Identification of MicroRNA-Related Tumorigenesis Variants and Genes in the Cancer Genome Atlas (TCGA) Data
Abstract
1. Introduction
2. Materials and Methods
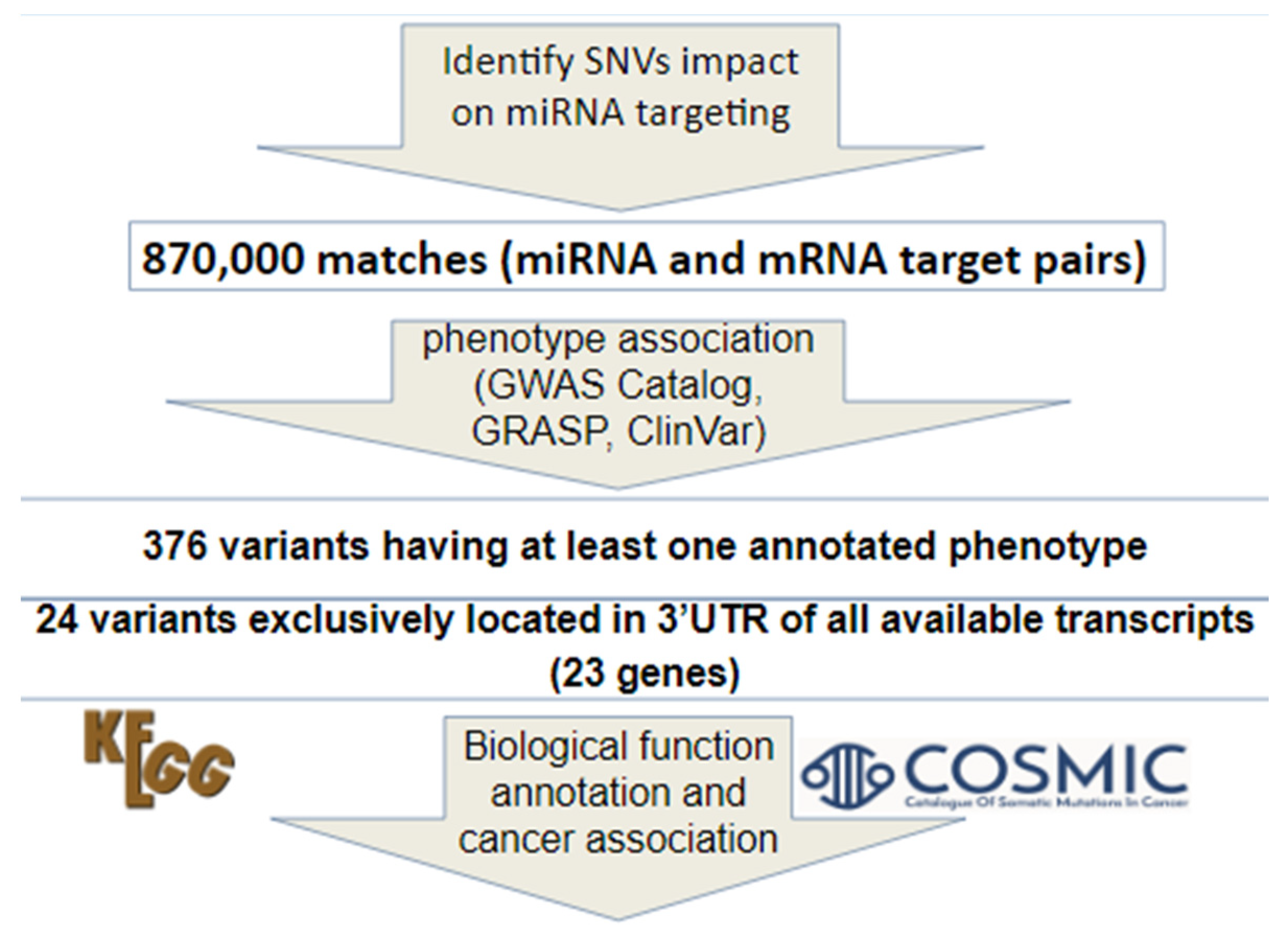
2.1. Retrieval and Screening of miRNA Target Site SNVs Using dbMTS
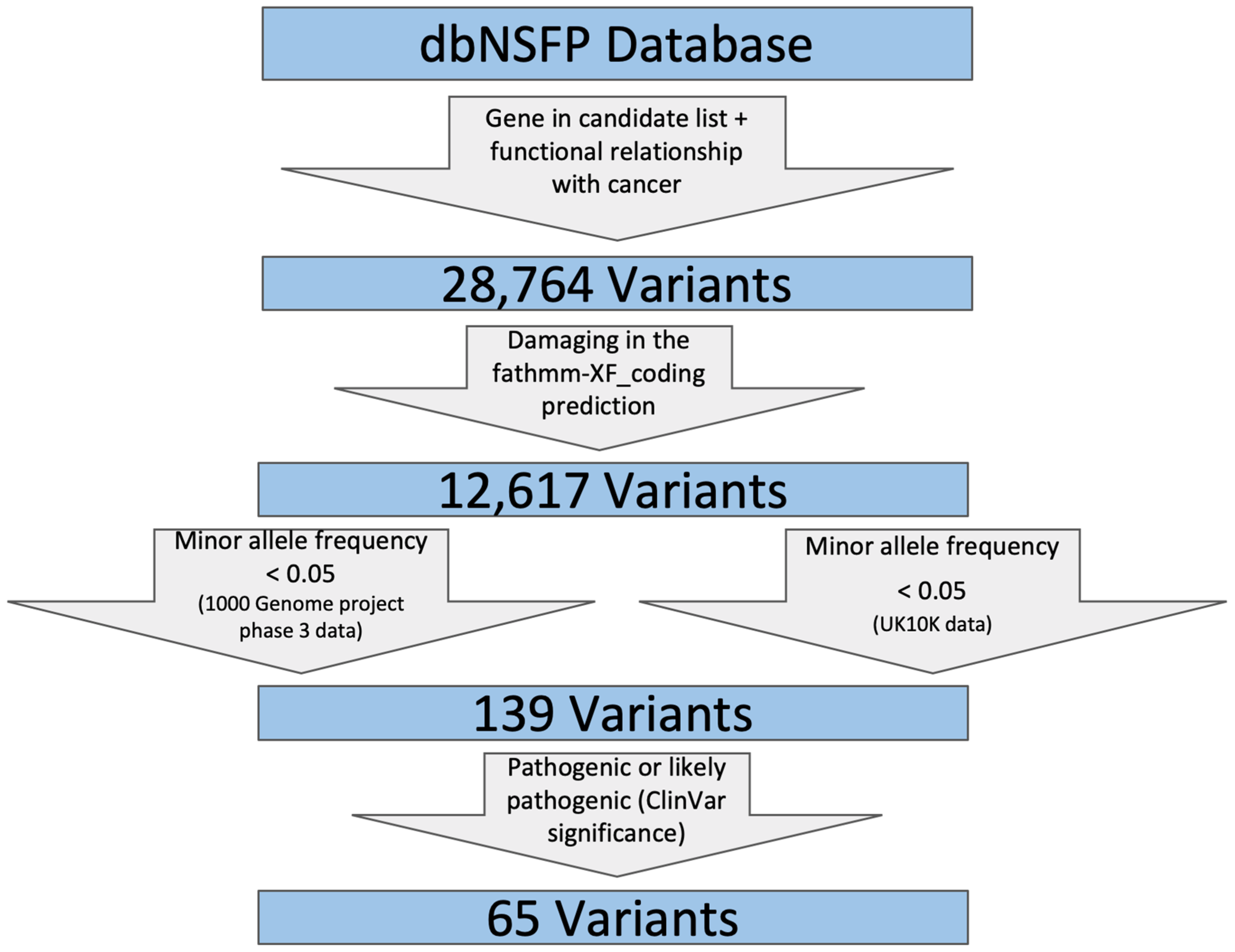
2.2. Retrieval and Screening of Nonsynonymous SNVs
- (1)
- SNVs located in genes that are included in our candidate gene list;
- (2)
- SNVs that include one of the following strings reported for ClinVar phenotype in dbNSFP to ensure their functional relationship with cancer: (‘carcinoma’, ‘sarcoma’, ‘leukemia’, ‘tumor’, ‘cancer’, ‘lymphoma’, ‘myeloma’);
- (3)
- FATHMM was used to filter for damaging variants by removing those SNVs that are predicted as ‘Neutral’;
- (4)
- SNV allele frequency should not be high (minor allele frequency <0.05 in both 1000Gp3_AF and UK10K_AF) because pathogenic SNVs are not likely to be prevalent in the general population;
- (5)
- SNVs that are pathogenic or likely pathogenic as reported by ClinVar clinical significance. These steps were achieved using a custom Python script.
2.3. Functional and Pathway Analyses
2.4. Survival Analyses
3. Results and Discussion
3.1. Twenty four Non-Coding SNVs were Identified at Potential miRNA Target Sites
3.2. Two Hundred and Thirty Three Somatic SNVs were Identified in Experimentally Identified miRNA Target Sites
3.3. Sixty Five Nonsynonymous SNVs were Identified in the Candidate Gene List
3.4. Biological Pathway Analysis
3.5. Survival Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nicoloso, M.S.; Sun, H.; Spizzo, R.; Kim, H.; Wickramasinghe, P.; Shimizu, M.; Wojcik, S.E.; Ferdin, J.; Kunej, T.; Xiao, L.; et al. Single-Nucleotide Polymorphisms inside MicroRNA Target Sites Influence Tumor Susceptibility. Cancer Res. 2010, 70, 2789–2798. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Ding, L.; Liu, H.; Xu, Z.; Jiang, H.; Handelman, S.K.; Bai, Y. Identifying Interaction Clusters for MiRNA and MRNA Pairs in TCGA Network. Genes 2019, 10, 702. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Mou, C.; Swartz, M.D.; Yu, B.; Bai, Y.; Tu, Y.; Liu, X. DbMTS: A Comprehensive Database of Putative Human MicroRNA Target Site SNVs and Their Functional Predictions. Hum. Mutat. 2020, 41, 1123–1130. [Google Scholar] [CrossRef]
- Liu, X.; Jian, X.; Boerwinkle, E. DbNSFP: A Lightweight Database of Human Nonsynonymous SNPs and Their Functional Predictions. Hum. Mutat. 2011, 32, 894–899. [Google Scholar] [CrossRef]
- Buniello, A.; Macarthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of Published Genome-Wide Association Studies, Targeted Arrays and Summary Statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [PubMed]
- Eicher, J.D.; Landowski, C.; Stackhouse, B.; Sloan, A.; Chen, W.; Jensen, N.; Lien, J.P.; Leslie, R.; Johnson, A.D. GRASP v2.0: An Update on the Genome-Wide Repository of Associations between SNPs and Phenotypes. Nucleic Acids Res. 2015, 43, D799–D804. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: Public Archive of Relationships among Sequence Variation and Human Phenotype. Nucleic Acids Res. 2014, 42, D980–D985. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Cui, Y. SomamiR 2.0: A Database of Cancer Somatic Mutations Altering MicroRNA-CeRNA Interactions. Nucleic Acids Res. 2016, 44, D1005–D1010. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Anaya, J. OncoLnc: Linking TCGA Survival Data to MRNAs, MiRNAs, and LncRNAs. PeerJ Comput. Sci. 2016, 2, e67. [Google Scholar] [CrossRef]
- Jia, X.; Wang, C.; Huang, H.; Zhang, P.; Yao, Z.; Xu, L. FLAD1 Is Overexpressed in Breast Cancer and Is a Potential Predictor of Prognosis and Treatment. Int. J. Clin. Exp. Med. 2019, 12, 3138–3152. [Google Scholar]
- Thammaiah, C.K.; Jayaram, S. Role of Let-7 Family MicroRNA in Breast Cancer. Non-Coding RNA Res. 2016, 1, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Wang, Y.; Wang, L.; Huang, Y.; Xu, Y.; Xu, L.; Guo, Y.; Lu, J.; Li, X.; Zhu, M.; et al. MicroRNA-30b Targets Snail to Impede Epithelial-Mesenchymal Transition in Pancreatic Cancer Stem Cells. J. Cancer 2018, 9, 2147–2159. [Google Scholar] [CrossRef] [PubMed]
- Leslie, R.; O’Donnell, C.J.; Johnson, A.D. GRASP: Analysis of Genotype-Phenotype Results from 1390 Genome-Wide Association Studies and Corresponding Open Access Database. Bioinformatics 2014, 30, i185–i194. [Google Scholar] [CrossRef] [PubMed]
- Brosens, L.A.A.; Langeveld, D.; van Hattem, W.A.; Giardiello, F.M.; Offerhaus, G.J.A. Juvenile Polyposis Syndrome. World J. Gastroenterol. 2011, 17, 4839–4844. [Google Scholar] [CrossRef]
- Cao, X.; Eu, K.W.; Kumarasinghe, M.P.; Li, H.H.; Loi, C.; Cheah, P.Y. Mapping of Hereditary Mixed Polyposis Syndrome (HMPS) to Chromosome 10q23 by Genomewide High-Density Single Nucleotide Polymorphism (SNP) Scan and Identification of BMPR1A Loss of Function. J. Med. Genet. 2005, 43, e13. [Google Scholar] [CrossRef]
- Rigaud, S.; Fondanèche, M.C.; Lambert, N.; Pasquier, B.; Mateo, V.; Soulas, P.; Galicier, L.; Le Deist, F.; Rieux-Laucat, F.; Revy, P.; et al. XIAP Deficiency in Humans Causes an X-Linked Lymphoproliferative Syndrome. Nature 2006, 444, 110–114. [Google Scholar] [CrossRef]
- Ni, Y.; Zbuk, K.M.; Sadler, T.; Patocs, A.; Lobo, G.; Edelman, E.; Platzer, P.; Orloff, M.S.; Waite, K.A.; Eng, C. Germline Mutations and Variants in the Succinate Dehydrogenase Genes in Cowden and Cowden-like Syndromes. Am. J. Hum. Genet. 2008, 83, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Parenti, G.; Zampetti, B.; Rapizzi, E.; Ercolino, T.; Giach, V.; Mannelli, M. Updated and New Perspectives on Diagnosis, Prognosis, and Therapy of Malignant Pheochromocytoma/Paraganglioma. J. Oncol. 2012, 2012, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Borkowska, J.; Schwartz, R.A.; Kotulska, K.; Jozwiak, S. Tuberous Sclerosis Complex: Tumors and Tumorigenesis. Int. J. Dermatol. 2010, 50, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Cleaver, J.E.; Lam, E.T.; Revet, I. Disorders of Nucleotide Excision Repair: The Genetic and Molecular Basis of Heterogeneity. Nat. Rev. Genet. 2009, 10, 756–768. [Google Scholar] [CrossRef] [PubMed]
- Menko, F.H.; Van Steensel, M.; Giraud, S.; Friis-Hansen, L.J.; Richard, S.; Ungari, S.; Nordenskjöld, M.; Hansen, T.V.; Solly, J.; Maher, E. Birt-Hogg-Dubé Syndrome: Diagnosis and Management. Lancet Oncol. 2009, 10, 1199–1206. [Google Scholar] [CrossRef]
- Bachinski, L.L.; Olufemi, S.E.; Zhou, X.; Wu, C.C.; Yip, L.; Shete, S.; Lozano, G.; Amos, C.I.; Strong, L.C.; Krahe, R. Genetic Mapping of a Third Li-Fraumeni Syndrome Predisposition Locus to Human Chromosome 1q23. Cancer Res. 2005, 65, 427–431. [Google Scholar] [PubMed]
- Lynch, H.T.; Riegert-Johnson, D.L.; Snyder, C.; Lynch, J.F.; Hagenkord, J.; Boland, C.R.; Rhees, J.; Thibodeau, S.N.; Boardman, L.A.; Davies, J.; et al. Lynch Syndrome-Associated Extracolonic Tumors Are Rare in Two Extended Families with the Same EPCAM Deletion. Am. J. Gastroenterol. 2011, 106, 1829–1836. [Google Scholar] [CrossRef]
- Half, E.; Bercovich, D.; Rozen, P. Familial Adenomatous Polyposis. Orphanet J. Rare Dis. 2009, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Waltes, R.; Kalb, R.; Gatei, M.; Kijas, A.W.; Stumm, M.; Sobeck, A.; Wieland, B.; Varon, R.; Lerenthal, Y.; Lavin, M.F.; et al. Human RAD50 Deficiency in a Nijmegen Breakage Syndrome-like Disorder. Am. J. Hum. Genet. 2009, 84, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, C.; Kanazawa, N.; Yoshikawa, Y.; Yoshikawa, R.; Saitoh, Y.; Chiyo, H.; Tanizawa, T.; Hashimoto-Tamaoki, T.; Nakano, Y. Germline PTCH1 Mutations in Japanese Basal Cell Nevus Syndrome Patients. J. Hum. Genet. 2009, 54, 403–408. [Google Scholar] [CrossRef] [PubMed][Green Version]


| Gene Symbol | Cancer Type * | Disrupted miRNAs |
|---|---|---|
| ACVR2B | BRCA, HNSC, STAD | has-miR-4670 |
| ADAM12 | THCA, BRCA, HNSC, LUAD | has-miR-4726 |
| BMPR1A | LUAD, KICH, KIRC | has-miR-520f |
| CD93 | BRCA, LIHC, LUAD, LUSC, STAD | has-miR-1179 |
| CDS1 | BRCA, STAD, UCEC, KIRC, KIRP, LUAD | has-miR-411 |
| CHST3 | KIRP, LUAD, STAD | has-miR-652 |
| ELOVL4 | BRCA, UCEC, KIRC | has-miR-185 |
| FAM20B | KIRP, LUAD, UCEC, BRCA, STAD | has-miR-649 |
| FLRT2 | STAD, BRCA, KIRC, LUAD | has-miR-5688 |
| GIPC2 | LUAD, STAD | has-miR-4765 |
| GPR143 | KIRC, KIRP, LIHC | has-miR-520f |
| HEMK1 | BRCA, STAD, KIRC, KIRP, LUAD | has-miR-3911 |
| MAFB | LUAD, PRAD, STAD, BRCA, HNSC, KICH, KIRC | has-miR-1266 |
| PGR | LUAD, PRAD, BRCA, KIRC | has-miR-182 |
| PRKCA | BRCA, KIRC, KIRP, LUAD, PRAD | has-miR-296 |
| PTCHD1 | STAD, THCA, BRCA, HNSC | has-miR-185 |
| SCN2B | LUAD, PRAD, STAD, BRCA | has-miR-141 |
| SLC30A7 | LUAD, PRAD, KICH, KIRC, KIRP, STAD, BRCA | has-miR-152 |
| SLC39A8 | LUAD, LUSC, PRAD | has-miR-141 |
| TLL2 | LUAD, STAD, BRCA, COAD, KIRC, KIRP | has-miR-29a |
| TRAM2 | LUAD, PRAD, STAD, BRCA | has-miR-625 |
| XIAP | BRCA, KIRP, LUAD, PRAD | has-miR-580 |
| ZDHHC15 | STAD, BRCA, KIRC, LUAD, PRAD | has-miR-367 |
| Chromosome | Position | Ref. Allele | Alt. Allele | Gene Symbol | MiRNA | Cancer Classification ((Tissue)(Histology)) |
|---|---|---|---|---|---|---|
| 1 | 154992969 | G | C | FLAD1 | let-7b | (breast)(carcinoma) |
| 3 | 49015647 | G | A | WDR6 | miR-222 | (large_intestine)(carcinoma) |
| 3 | 57559757 | A | G | PDE12 | miR-484 | (skin)(malignant_melanoma) |
| 5 | 93593940 | G | A | NR2F1 | miR-149 | (oesophagus)(carcinoma) |
| 5 | 131204977 | A | T | LYRM7 | miR-30e | (liver)(carcinoma) |
| 6 | 30724942 | T | C | TUBB | miR-708 | (kidney)(neoplasm) |
| 6 | 43504866 | A | T | TJAP1 | miR-744 | (ovary)(neoplasm) |
| 7 | 56063723 | T | C | CCT6A | miR-3663-3p | (ovary)(neoplasm) |
| 9 | 128695535 | G | T | SET | miR-185 | (ovary)(neoplasm) |
| 16 | 68835430 | C | T | CDH1 | miR-30c | (pancreas)(carcinoma) |
| 19 | 29675270 | C | A | PLEKHF1 | miR-331-3p | (liver)(carcinoma) |
| Gene Symbol | Tissue Related to Cancer |
|---|---|
| APC | large intestine, stomach, lung, thyroid, urinary tract |
| ATM | lymphoid, lung, thyroid, large intestine |
| CHEK2 | large intestine, urinary tract |
| MET | endometrium, autonomic ganglia, pancreas, lung, large intestine vulva, cervix, kidney, thyroid, ovary, adrenal gland |
| NF1 | small intestine |
| PTCH1 | ovary, skin |
| RET | thyroid, lung, lymphoid, adrenal gland |
| TSC2 | central nervous system, stomach, cervix |
| WT1 | skin, kidney, lymphoid |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Wu, B.; Han, H.; Zhao, J.; Bai, Y.; Liu, X. Identification of MicroRNA-Related Tumorigenesis Variants and Genes in the Cancer Genome Atlas (TCGA) Data. Genes 2020, 11, 953. https://doi.org/10.3390/genes11090953
Li C, Wu B, Han H, Zhao J, Bai Y, Liu X. Identification of MicroRNA-Related Tumorigenesis Variants and Genes in the Cancer Genome Atlas (TCGA) Data. Genes. 2020; 11(9):953. https://doi.org/10.3390/genes11090953
Chicago/Turabian StyleLi, Chang, Brian Wu, Han Han, Jeff Zhao, Yongsheng Bai, and Xiaoming Liu. 2020. "Identification of MicroRNA-Related Tumorigenesis Variants and Genes in the Cancer Genome Atlas (TCGA) Data" Genes 11, no. 9: 953. https://doi.org/10.3390/genes11090953
APA StyleLi, C., Wu, B., Han, H., Zhao, J., Bai, Y., & Liu, X. (2020). Identification of MicroRNA-Related Tumorigenesis Variants and Genes in the Cancer Genome Atlas (TCGA) Data. Genes, 11(9), 953. https://doi.org/10.3390/genes11090953






