Negative Regulation of Serine Threonine Kinase 11 (STK11) through miR-100 in Head and Neck Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Nucleic Acid Isolation and Assessment
2.3. HPV Detection and Genotyping
2.4. Bisulfite Conversion and STK11 Gene Methylation
2.5. Real-Time PCR Analysis
2.6. Cell Culture and Transfection
2.7. Luciferase Reporter Assay
2.8. Patient Survival Analysis of STK11, miR-100-3p, and miR-100-5p
2.9. Statistical Analysis
3. Results
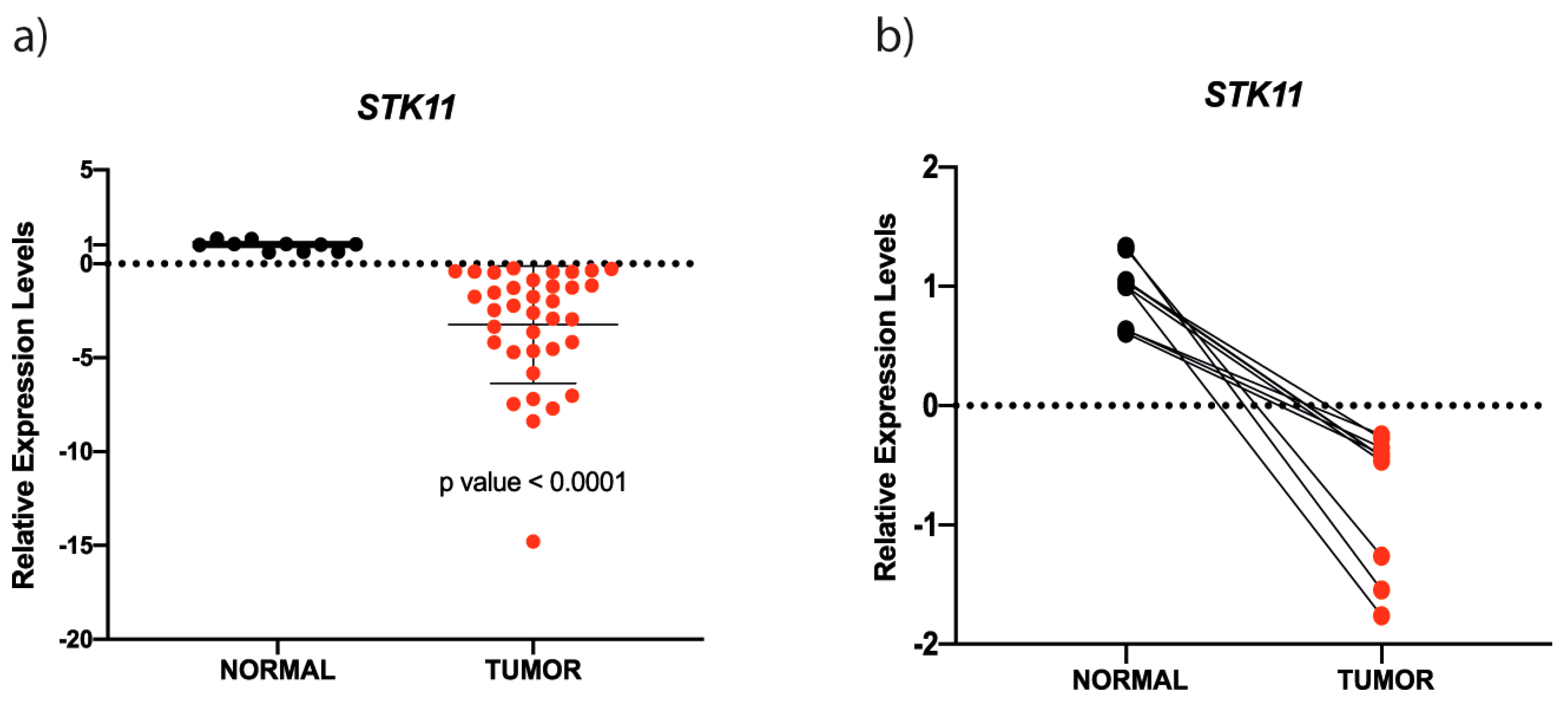
3.1. STK11 mRNA is Down-Regulated in HNC
3.2. STK11 Promoter Methylation Status
3.3. miR-100-3p Interacts with STK11 mRNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Shin, H.-R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 2010, 127, 2893–2917. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Siegel, R.; Ward, E.; Murray, T.; Xu, J.; Smigal, C.; Thun, M.J. Cancer statistics, 2006. CA Cancer J. Clin. 2006, 56, 106–130. [Google Scholar] [CrossRef] [PubMed]
- Leemans, C.R.; Braakhuis, B.J.M.; Brakenhoff, R.H. The molecular biology of head and neck cancer. Nat. Rev. Cancer 2011, 11, 9–22. [Google Scholar] [CrossRef]
- Jethwa, A.R.; Khariwala, S.S. Tobacco-related carcinogenesis in head and neck cancer. Cancer Metastasis Rev. 2017, 36, 411–423. [Google Scholar] [CrossRef]
- Ragin, C.C.R.; Modugno, F.; Gollin, S.M. The epidemiology and risk factors of head and neck cancer: A focus on human papillomavirus. J. Dent. Res. 2007, 86, 104–114. [Google Scholar] [CrossRef]
- Gillison, M.L.; Koch, W.M.; Capone, R.B.; Spafford, M.; Westra, W.H.; Wu, L.; Zahurak, M.L.; Daniel, R.W.; Viglione, M.; Symer, D.E.; et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 2000, 92, 709–720. [Google Scholar] [CrossRef]
- Puram, S.V.; Rocco, J.W. Molecular aspects of head and neck cancer therapy. Hematol. Oncol. Clin. N. Am. 2015, 29, 971–992. [Google Scholar] [CrossRef]
- Gary, C.; Hajek, M.; Biktasova, A.; Bellinger, G.; Yarbrough, W.G.; Issaeva, N. Selective antitumor activity of roscovitine in head and neck cancer. Oncotarget 2016, 7, 38598–38611. [Google Scholar] [CrossRef]
- Karuman, P.; Gozani, O.; Odze, R.D.; Zhou, X.C.; Zhu, H.; Shaw, R.; Brien, T.P.; Bozzuto, C.D.; Ooi, D.; Cantley, L.C.; et al. The Peutz-Jegher gene product LKB1 is a mediator of p53-dependent cell death. Mol. Cell 2001, 7, 1307–1319. [Google Scholar] [CrossRef]
- Hawley, S.A.; Boudeau, J.; Reid, J.L.; Mustard, K.J.; Udd, L.; Makela, T.P.; Alessi, D.R.; Hardie, D.G. Complexes between the LKB1 tumor suppressor, STRAD α/β and MO25 α/β are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2003, 2, 28. [Google Scholar] [CrossRef]
- Thomson, D.M.; Brown, J.D.; Fillmore, N.; Condon, B.M.; Kim, H.-J.; Barrow, J.R.; Winder, W.W. LKB1 and the regulation of malonyl-CoA and fatty acid oxidation in muscle. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E1572–E1579. [Google Scholar] [CrossRef] [PubMed]
- Tiainen, M.; Ylikorkala, A.; Makela, T.P. Growth suppression by Lkb1 is mediated by a G(1) cell cycle arrest. Proc. Natl. Acad. Sci. USA 1999, 96, 9248–9251. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Huang, D.; Lu, N.; Luo, L. Role of the LKB1/AMPK pathway in tumor invasion and metastasis of cancer cells (Review). Oncol. Rep. 2015, 34, 2821–2826. [Google Scholar] [CrossRef] [PubMed]
- Mehenni, H.; Gehrig, C.; Nezu, J.; Oku, A.; Shimane, M.; Rossier, C.; Guex, N.; Blouin, J.L.; Scott, H.S.; Antonarakis, S.E. Loss of LKB1 kinase activity in Peutz-Jeghers syndrome, and evidence for allelic and locus heterogeneity. Am. J. Hum. Genet. 1998, 63, 1641–1650. [Google Scholar] [CrossRef]
- Avizienyte, E.; Loukola, A.; Roth, S.; Hemminki, A.; Tarkkanen, M.; Salovaara, R.; Arola, J.; Butzow, R.; Husgafvel-Pursiainen, K.; Kokkola, A.; et al. LKB1 somatic mutations in sporadic tumors. Am. J. Pathol. 1999, 154, 677–681. [Google Scholar] [CrossRef]
- Sanchez-Cespedes, M.; Parrella, P.; Esteller, M.; Nomoto, S.; Trink, B.; Engles, J.M.; Westra, W.H.; Herman, J.G.; Sidransky, D. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002, 62, 3659–3662. [Google Scholar]
- Bignell, G.R.; Barfoot, R.; Seal, S.; Collins, N.; Warren, W.; Stratton, M.R. Low frequency of somatic mutations in the LKB1/Peutz-Jeghers syndrome gene in sporadic breast cancer. Cancer Res. 1998, 58, 1384–1386. [Google Scholar]
- Nakanishi, C.; Yamaguchi, T.; Iijima, T.; Saji, S.; Toi, M.; Mori, T.; Miyaki, M. Germline mutation of the LKB1/STK11 gene with loss of the normal allele in an aggressive breast cancer of Peutz-Jeghers syndrome. Oncology 2004, 67, 476–479. [Google Scholar] [CrossRef]
- Petersen, G.M. Familial pancreatic cancer. Semin. Oncol. 2016, 43, 548–553. [Google Scholar] [CrossRef]
- Su, G.H.; Hruban, R.H.; Bansal, R.K.; Bova, G.S.; Tang, D.J.; Shekher, M.C.; Westerman, A.M.; Entius, M.M.; Goggins, M.; Yeo, C.J.; et al. Germline and somatic mutations of the STK11/LKB1 Peutz-Jeghers gene in pancreatic and biliary cancers. Am. J. Pathol. 1999, 154, 1835–1840. [Google Scholar] [CrossRef]
- Guldberg, P.; thor Straten, P.; Ahrenkiel, V.; Seremet, T.; Kirkin, A.F.; Zeuthen, J. Somatic mutation of the Peutz-Jeghers syndrome gene, LKB1/STK11, in malignant melanoma. Oncogene 1999, 18, 1777–1780. [Google Scholar] [CrossRef] [PubMed]
- Rowan, A.; Bataille, V.; MacKie, R.; Healy, E.; Bicknell, D.; Bodmer, W.; Tomlinson, I. Somatic mutations in the Peutz-Jeghers (LKB1/STKII) gene in sporadic malignant melanomas. J. Investig. Dermatol. 1999, 112, 509–511. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.J.; Cho, Y.G.; Park, J.Y.; Kim, T.Y.; Lee, J.H.; Kim, H.S.; Lee, J.W.; Song, Y.H.; Nam, S.W.; Lee, S.H.; et al. Genetic analysis of the LKB1/STK11 gene in hepatocellular carcinomas. Eur. J. Cancer 2004, 40, 136–141. [Google Scholar] [CrossRef]
- Qiu, W.; Schonleben, F.; Thaker, H.M.; Goggins, M.; Su, G.H. A novel mutation of STK11/LKB1 gene leads to the loss of cell growth inhibition in head and neck squamous cell carcinoma. Oncogene 2006, 25, 2937–2942. [Google Scholar] [CrossRef] [PubMed]
- Zeng, P.-Y.; Berger, S.L. LKB1 is recruited to the p21/WAF1 promoter by p53 to mediate transcriptional activation. Cancer Res. 2006, 66, 10701–10708. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Choi, J.E.; Na, Y.K.; Lee, E.J.; Lee, W.K.; Choi, Y.Y.; Yoon, G.S.; Jeon, H.-S.; Kim, D.S.; Park, J.Y. Genetic and epigenetic alterations of the LKB1 gene and their associations with mutations in TP53 and EGFR pathway genes in Korean non-small cell lung cancers. Lung Cancer 2013, 81, 194–199. [Google Scholar] [CrossRef]
- Kullmann, L.; Krahn, M.P. Controlling the master-upstream regulation of the tumor suppressor LKB1. Oncogene 2018, 37, 3045–3057. [Google Scholar] [CrossRef]
- Zheng, F.; Yuan, X.; Chen, E.; Ye, Y.; Li, X.; Dai, Y. Methylation of STK11 promoter is a risk factor for tumor stage and survival in clear cell renal cell carcinoma. Oncol. Lett. 2017, 14, 3065–3070. [Google Scholar] [CrossRef]
- Trojan, J.; Brieger, A.; Raedle, J.; Esteller, M.; Zeuzem, S. 5’-CpG island methylation of the LKB1/STK11 promoter and allelic loss at chromosome 19p13.3 in sporadic colorectal cancer. Gut 2000, 47, 272–276. [Google Scholar] [CrossRef]
- Lee, C.G.; Kim, Y.W.; Kim, E.H.; Meng, Z.; Huang, W.; Hwang, S.J.; Kim, S.G. Farnesoid X receptor protects hepatocytes from injury by repressing miR-199a-3p, which increases levels of LKB1. Gastroenterology 2012, 142, 1206–1217.e7. [Google Scholar] [CrossRef]
- Chen, H.; Untiveros, G.M.; McKee, L.A.K.; Perez, J.; Li, J.; Antin, P.B.; Konhilas, J.P. Micro-RNA-195 and -451 regulate the LKB1/AMPK signaling axis by targeting MO25. PLoS ONE 2012, 7, e41574. [Google Scholar] [CrossRef] [PubMed]
- Lao, G.; Liu, P.; Wu, Q.; Zhang, W.; Liu, Y.; Yang, L.; Ma, C. Mir-155 promotes cervical cancer cell proliferation through suppression of its target gene LKB1. Tumour. Biol. 2014, 35, 11933–11938. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.-N.; Jiang, M.-J.; Mei, Z.; Dai, J.-J.; Dai, C.-Y.; Fang, C.; Huang, Q.; Tian, L. microRNA-7 impairs autophagy-derived pools of glucose to suppress pancreatic cancer progression. Cancer Lett. 2017, 400, 69–78. [Google Scholar] [CrossRef]
- Sotlar, K.; Diemer, D.; Dethleffs, A.; Hack, Y.; Stubner, A.; Vollmer, N.; Menton, S.; Menton, M.; Dietz, K.; Wallwiener, D.; et al. Detection and typing of human papillomavirus by e6 nested multiplex PCR. J. Clin. Microbiol. 2004, 42, 3176–3184. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-C.; Dahiya, R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef]
- Dweep, H.; Gretz, N. MiRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef]
- Krü, J.; Rehmsmeier, M. RNAhybrid: Microrna target prediction easy, fast and flexible. Nucleic Acids Res. 2006. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Sougnez, C.; Lichtenstein, L.; Cibulskis, K.; Lander, E.; Gabriel, S.B.; Getz, G.; Ally, A.; Balasundaram, M.; Birol, I.; et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2015, 44, 71. [Google Scholar] [CrossRef]
- Walboomers, J.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.; Peto, J.; Meijer, C.J.; Munoz, N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Khode, S.R.; Dwivedi, R.C.; Rhys-Evans, P.; Kazi, R. Exploring the link between human papilloma virus and oral and oropharyngeal cancers. J. Cancer Res. Ther. 2014, 10, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Paz, I.B.; Cook, N.; Odom-Maryon, T.; Xie, Y.; Wilczynski, S.P. Human papillomavirus (HPV) in head and neck cancer. An association of HPV 16 with squamous cell carcinoma of Waldeyer’s tonsillar ring. Cancer 1997, 79, 595–604. [Google Scholar] [CrossRef]
- Haraf, D.J.; Nodzenski, E.; Brachman, D.; Mick, R.; Montag, A.; Graves, D.; Vokes, E.E.; Weichselbaum, R.R. Human papilloma virus and p53 in head and neck cancer: Clinical correlates and survival. Clin. Cancer Res. 1996, 2, 755–762. [Google Scholar] [PubMed]
- Schwartz, S.M.; Daling, J.R.; Doody, D.R.; Wipf, G.C.; Carter, J.J.; Madeleine, M.M.; Mao, E.J.; Fitzgibbons, E.D.; Huang, S.; Beckmann, A.M.; et al. Oral cancer risk in relation to sexual history and evidence of human papillomavirus infection. J. Natl. Cancer Inst. 1998, 90, 1626–1636. [Google Scholar] [CrossRef]
- Fouret, P.; Monceaux, G.; Temam, S.; Lacourreye, L.; St Guily, J.L. Human papillomavirus in head and neck squamous cell carcinomas in nonsmokers. Arch. Otolaryngol. Head Neck Surg. 1997, 123, 513–516. [Google Scholar] [CrossRef]
- Gillison, M.L.; Castellsague, X.; Chaturvedi, A.; Goodman, M.T.; Snijders, P.; Tommasino, M.; Arbyn, M.; Franceschi, S. Eurogin Roadmap: Comparative epidemiology of HPV infection and associated cancers of the head and neck and cervix. Int. J. Cancer 2014, 134, 497–507. [Google Scholar] [CrossRef]
- Kreimer, A.R.; Clifford, G.M.; Boyle, P.; Franceschi, S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: A systematic review. Cancer Epidemiol. Biomark. Prev. 2005, 14, 467–475. [Google Scholar] [CrossRef]
- Resta, N.; Pierannunzio, D.; Lenato, G.M.; Stella, A.; Capocaccia, R.; Bagnulo, R.; Lastella, P.; Susca, F.C.; Bozzao, C.; Loconte, D.C.; et al. Cancer risk associated with STK11/LKB1 germline mutations in Peutz-Jeghers syndrome patients: Results of an Italian multicenter study. Dig. Liver Dis. 2013, 45, 606–611. [Google Scholar] [CrossRef]
- Rowan, A.; Churchman, M.; Jefferey, R.; Hanby, A.; Poulsom, R.; Tomlinson, I. In situ analysis of LKB1/STK11 mRNA expression in human normal tissues and tumours. J. Pathol. 2000, 192, 203–206. [Google Scholar] [CrossRef]
- Ekizoglu, S.; Dalay, N.; Karaman, E.; Akdeniz, D.; Ozaydin, A.; Buyru, N. LKB1 downregulation may be independent of promoter methylation or FOXO3 expression in head and neck cancer. Transl. Res. 2013, 162, 122–129. [Google Scholar] [CrossRef]
- Dai, W.; Teodoridis, J.M.; Zeller, C.; Graham, J.; Hersey, J.; Flanagan, J.M.; Stronach, E.; Millan, D.W.; Siddiqui, N.; Paul, J.; et al. Systematic CpG islands methylation profiling of genes in the wnt pathway in epithelial ovarian cancer identifies biomarkers of progression-free survival. Clin. Cancer Res. 2011, 17, 4052–4062. [Google Scholar] [CrossRef] [PubMed]
- Claus, R.; Wilop, S.; Hielscher, T.; Sonnet, M.; Dahl, E.; Galm, O.; Jost, E.; Plass, C. A systematic comparison of quantitative high-resolution DNA methylation analysis and methylation-specific PCR. Epigenetics 2012, 7, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Summers, T.; Langan, R.C.; Nissan, A.; Brucher, B.L.D.M.; Bilchik, A.J.; Protic, M.; Daumer, M.; Avital, I.; Stojadinovic, A. Serum-based DNA methylation biomarkers in colorectal cancer: Potential for screening and early detection. J. Cancer 2013, 4, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, T.; Sheng, Y.; Zhang, C.; Peng, Y.; Wang, X.; Zhang, C. Methylation Profiling of Multiple Tumor Suppressor Genes in Hepatocellular Carcinoma and the Epigenetic Mechanism of 3OST2 Regulation. J. Cancer 2015, 6, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Sartor, M.A.; Dolinoy, D.C.; Jones, T.R.; Colacino, J.A.; Prince, M.E.P.; Carey, T.E.; Rozek, L.S. Genome-wide methylation and expression differences in HPV(+) and HPV(-) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics 2011, 6, 777–787. [Google Scholar] [CrossRef]
- Henson, B.J.; Bhattacharjee, S.; O’Dee, D.M.; Feingold, E.; Gollin, S.M. Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosomes Cancer 2009, 48, 569–582. [Google Scholar] [CrossRef]
- Peng, D.-X.; Luo, M.; Qiu, L.-W.; He, Y.-L.; Wang, X.-F. Prognostic implications of microRNA-100 and its functional roles in human epithelial ovarian cancer. Oncol. Rep. 2012, 27, 1238–1244. [Google Scholar] [CrossRef]
- Xu, C.; Zeng, Q.; Xu, W.; Jiao, L.; Chen, Y.; Zhang, Z.; Wu, C.; Jin, T.; Pan, A.; Wei, R.; et al. miRNA-100 inhibits human bladder urothelial carcinogenesis by directly targeting mTOR. Mol. Cancer Ther. 2013, 12, 207–219. [Google Scholar] [CrossRef]
- Ge, Y.-Y.; Shi, Q.; Zheng, Z.-Y.; Gong, J.; Zeng, C.; Yang, J.; Zhuang, S.-M. MicroRNA-100 promotes the autophagy of hepatocellular carcinoma cells by inhibiting the expression of mTOR and IGF-1R. Oncotarget 2014, 5, 6218–6228. [Google Scholar] [CrossRef]





| Primer/Cocktail | HPV Genotype | Amplicon (bp) | Sequence (5′-3′) |
|---|---|---|---|
| GP-E7-5B | CTG AGC TGT CAR NTA ATT GCT CA | ||
| GP-E6-3F | 630 | GGG WGK KAC TGA AAT CGG T | |
| GP-E7-6B | TCC TCT GAG TYG YCP AAT TGC TC | ||
| Cocktail I | 16 | 457 | Forward CAC AGT TATGCA CAG AGC TGC Reverse CAT ATA TTC ATG CAA TGT AGG TGT A |
| 18 | 322 | Forward CAC TTC ACT GCA AGA CAT AGA Reverse GTT GTG AAA TCGTCGTTT TTC A | |
| 31 | 263 | Forward GAA ATT GCA TGA ACT AAG CTC G Reverse CAC ATA TAC CTT TGTTTG TCA A | |
| 59 | 215 | Forward CAA AGG GGA ACT GCA AGA AAG Reverse TAT AAC AGC GTA TCA GCA GC | |
| 45 | 151 | Forward GTG GAA AAG TGC ATT ACA GG Reverse ACC TCT GTG GGT CCC AAT GT | |
| Cocktail II | 33 | 398 | Forward ACT ATA CAC AAC ATT GAA CTA Reverse GTT TTT ACA CGT CAC AGT GCA |
| 6/11 | 334 | Forward TGC AAG AAT GCA CTG ACC AC Reverse TGC ATG TTG TCC AGC AGT GT | |
| 58 | 274 | Forward GTA AAG TGT GCT TAC GAT TGC Reverse GTTGTTACA GGT TAC ACT TGT | |
| 52 | 229 | Forward TAA GGC TCG AGT GTG TGC AG Reverse CTAATA GTT ATT TCA CTT AAT GGT |
| Clinical Parameters | Patients n = 59 (100%) | STK11 Promoter Methylation n = 59 | STK11 Expression n = 36 | miRNA Expression n = 20 |
|---|---|---|---|---|
| Gender | ||||
| Male | 40 (67.8%) | |||
| Female | 19 (32.2%) | |||
| Age | ||||
| <50 | 10 (16.9%) | |||
| ≥50 | 49 (83.1%) | |||
| Clinical stage | ||||
| II | 5 (8.5%) | 5 (8.5%) | 4 (11.1%) | 2 (10%) |
| III | 11 (18.6%) | 11, 3a (18.6%) | 6, 3a (16.7%) | 4, 3a (20%) |
| IVA | 28 (47.5%) | 28, 6a (47.5%) | 15, 6a (41.7%) | 8, 6a (40%) |
| IVB | 10 (16.9%) | 10 (16.9%) | 8 (22.2%) | 4 (20%) |
| IVC | 5 (8.5%) | 5, 1a (8.5%) | 3, 1a (8.3%) | 2, 1a (10%) |
| Anatomic region | ||||
| Lip and oral cavity | 50 (84.7%) | |||
| Larynx | 1 (1.7%) | |||
| Pharynx | 7 (11.9%) | |||
| Nasal cavity | 1 (1.7%) | |||
| Histologic grade | ||||
| Low | 10 (16.9%) | |||
| Moderate | 48 (81.4%) | |||
| High | 1 (1.7%) | |||
| High-risk HPV | ||||
| 16 | 4 (6.8%) | |||
| Negative for HPV | 55 (93.2%) | |||
| Smoking Habit | ||||
| Positive | 34 (57.6%) | |||
| Negative | 25 (42.4%) | |||
| Alcoholism | ||||
| Positive | 34 (57.6%) | |||
| Negative | 25 (42.4%) | |||
| Tumor size | ||||
| >5 cm | 29 (49.2%) | |||
| <5 cm | 30 (50.8%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Figueroa-González, G.; Carrillo-Hernández, J.F.; Perez-Rodriguez, I.; Cantú de León, D.; Campos-Parra, A.D.; Martínez-Gutiérrez, A.D.; Coronel-Hernández, J.; García-Castillo, V.; López-Camarillo, C.; Peralta-Zaragoza, O.; et al. Negative Regulation of Serine Threonine Kinase 11 (STK11) through miR-100 in Head and Neck Cancer. Genes 2020, 11, 1058. https://doi.org/10.3390/genes11091058
Figueroa-González G, Carrillo-Hernández JF, Perez-Rodriguez I, Cantú de León D, Campos-Parra AD, Martínez-Gutiérrez AD, Coronel-Hernández J, García-Castillo V, López-Camarillo C, Peralta-Zaragoza O, et al. Negative Regulation of Serine Threonine Kinase 11 (STK11) through miR-100 in Head and Neck Cancer. Genes. 2020; 11(9):1058. https://doi.org/10.3390/genes11091058
Chicago/Turabian StyleFigueroa-González, Gabriela, José F. Carrillo-Hernández, Itzel Perez-Rodriguez, David Cantú de León, Alma D. Campos-Parra, Antonio D. Martínez-Gutiérrez, Jossimar Coronel-Hernández, Verónica García-Castillo, César López-Camarillo, Oscar Peralta-Zaragoza, and et al. 2020. "Negative Regulation of Serine Threonine Kinase 11 (STK11) through miR-100 in Head and Neck Cancer" Genes 11, no. 9: 1058. https://doi.org/10.3390/genes11091058
APA StyleFigueroa-González, G., Carrillo-Hernández, J. F., Perez-Rodriguez, I., Cantú de León, D., Campos-Parra, A. D., Martínez-Gutiérrez, A. D., Coronel-Hernández, J., García-Castillo, V., López-Camarillo, C., Peralta-Zaragoza, O., Jacobo-Herrera, N. J., Guardado-Estrada, M., & Pérez-Plasencia, C. (2020). Negative Regulation of Serine Threonine Kinase 11 (STK11) through miR-100 in Head and Neck Cancer. Genes, 11(9), 1058. https://doi.org/10.3390/genes11091058









