Maternal Transmission Ratio Distortion in Two Iberian Pig Varieties
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Data, and Ethics Statements
2.2. Transmission Ratio Distortion Analysis
2.3. Analysis of Gene Enrichment
2.4. Data Availability
3. Results and Discussion
3.1. Estimates of Maternal Transmission Ratio Distortion Loci
3.2. Possible Biological Implications of Transmission Ratio Distortion Loci
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lyttle, T.W. Segregation distorters. Annu. Rev. Genet. 1991, 25, 511–557. [Google Scholar] [CrossRef] [PubMed]
- Silver, L.M. The peculiar journey of a selfish chromosome: Mouse t haplotypes and meiotic drive. Trends Genet. TIG 1993, 9, 250–254. [Google Scholar] [CrossRef]
- Villena, F.P.-M.D.; Casa-Esperón, E.D.L.; Briscoe, T.L.; Sapienza, C. A Genetic test to determine the origin of maternal transmission ratio distortion: Meiotic drive at the mouse om locus. Genetics 2000, 154, 333–342. [Google Scholar]
- Paz-Miguel, J.E.; Pardo-Manuel de Villena, F.; Sánchez-Velasco, P.; Leyva-Cobián, F. H2-haplotype-dependent unequal transmission of the 1716 translocation chromosome from Ts65Dn females. Mamm. Genome 2001, 12, 83–85. [Google Scholar] [CrossRef]
- Meyer, W.K.; Arbeithuber, B.; Ober, C.; Ebner, T.; Tiemann-Boege, I.; Hudson, R.R.; Przeworski, M. Evaluating the evidence for transmission distortion in human pedigrees. Genetics 2012, 191, 215–232. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, L.; Xu, S.; Hu, L.; Hurst, L.D.; Kong, X. Identification of two maternal transmission ratio distortion loci in pedigrees of the framingham heart study. Sci. Rep. 2013, 3, 2147. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Hao, L.; Han, Z.; Gao, S.; Latham, K.E.; de Villena, F.P.-M.; Sapienza, C. Maternal transmission ratio distortion at the mouse om locus results from meiotic drive at the second meiotic division. Genetics 2005, 170, 327–334. [Google Scholar] [CrossRef][Green Version]
- Nur, U. Maintenance of a “Parasitic” B Chromosome in the Grasshopper Melanoplus Femur-Rubrum. Genetics 1977, 87, 499–512. [Google Scholar]
- Solignac, M.; Vautrin, D.; Baudry, E.; Mougel, F.; Loiseau, A.; Cornuet, J.-M. A Microsatellite-Based linkage map of the honeybee, Apis mellifera L. Genetics 2004, 167, 253–262. [Google Scholar] [CrossRef]
- Vongs, A.; Kakutani, T.; Martienssen, R.A.; Richards, E.J. Arabidopsis thaliana DNA methylation mutants. Science 1993, 260, 1926–1928. [Google Scholar] [CrossRef] [PubMed]
- Koide, Y.; Onishi, K.; Nishimoto, D.; Baruah, A.R.; Kanazawa, A.; Sano, Y. Sex-independent transmission ratio distortion system responsible for reproductive barriers between Asian and African rice species. New Phytol. 2008, 179, 888–900. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi, N. A genetically determined incompatibility system between spermatozoa and eggs leading to embryonic death in mice. J. Reprod. Fertil. 1974, 41, 85–96. [Google Scholar] [CrossRef]
- Agulnik, S.I.; Agulnik, A.I.; Ruvinsky, A.O. Meiotic drive in female mice heterozygous for the HSR inserts on chromosome 1. Genet. Res. 1990, 55, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Fishman, L.; Kelly, A.J.; Morgan, E.; Willis, J.H. A genetic map in the mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 2001, 159, 1701–1716. [Google Scholar] [PubMed]
- Dyer, K.A.; Charlesworth, B.; Jaenike, J. Chromosome-wide linkage disequilibrium as a consequence of meiotic drive. Proc. Natl. Acad. Sci. USA 2007, 104, 1587–1592. [Google Scholar] [CrossRef] [PubMed]
- Fishman, L.; McIntosh, M. Standard Deviations: The biological bases of transmission ratio distortion. Annu. Rev. Genet. 2019, 53, 347–372. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.O.; Labbe, A.; Infante-Rivard, C. Transmission ratio distortion: Review of concept and implications for genetic association studies. Hum. Genet. 2013, 132, 245–263. [Google Scholar] [CrossRef] [PubMed]
- Lorieux, M.; Goffinet, B.; Perrier, X.; de León, D.G.; Lanaud, C. Maximum-likelihood models for mapping genetic markers showing segregation distortion. 1. Backcross populations. TAG Theor. Appl. Genet. Theor. Angew. Genet. 1995, 90, 73–80. [Google Scholar] [CrossRef]
- Xu, S. Theoretical Basis of the Beavis Effect. Genetics 2003, 165, 2259–2268. [Google Scholar]
- Philipsen, M.; Kristensen, B. Preliminary evidence of segregation distortion in the SLA system. Anim. Blood Groups Biochem. Genet. 1985, 16, 125–133. [Google Scholar] [CrossRef]
- Jeon, J.-T.; Carlborg, Ö.; Törnsten, A.; Giuffra, E.; Amarger, V.; Chardon, P.; Andersson-Eklund, L.; Andersson, K.; Hansson, I.; Lundström, K.; et al. A paternally expressed QTL affecting skeletal and cardiac muscle mass in pigs maps to the IGF2 locus. Nat. Genet. 1999, 21, 157–158. [Google Scholar] [CrossRef] [PubMed]
- Pinton, A.; Calgaro, A.; Bonnet, N.; Ferchaud, S.; Billoux, S.; Dudez, A.M.; Mary, N.; Massip, K.; Bonnet-Garnier, A.; Yerle, M.; et al. Influence of sex on the meiotic segregation of a t(13;17) Robertsonian translocation: A case study in the pig. Hum. Reprod. 2009, 24, 2034–2043. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Casellas, J.; Manunza, A.; Mercader, A.; Quintanilla, R.; Amills, M. A flexible bayesian model for testing for transmission ratio distortion. Genetics 2014, 198, 1357–1367. [Google Scholar] [CrossRef]
- Casellas, J.; Gularte, R.J.; Farber, C.R.; Varona, L.; Mehrabian, M.; Schadt, E.E.; Lusis, A.J.; Attie, A.D.; Yandell, B.S.; Medrano, J.F. Genome scans for transmission ratio distortion regions in mice. Genetics 2012, 191, 247–259. [Google Scholar] [CrossRef][Green Version]
- Shendure, J.; Melo, J.A.; Pociask, K.; Derr, R.; Silver, L.M. Sex-restricted non-Mendelian inheritance of mouse chromosome 11 in the offspring of crosses between C57BL/6J and (C57BL/6J x DBA/2J)F1 mice. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 1998, 9, 812–815. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.O.; Infante-Rivard, C.; Labbe, A. Analysis of case-parent trios using a loglinear model with adjustment for transmission ratio distortion. Front. Genet. 2016, 7, 155. [Google Scholar] [CrossRef]
- Lopez-Bote, C.J. Sustained utilization of the Iberian pig breed. Meat Sci. 1998, 49, S17–S27. [Google Scholar] [CrossRef]
- Silió, L. Iberian pig breeding programme. Dev. Breed. Strateg. Low. Input Anim. Prod. Environ. 2000, 511–519. [Google Scholar]
- Ibáñez-Escriche, N.; Varona, L.; Magallón, E.; Noguera, J.L. Crossbreeding effects on pig growth and carcass traits from two Iberian strains. Animal 2014, 8, 1569–1576. [Google Scholar] [CrossRef]
- FAO. Global Plan of Action for Animal Genetic Resources and Interlaken Declaration; International Technical Conference on Animal Genetic Resources for Food and Agriculture: Interlaken, Switzerland, 2007. [Google Scholar]
- Esteve-Codina, A.; Kofler, R.; Himmelbauer, H.; Ferretti, L.; Vivancos, A.P.; Groenen, M.A.M.; Folch, J.M.; Rodríguez, M.C.; Pérez-Enciso, M. Partial short-read sequencing of a highly inbred Iberian pig and genomics inference thereof. Heredity 2011, 107, 256–264. [Google Scholar] [CrossRef][Green Version]
- Vázquez-Gómez, M.; García-Contreras, C.; Astiz, S.; Torres-Rovira, L.; Fernández-Moya, E.; Olivares, Á.; Daza, A.; Óvilo, C.; González-Bulnes, A.; Isabel, B. Piglet birthweight and sex affect growth performance and fatty acid composition in fatty pigs. Anim. Prod. Sci. 2020, 60, 573–583. [Google Scholar] [CrossRef]
- Laval, G.; Iannuccelli, N.; Legault, C.; Milan, D.; Groenen, M.A.; Giuffra, E.; Andersson, L.; Nissen, P.H.; Jørgensen, C.B.; Beeckmann, P.; et al. Genetic diversity of eleven European pig breeds. Genet. Sel. Evol. 2000, 32, 187. [Google Scholar] [CrossRef] [PubMed]
- Fabuel, E.; Barragán, C.; Silió, L.; Rodríguez, M.C.; Toro, M.A. Analysis of genetic diversity and conservation priorities in Iberian pigs based on microsatellite markers. Heredity 2004, 93, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Alonso, I.; Ibáñez-Escriche, N.; Noguera, J.L.; Casellas, J.; Hijas-Villalba, M.M.D.; Gracia-Santana, M.J.; Varona, L. Genomic differentiation among varieties of Iberian pig. Span. J. Agric. Res. 2020, 18, 0401. [Google Scholar] [CrossRef]
- Ibáñez-Escriche, N.; Magallón, E.; Gonzalez, E.; Tejeda, J.F.; Noguera, J.L. Genetic parameters and crossbreeding effects of fat deposition and fatty acid profiles in Iberian pig lines1. J. Anim. Sci. 2016, 94, 28–37. [Google Scholar] [CrossRef]
- Pena, R.N.; Noguera, J.L.; García-Santana, M.J.; González, E.; Tejeda, J.F.; Ros-Freixedes, R.; Ibáñez-Escriche, N. Five genomic regions have a major impact on fat composition in Iberian pigs. Sci. Rep. 2019, 9, 2031. [Google Scholar] [CrossRef]
- Noguera, J.L.; Ibáñez-Escriche, N.; Casellas, J.; Rosas, J.P.; Varona, L. Genetic parameters and direct, maternal and heterosis effects on litter size in a diallel cross among three commercial varieties of Iberian pig. Anim. Int. J. Anim. Biosci. 2019, 13, 2765–2772. [Google Scholar] [CrossRef]
- Casellas, J.; Ibáñez-Escriche, N.; Varona, L.; Rosas, J.P.; Noguera, J.L. Inbreeding depression load for litter size in Entrepelado and Retinto Iberian pig varieties. J. Anim. Sci. 2019, 97, 1979–1986. [Google Scholar] [CrossRef]
- Nelson, W.A. Statistical Methods. In Encyclopedia of Ecology; Jørgensen, S.E., Fath, B.D., Eds.; Academic Press: Oxford, UK, 2008; pp. 3350–3362. ISBN 978-0-08-045405-4. [Google Scholar]
- Spielman, R.S.; McGinnis, R.E.; Ewens, W.J. Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am. J. Hum. Genet. 1993, 52, 506–516. [Google Scholar]
- Weinberg, C.R.; Wilcox, A.J.; Lie, R.T. A Log-Linear approach to case-parent–triad data: Assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am. J. Hum. Genet. 1998, 62, 969–978. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Haider, S.; Ballester, B.; Smedley, D.; Zhang, J.; Rice, P.; Kasprzyk, A. BioMart Central Portal—unified access to biological data. Nucleic Acids Res. 2009, 37, W23–W27. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2016, 45, D183–D189. [Google Scholar] [CrossRef] [PubMed]
- Online Mendelian Inheritance in Man OMIM®. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD). Available online: https://omim.org/ (accessed on 10 July 2020).
- Online Mendelian Inheritance in Animals OMIA. Sydney School of Veterinary Science. Available online: https://omia.org/ (accessed on 10 July 2020).
- Kristensen, T.N.; Sørensen, A.C. Inbreeding–lessons from animal breeding, evolutionary biology and conservation genetics. Anim. Sci. 2005, 80, 121–133. [Google Scholar] [CrossRef]
- Bosse, M.; Megens, H.-J.; Madsen, O.; Paudel, Y.; Frantz, L.A.F.; Schook, L.B.; Crooijmans, R.P.M.A.; Groenen, M.A.M. Regions of homozygosity in the porcine genome: Consequence of demography and the recombination landscape. PLoS Genet. 2012, 8, e1003100. [Google Scholar] [CrossRef]
- Gutiérrez, J.B. Jamón Curado: Aspectos Científicos y Tecnológicos; Ed. Díaz de Santos: Madrid, Spain, 2012; ISBN 978-84-9969-076-6. [Google Scholar]
- Silio, L.; Rodriguez, M.C.; Fernandez, A.; Barragan, C.; Benitez, R.; Ovilo, C.; Fernandez, A.I. Measuring inbreeding and inbreeding depression on pig growth from pedigree or SNP-derived metrics. J. Anim. Breed. Genet. 2013, 130, 349–360. [Google Scholar] [CrossRef]
- Eaves, I.A.; Bennett, S.T.; Forster, P.; Ferber, K.M.; Ehrmann, D.; Wilson, A.J.; Bhattacharyya, S.; Ziegler, A.-G.; Brinkmann, B.; Todd, J.A. Transmission ratio distortion at the INS - IGF2 VNTR. Nat. Genet. 1999, 22, 324–325. [Google Scholar] [CrossRef]
- Pardo-Manuel de Villena, F.; Sapienza, C. Transmission ratio distortion in offspring of heterozygous female carriers of Robertsonian translocations. Hum. Genet. 2001, 108, 31–36. [Google Scholar] [CrossRef]
- Saura, M.; Fernández, A.; Varona, L.; Fernández, A.I.; de Cara, M.Á.R.; Barragán, C.; Villanueva, B. Detecting inbreeding depression for reproductive traits in Iberian pigs using genome-wide data. Genet. Sel. Evol. 2015, 47, 1. [Google Scholar] [CrossRef]
- Hunt, S.E.; McLaren, W.; Gil, L.; Thormann, A.; Schuilenburg, H.; Sheppard, D.; Parton, A.; Armean, I.M.; Trevanion, S.J.; Flicek, P.; et al. Ensembl variation resources. Database 2018, 2018. [Google Scholar] [CrossRef]
- Labbe, A.; Huang, L.O.; Infante-Rivard, C. Transmission Ratio Distortion: A Neglected Phenomenon with Many Consequences in Genetic Analysis and Population Genetics. In Epigenetics and Complex Traits; Naumova, A.K., Greenwood, C.M.T., Eds.; Springer: New York, NY, USA, 2013; pp. 265–285. ISBN 978-1-4614-8078-5. [Google Scholar]
- Kido, T.; Sikora-Wohlfeld, W.; Kawashima, M.; Kikuchi, S.; Kamatani, N.; Patwardhan, A.; Chen, R.; Sirota, M.; Kodama, K.; Hadley, D.; et al. Are minor alleles more likely to be risk alleles? BMC Med. Genomics 2018, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Balick, D.J.; Do, R.; Cassa, C.A.; Reich, D.; Sunyaev, S.R. Dominance of deleterious alleles controls the response to a population bottleneck. PLoS Genet. 2015, 11, e1005436. [Google Scholar] [CrossRef]
- Plough, L.V.; Hedgecock, D. Quantitative trait locus analysis of stage-specific inbreeding depression in the Pacific oyster Crassostrea gigas. Genetics 2011, 189, 1473–1486. [Google Scholar] [CrossRef] [PubMed]
- Xu, S. Quantitative trait locus mapping can benefit from segregation distortion. Genetics 2008, 180, 2201–2208. [Google Scholar] [CrossRef] [PubMed]
- Id-Lahoucine, S.; Cánovas, A.; Jaton, C.; Miglior, F.; Fonseca, P.A.S.; Sargolzaei, M.; Miller, S.; Schenkel, F.S.; Medrano, J.F.; Casellas, J. Implementation of Bayesian methods to identify SNP and haplotype regions with transmission ratio distortion across the whole genome: TRDscan v.1.0. J. Dairy Sci. 2019, 102, 3175–3188. [Google Scholar] [CrossRef] [PubMed]
- Schulz, R.; Underkoffler, L.A.; Collins, J.N.; Oakey, R.J. Nondisjunction and transmission ratio distortion of Chromosome 2 in a (2.8) Robertsonian translocation mouse strain. Mamm. Genome 2006, 17, 239–247. [Google Scholar] [CrossRef][Green Version]
- Eversley, C.D.; Clark, T.; Xie, Y.; Steigerwalt, J.; Bell, T.A.; de Villena, F.P.; Threadgill, D.W. Genetic mapping and developmental timing of transmission ratio distortion in a mouse interspecific backcross. BMC Genet. 2010, 11, 98. [Google Scholar] [CrossRef]
- Whetstine, J.R. Chapter 287 - Histone Methylation: Chemically Inert but Chromatin Dynamic. In Handbook of Cell Signaling, 2nd ed.; Bradshaw, R.A., Dennis, E.A., Eds.; Academic Press: San Diego, CA, USA, 2010; pp. 2389–2397. ISBN 978-0-12-374145-5. [Google Scholar]
- Rugg-Gunn, P.J.; Cox, B.J.; Ralston, A.; Rossant, J. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc. Natl. Acad. Sci. USA 2010, 107, 10783–10790. [Google Scholar] [CrossRef]
- Canovas, S.; Ross, P.J. Epigenetics in preimplantation mammalian development. Theriogenology 2016, 86, 69–79. [Google Scholar] [CrossRef]
- Jambhekar, A.; Dhall, A.; Shi, Y. Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 2019, 20, 625–641. [Google Scholar] [CrossRef]
- Gonzalez-Bulnes, A.; Astiz, S.; Ovilo, C.; Lopez-Bote, C.J.; Torres-Rovira, L.; Barbero, A.; Ayuso, M.; Garcia-Contreras, C.; Vazquez-Gomez, M. Developmental Origins of Health and Disease in swine: Implications for animal production and biomedical research. Theriogenology 2016, 86, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Biochemistry, 5th ed.; W. H. Freeman and Company: NewYork, NY, USA, 2002. [Google Scholar]
- Ishibashi, Y.; Kohyama-Koganeya, A.; Hirabayashi, Y. New insights on glucosylated lipids: Metabolism and functions. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2013, 1831, 1475–1485. [Google Scholar] [CrossRef] [PubMed]
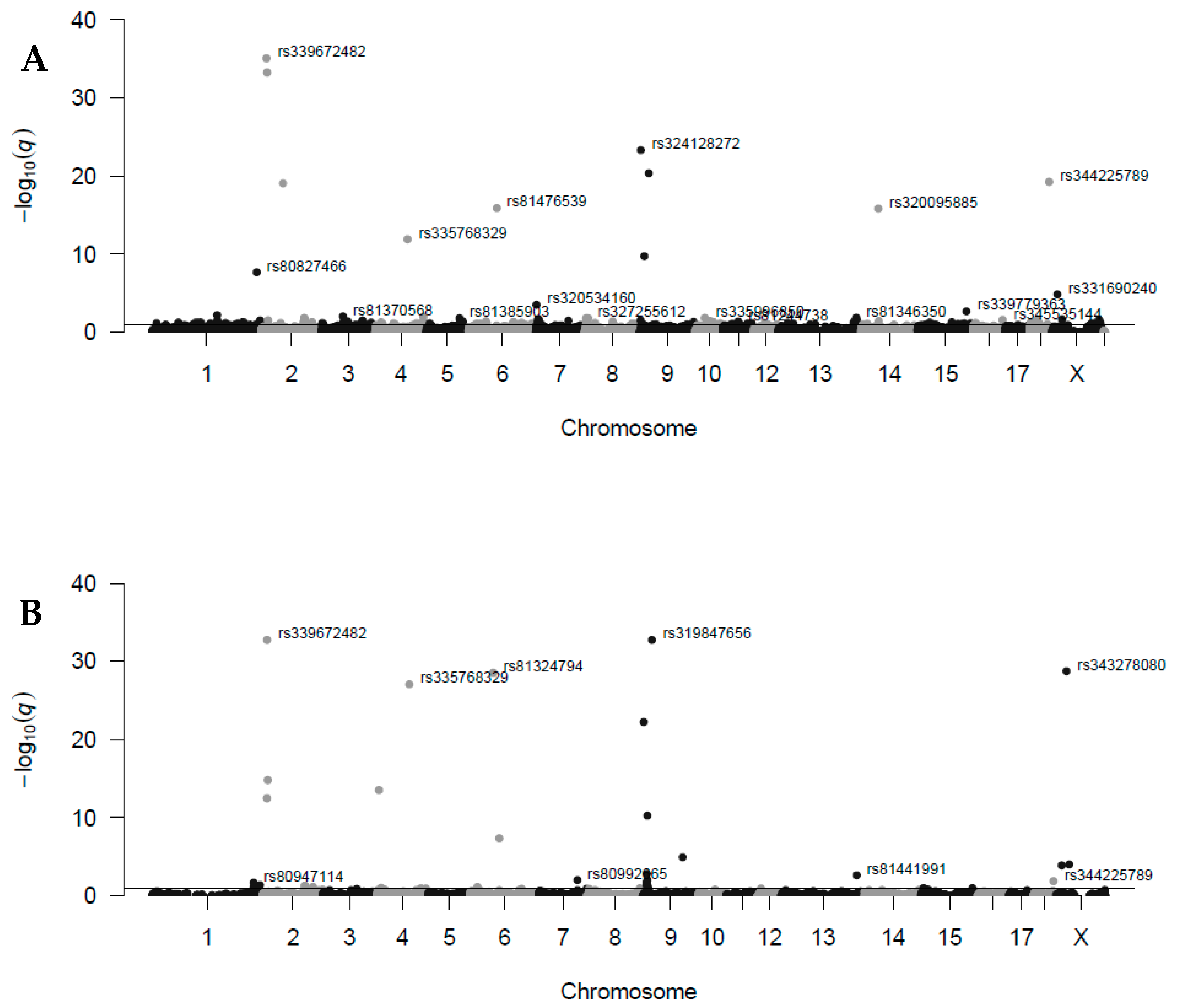
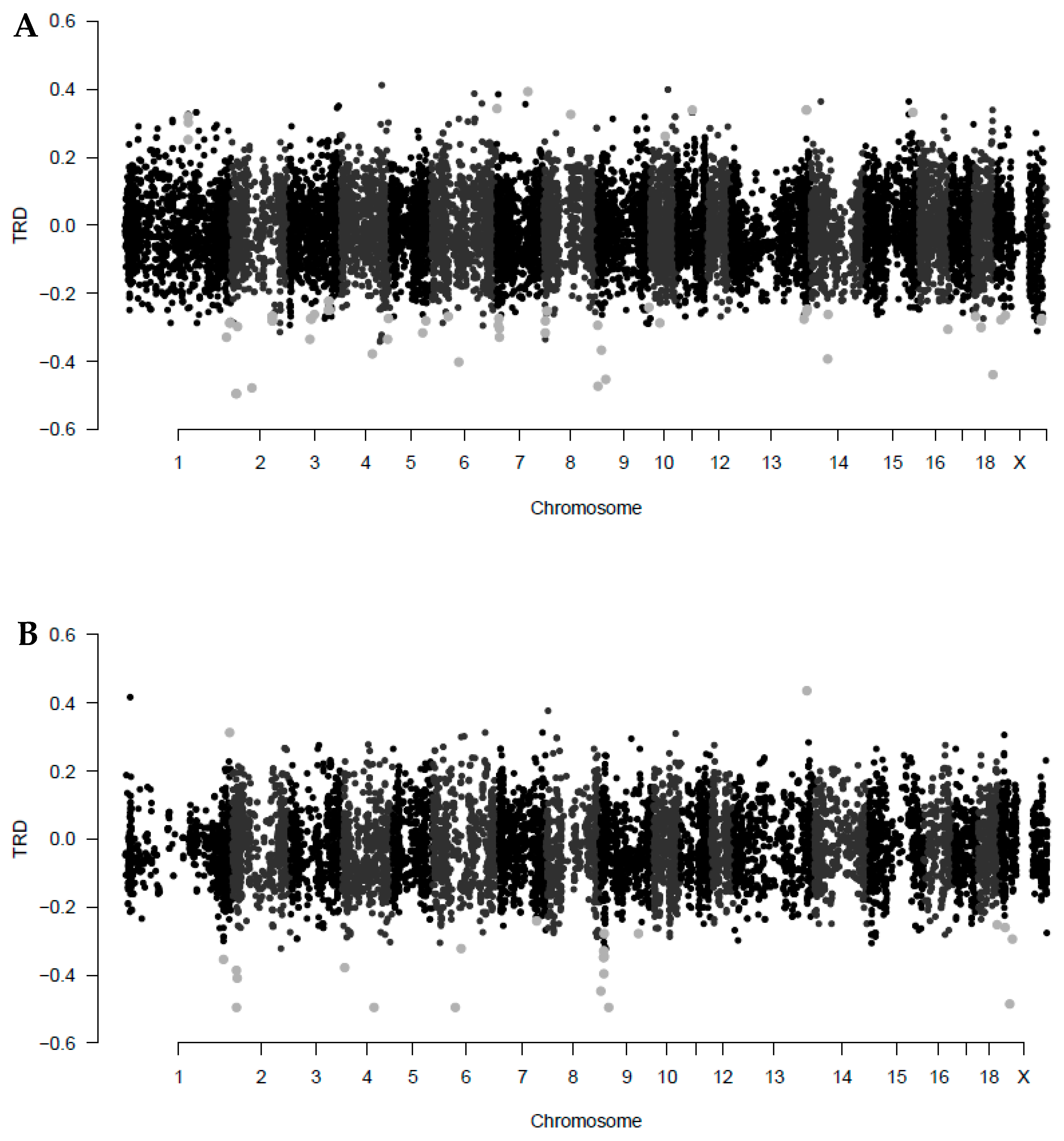
| ID Marker | Chr | Pos (Mb) | TRD_E | q-val_E | CallR_E | TRD_R | q-val_R | CallR_R | Gene with TRD Locus |
|---|---|---|---|---|---|---|---|---|---|
| rs339672482 | 2 | 12.698 | −0.495 | 8.5 × 10−36 | 100.0 | −0.495 | 1.8 × 10−33 | 100.0 | ENSSSCG00000031496 |
| rs343381067 | 2 | 14.467 | −0.495 | 5.3 × 10−34 | 98.9 | −0.409 | 1.4 × 10−15 | 99.4 | ENSSSCG00000031436 |
| rs335768329 | 4 | 83.114 | −0.378 | 1.3 × 10−12 | 96.6 | −0.495 | 8.8 × 10−28 | 94.5 | ADCY10 |
| rs81476539 | 6 | 72.737 | −0.402 | 1.3 × 10−16 | 100.0 | −0.322 | 4.1 × 10−08 | 100.0 | |
| rs324128272 | 9 | 2.722 | −0.473 | 4.8 × 10−24 | 99.4 | −0.447 | 5.5 × 10−23 | 99.4 | |
| rs346413844 | 9 | 11.795 | −0.367 | 1.8 × 10−10 | 93.7 | −0.346 | 5.2 × 10−11 | 97.8 | |
| rs319847656 | 9 | 23.041 | −0.453 | 4.3 × 10−21 | 99.4 | −0.495 | 1.8 × 10−33 | 100.0 | ENSSSCG00000049020 |
| rs344225789 | 18 | 49.209 | −0.439 | 5.5 × 10−20 | 99.4 | −0.252 | 0.0131 | 99.4 | |
| rs331690240 | 20 | 14.2345 | −0.278 | 1.4 × 10−05 | 100.0 | −0.26 | 1.2 × 10−04 | 99.4 | ENSSSCG00000050465 |
| rs343278080 | 20 | 26.079 | −0.265 | 0.0231 | 100.0 | −0.485 | 1.8 × 10−29 | 100.0 |
| ID Marker | Chromosome | Position (b) | Gene with TRDL | Iberian Variety | Marker Call Rate |
|---|---|---|---|---|---|
| rs337916686 | 1 | 162,977,295 | ATP8B1 | Entrepelado | 100.0 |
| rs80864027 | 1 | 163,382,446 | IGDCC4 | Entrepelado | 100.0 |
| rs81305791 | 1 | 271,207,043 | NUP214 | Entrepelado | 100.0 |
| rs339672482 | 2 | 12,697,523 | ENSSSCG00000031496 | Entrepelado | 100.0 |
| rs343381067 | 2 | 14,467,327 | ENSSSCG00000031436 | Entrepelado | 98.9 |
| rs81221692 | 3 | 102,885,453 | PRKD3 | Entrepelado | 100.0 |
| rs335768329 | 4 | 83,113,768 | ADCY10 | Entrepelado | 96.6 |
| rs81385903 | 5 | 83,334,157 | ANO4 | Entrepelado | 100.0 |
| rs55618893 | 5 | 91,751,676 | LUM | Entrepelado | 100.0 |
| rs320534160 | 7 | 1,148,169 | GMDS | Entrepelado | 100.0 |
| rs80939667 | 7 | 7,368,476 | GCNT2 | Entrepelado | 100.0 |
| rs338044350 | 7 | 7,378,724 | GCNT2 | Entrepelado | 100.0 |
| rs327255612 | 8 | 4,896,260 | EVC2 | Entrepelado | 100.0 |
| rs81420408 | 9 | 2,210,850 | SYT9 | Entrepelado | 100.0 |
| rs319847656 | 9 | 23,040,656 | ENSSSCG00000049020 | Entrepelado | 99.4 |
| rs81262274 | 9 | 135,247,805 | ENSSSCG00000031141 | Entrepelado | 100.0 |
| rs335996850 | 10 | 24,233,693 | RNPEP | Entrepelado | 100.0 |
| rs320095885 | 14 | 42,820,750 | SGSM1 | Entrepelado | 100.0 |
| rs80994847 | 14 | 43,934,858 | SEZ6L | Entrepelado | 100.0 |
| rs334182161 | 18 | 3,613,926 | DPP6 | Entrepelado | 100.0 |
| rs327443567 | 18 | 17,971,591 | ENSSSCG00000051610 | Entrepelado | 100.0 |
| rs326744865 | 18 | 18,486,229 | CPA1 | Entrepelado | 100.0 |
| rs331690240 | 20 | 14,234,646 | ENSSSCG00000050465 | Entrepelado | 100.0 |
| rs320767193 | 20 | 120,168,429 | ENSSSCG00000042120 | Entrepelado | 100.0 |
| rs80947114 | 1 | 252,918,477 | SUSD1 | Retinto | 100.0 |
| rs333078973 | 1 | 268,840,777 | CERCAM | Retinto | 100.0 |
| rs339672482 | 2 | 12,697,523 | ENSSSCG00000031496 | Retinto | 100.0 |
| rs343381067 | 2 | 14,467,327 | ENSSSCG00000031436 | Retinto | 99.4 |
| rs323787335 | 4 | 7,369,788 | ZFAT | Retinto | 100.0 |
| rs335768329 | 4 | 83,113,768 | ADCY10 | Retinto | 94.5 |
| rs81414835 | 9 | 9,862,005 | MAP6 | Retinto | 99.4 |
| rs81414870 | 9 | 9,898,353 | MAP6 | Retinto | 99.4 |
| rs342178816 | 9 | 11,759,440 | ENSSSCG00000014877 | Retinto | 100.0 |
| rs319847656 | 9 | 23,040,656 | ENSSSCG00000049020 | Retinto | 100.0 |
| rs81280147 | 9 | 99,713,062 | CD36 | Retinto | 100.0 |
| rs331690240 | 20 | 14,234,646 | ENSSSCG00000050465 | Retinto | 99.4 |
| GO ID | Term | Genes Found | Fold Enrichment | p-Value Corrected |
|---|---|---|---|---|
| GO:0030307 | Cell growth | 1 | 56.78 | 0.0217 |
| GO:0097061 | Dendrite | 1 | 56.78 | 0.0217 |
| GO:0043542 | Endothelial cell migration | 1 | 45.42 | 0.026 |
| GO:0048332 | Mesoderm morphogenesis | 1 | 22.71 | 0.0472 |
| GO:0045216 | Cell-cell junction | 2 | 18.17 | 0.00622 |
| GO:0050770 | Axonogenesis | 2 | 10.32 | 0.0174 |
| GO:0010975 | Neuron development | 2 | 6.88 | 0.0359 |
| GO:0000904 | Cell morphogenesis | 4 | 5.05 | 0.00881 |
| GO:0000902 | Cell morphogenesis | 5 | 4.38 | 0.00617 |
| GO:0009653 | Morphogenesis | 6 | 2.98 | 0.0163 |
| GO ID | Term | Entrepelado Variety | Retinto Variety | ||||
|---|---|---|---|---|---|---|---|
| Genes Found | Fold Enrichment | p-Value Corrected | Genes Found | Fold Enrichment | p-Value Corrected | ||
| GO:0060048 | Cardiac muscle | 3 | 8.91 | 0.00649 | |||
| GO:0048813 | Dendrite | 2 | 8.41 | 0.0294 | |||
| GO:0002088 | Eye | 3 | 7.57 | 0.00966 | |||
| GO:0050953 | Light stimulus | 3 | 7.57 | 0.00966 | |||
| GO:0034968 | Histone-lysine methylation | 3 | 7.21 | 0.0109 | |||
| GO:0050803 | Synapse structure | 3 | 6.88 | 0.0122 | 2 | 9.47 | 0.0216 |
| GO:0045216 | Cell-cell junction | 3 | 6.06 | 0.0166 | 2 | 8.33 | 0.027 |
| GO:0017156 | Calcium | 3 | 5.41 | 0.0219 | |||
| GO:0051592 | Calcium | 3 | 4.73 | 0.0301 | |||
| GO:0050770 | Axonogenesis | 4 | 4.59 | 0.014 | 3 | 7.1 | 0.0101 |
| GO:0015850 | Compound transport | 4 | 4.49 | 0.015 | |||
| GO:0055002 | Striated muscle | 3 | 4.45 | 0.0348 | |||
| GO:0010975 | Neuron development | 5 | 3.82 | 0.0123 | 3 | 4.74 | 0.0281 |
| GO:0099504 | Synapse structure | 4 | 3.11 | 0.0453 | |||
| GO:0000904 | Cell morphogenesis | 8 | 2.24 | 0.031 | 5 | 2.89 | 0.0322 |
| GO:0009653 | Morphogenesis | 19 | 2.1 | 0.00348 | 7 | 1.6 | 0.218 |
| GO:0000902 | Cell morphogenesis | 10 | 1.95 | 0.0432 | 6 | 2.41 | 0.0416 |
| GO:0030307 | Cell growth | 1 | 26.05 | 0.0466 | |||
| GO:0097061 | Dendrite | 1 | 26.05 | 0.0466 | |||
| GO:0009247 | Lipid biosynthesis | 4 | 13.02 | 0.000369 | |||
| GO:0044255 | Lipid metabolism | 9 | 3.14 | 0.00285 | |||
| GO:0007606 | Chemical stimulus | 8 | 3.35 | 0.0033 | |||
| GO:0046467 | Lipid biosynthesis | 4 | 6.72 | 0.00361 | |||
| GO:0030148 | Lipid biosynthesis | 3 | 8.93 | 0.0056 | |||
| GO:0007186 | Receptor | 16 | 1.94 | 0.0117 | |||
| GO:0031623 | Receptor | 2 | 10.42 | 0.0183 | |||
| GO:0007584 | Response to nutrient | 1 | 26.05 | 0.0466 | |||
| GO:0015909 | Lipid transport | 1 | 26.05 | 0.0466 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez-Gómez, M.; Hijas-Villalba, M.M.d.; Varona, L.; Ibañez-Escriche, N.; Rosas, J.P.; Negro, S.; Noguera, J.L.; Casellas, J. Maternal Transmission Ratio Distortion in Two Iberian Pig Varieties. Genes 2020, 11, 1050. https://doi.org/10.3390/genes11091050
Vázquez-Gómez M, Hijas-Villalba MMd, Varona L, Ibañez-Escriche N, Rosas JP, Negro S, Noguera JL, Casellas J. Maternal Transmission Ratio Distortion in Two Iberian Pig Varieties. Genes. 2020; 11(9):1050. https://doi.org/10.3390/genes11091050
Chicago/Turabian StyleVázquez-Gómez, Marta, Melani Martín de Hijas-Villalba, Luis Varona, Noelia Ibañez-Escriche, Juan Pablo Rosas, Sara Negro, José Luis Noguera, and Joaquim Casellas. 2020. "Maternal Transmission Ratio Distortion in Two Iberian Pig Varieties" Genes 11, no. 9: 1050. https://doi.org/10.3390/genes11091050
APA StyleVázquez-Gómez, M., Hijas-Villalba, M. M. d., Varona, L., Ibañez-Escriche, N., Rosas, J. P., Negro, S., Noguera, J. L., & Casellas, J. (2020). Maternal Transmission Ratio Distortion in Two Iberian Pig Varieties. Genes, 11(9), 1050. https://doi.org/10.3390/genes11091050





