol. Sci. 2019, 20, 4788. [Google Scholar] [CrossRef]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019, 15. [Google Scholar] [CrossRef]
- Liao, X.; Zhao, Y.; Kong, X.; Khan, A.; Zhou, B.; Liu, D.; Kashif, M.H.; Chen, P.; Wang, H.; Zhou, R. Complete sequence of kenaf (Hibiscus cannabinus) mitochondrial genome and comparative analysis with the mitochondrial genomes of other plants. Sci. Rep. 2018, 8, 12714. [Google Scholar] [CrossRef]
- Guo, W.; Zhu, A.; Fan, W.; Mower, J.P. Complete mitochondrial genomes from the ferns Ophioglossum californicum and Psilotum nudum are highly repetitive with the largest organellar introns. New Phytol. 2017, 213, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.G.; Francs-Small, C.D.C.; Ostersetzer, O. Group II intron splicing factors in plant mitochondria. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef]
- Horn, R.; Gupta, K.J.; Colombo, N. Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion 2014, 19, 198–205. [Google Scholar] [CrossRef]
- Makarenko, M.S.; Usatov, A.V.; Tatarinova, T.V.; Azarin, K.V.; Logacheva, M.D.; Gavrilova, V.A.; Kornienko, I.V.; Horn, R. Organization features of the mitochondrial genome of sunflower (Helianthus annuus L.) with ANN2-type male-sterile cytoplasm. Plants 2019, 8, 439. [Google Scholar] [CrossRef]
- Horn, R. Molecular diversity of male sterility inducing and male-fertile cytoplasms in the genus Helianthus. Appl. Genet. 2002, 104, 562–570. [Google Scholar] [CrossRef]
- Qiu, F.; Baack, E.J.; Whitney, K.D.; Bock, D.G.; Tetreault, H.M.; Rieseberg, L.H.; Ungerer, M.C. Phylogenetic trends and environmental correlates of nuclear genome size variation in Helianthus sunflowers. New Phytol. 2019, 221, 1609–1618. [Google Scholar] [CrossRef]
- Kane, N.C.; Burke, J.M.; Marek, L.; Seiler, G.; Vear, F.; Baute, G.; Knapp, S.J.; Vincourt, P.; Rieseberg, L.H. Sunflower genetic, genomic and ecological resources. Mol. Ecol. Resour. 2013, 13, 10–20. [Google Scholar] [CrossRef]
- Ostevik, K.L.; Samuk, K.; Rieseberg, L.H. Ancestral reconstruction of karyotypes reveals an exceptional rate of nonrandom chromosomal evolution in sunflower. Genetics 2020, 214, 1031–1045. [Google Scholar] [CrossRef]
- Stephens, J.D.; Rogers, W.L.; Mason, C.M.; Donovan, L.A.; Malmberg, R.L. Species tree estimation of diploid Helianthus (Asteraceae) using target enrichment. Am. J. Bot. 2015, 102, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Kesseli, R.; Ochoa, O.; Michelmore, R. Variation at RFLP loci in Lactuca spp. and origin of cultivated lettuce (L. sativa). Genome 1991, 34, 430–436. [Google Scholar] [CrossRef]
- Smith, D.R.; Keeling, P.J. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar] [CrossRef]
- Bergthorsson, U.; Adams, K.L.; Thomason, B.; Palmer, J.D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 2003, 424, 197–201. [Google Scholar] [CrossRef]
- Gandini, C.L.; Garcia, L.E.; Abbona, C.C.; Sanchez-Puerta, M.V. The complete organelle genomes of Physochlaina orientalis: Insights into short sequence repeats across seed plant mitochondrial genomes. Mol. Phylogenetics Evol. 2019, 137, 274–284. [Google Scholar] [CrossRef]
- Newton, K.J.; Gabay-Laughnan, S.; De Paepe, R. Mitochondrial Mutations in Plants. In Plant Mitochondria: From Genome to Function; Day, D.A., Millar, A.H., Whelan, J., Eds.; Advances in Photosynthesis and, Respiration; Springer: Dordrecht, The Netherlands, 2004; pp. 121–141. ISBN 978-1-4020-2400-9. [Google Scholar]
- Bock, R.; Knoop, V. (Eds.) Genomics of Chloroplasts and Mitochondria; Advances in Photosynthesis and Respiration; Springer: Dordrecht, The Netherlands, 2012; ISBN 978-94-007-2919-3. [Google Scholar]
- Bock, D.G.; Andrew, R.L.; Rieseberg, L.H. On the adaptive value of cytoplasmic genomes in plants. Mol. Ecol. 2014, 23, 4899–4911. [Google Scholar] [CrossRef]
- Nugent, J.M.; Palmer, J.D. Location, identity, amount and serial entry of chloroplast DNA sequences in crucifer mitochondrial DNAs. Curr. Genet. 1988, 14, 501–509. [Google Scholar] [CrossRef]
- Chen, L.; Liu, Y.-G. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 2014, 65, 579–606. [Google Scholar] [CrossRef] [PubMed]
- Zancani, M.; Braidot, E.; Filippi, A.; Lippe, G. Structural and functional properties of plant mitochondrial F-ATP synthase. Mitochondrion 2020, 53, 178–193. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Zhao, C.; Chen, F.; Liu, Y.; Zhang, S.; Wu, H.; Zhang, L.; Liu, Y. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genom. 2018, 19, 614. [Google Scholar] [CrossRef]
- Grimes, B.T.; Sisay, A.K.; Carroll, H.D.; Cahoon, A.B. Deep sequencing of the tobacco mitochondrial transcriptome reveals expressed ORFs and numerous editing sites outside coding regions. BMC Genom. 2014, 15, 31. [Google Scholar] [CrossRef]
- Tang, H.; Zheng, X.; Li, C.; Xie, X.; Chen, Y.; Chen, L.; Zhao, X.; Zheng, H.; Zhou, J.; Ye, S.; et al. Multi-step formation, evolution, and functionalization of new cytoplasmic male sterility genes in the plant mitochondrial genomes. Cell Res. 2017, 27, 130–146. [Google Scholar] [CrossRef]

| Position in Mitogenome of H. annuus, (MN171345.1) | H. annuus | H. grosseserratus | H. strumosus | Gene | Substitution Type |
|---|---|---|---|---|---|
| 37,865 | T | T | C | coxIII | synonymous |
| 43,358 | G | T | G | rpl5 | synonymous |
| 67,110 | C | T | C | ccmB | synonymous |
| 112,693 | T | G | T | nad5 | synonymous |
| 114,492 | C | T | C | atp9 | synonymous |
| 122,169 | C | C | T | rps4 | nonsynonymous—Lys167Arg |
| 122,990 | T | G | T | nonsynonymous—Ile292Arg | |
| 169,209 | A | A | G | nad6 | synonymous |
| 188,296 | G | G | T | cob | synonymous |
| 188,466 | C | C | A | nonsynonymous—Asn42Lys | |
| 188,475 | C | A | C | nonsynonymous—Phe45Leu | |
| 189,199 | A | C | A | nonsynonymous—Ile287Leu | |
| 230,114 | A | C | A | rpl16 | nonsynonymous—Lys32Gln |
| 250,522 | G | A | G | matR | nonsynonymous—Ala458Glu |
| 251,624 | C | C | T | nonsynonymous—Gly91Arg | |
| 269,036 | T | G | T | atp6 | nonsynonymous—Phe37Ter |
| 269,037 | T | A | T | ||
| 269,064 | G | C | C | nonsynonymous—Lys46Asn | |
| 269,155 | T | T | C | nonsynonymous—Tyr77His |
| Name | Presence in | BLASTp Similarity | ||
|---|---|---|---|---|
| H. annuus | H. grosseserratus | H. strumosus | ||
| orf117 | + | + | + | hypothetical protein |
| orf126 | − | − | + | hypothetical protein |
| orf139 | + | + | + | rps11 |
| orf148 | ± | − | + | hypothetical protein |
| orf184 | − | − | + | hypothetical protein |
| orf188 | ± | + | − | cox2 |
| orf207 | − | − | + | RNA polymerase |
| orf254 | + | − | + | hypothetical protein |
| orf259 | + | − | + | hypothetical protein |
| orf291 | + | − | + | hypothetical protein |
| orf298 | + | − | + | hypothetical protein |
| orf316 | + | − | + | cox2 |
| orf334 | orf334 | orf284 | orf365 | psaA |
| orf633 | − | + | − | atp1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makarenko, M.; Usatov, A.; Tatarinova, T.; Azarin, K.; Kovalevich, A.; Gavrilova, V.; Horn, R. The Investigation of Perennial Sunflower Species (Helianthus L.) Mitochondrial Genomes. Genes 2020, 11, 982. https://doi.org/10.3390/genes11090982
Makarenko M, Usatov A, Tatarinova T, Azarin K, Kovalevich A, Gavrilova V, Horn R. The Investigation of Perennial Sunflower Species (Helianthus L.) Mitochondrial Genomes. Genes. 2020; 11(9):982. https://doi.org/10.3390/genes11090982
Chicago/Turabian StyleMakarenko, Maksim, Alexander Usatov, Tatiana Tatarinova, Kirill Azarin, Alexey Kovalevich, Vera Gavrilova, and Renate Horn. 2020. "The Investigation of Perennial Sunflower Species (Helianthus L.) Mitochondrial Genomes" Genes 11, no. 9: 982. https://doi.org/10.3390/genes11090982
APA StyleMakarenko, M., Usatov, A., Tatarinova, T., Azarin, K., Kovalevich, A., Gavrilova, V., & Horn, R. (2020). The Investigation of Perennial Sunflower Species (Helianthus L.) Mitochondrial Genomes. Genes, 11(9), 982. https://doi.org/10.3390/genes11090982






