Transcriptomic Analysis Revealed the Common and Divergent Responses of Maize Seedling Leaves to Cold and Heat Stresses
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Stress Treatment
2.2. RNA Extraction, Library Preparation and RNA Sequencing
2.3. Data Filtering and Assessment
2.4. Gene Quantification and DEGs Identification
2.5. Expression Network Construction
2.6. Analysis of Quantitative Real-Time PCR (qRT-PCR)
3. Results
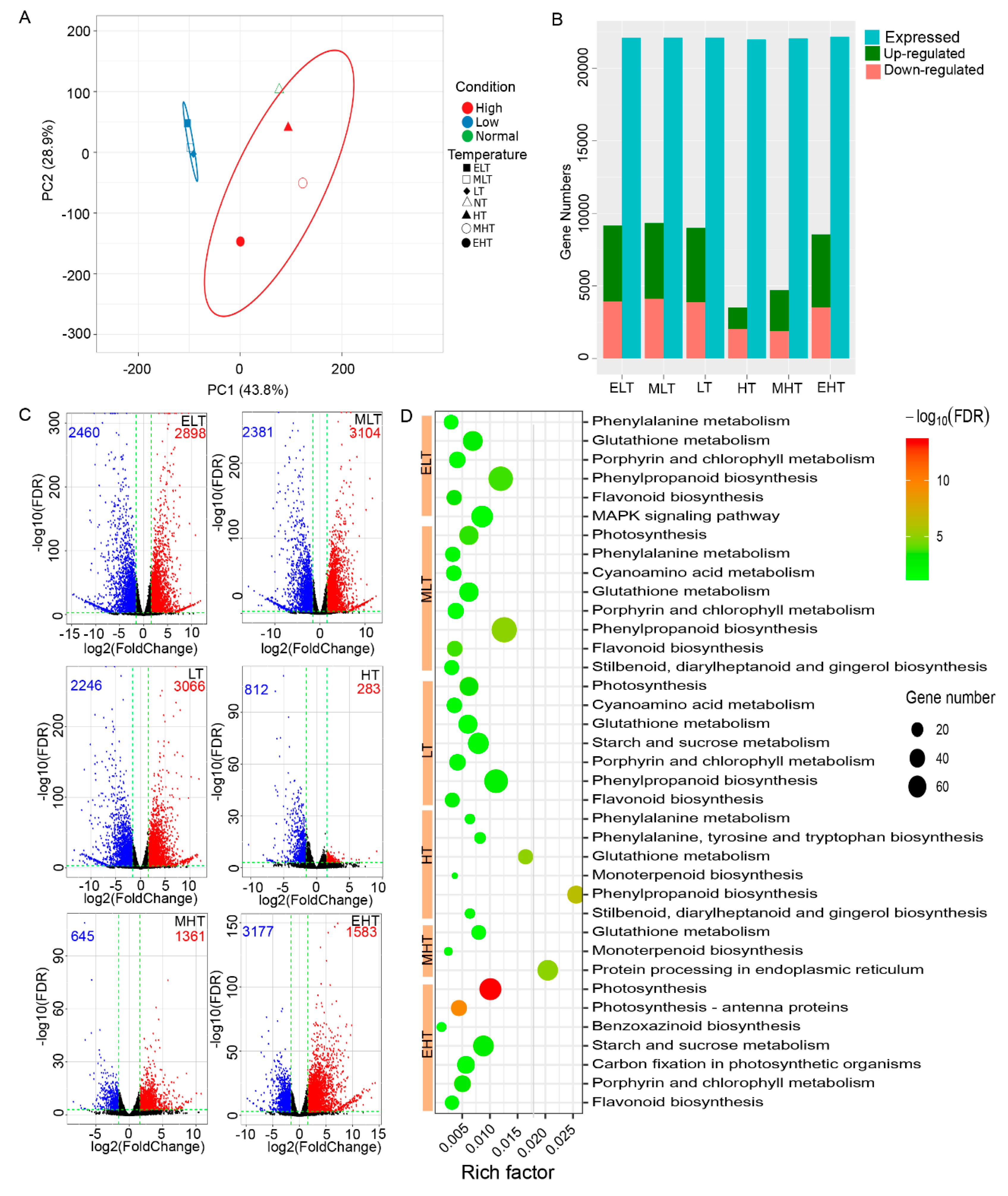
3.1. TS Inducing Large Amount Alterations in Transcriptome
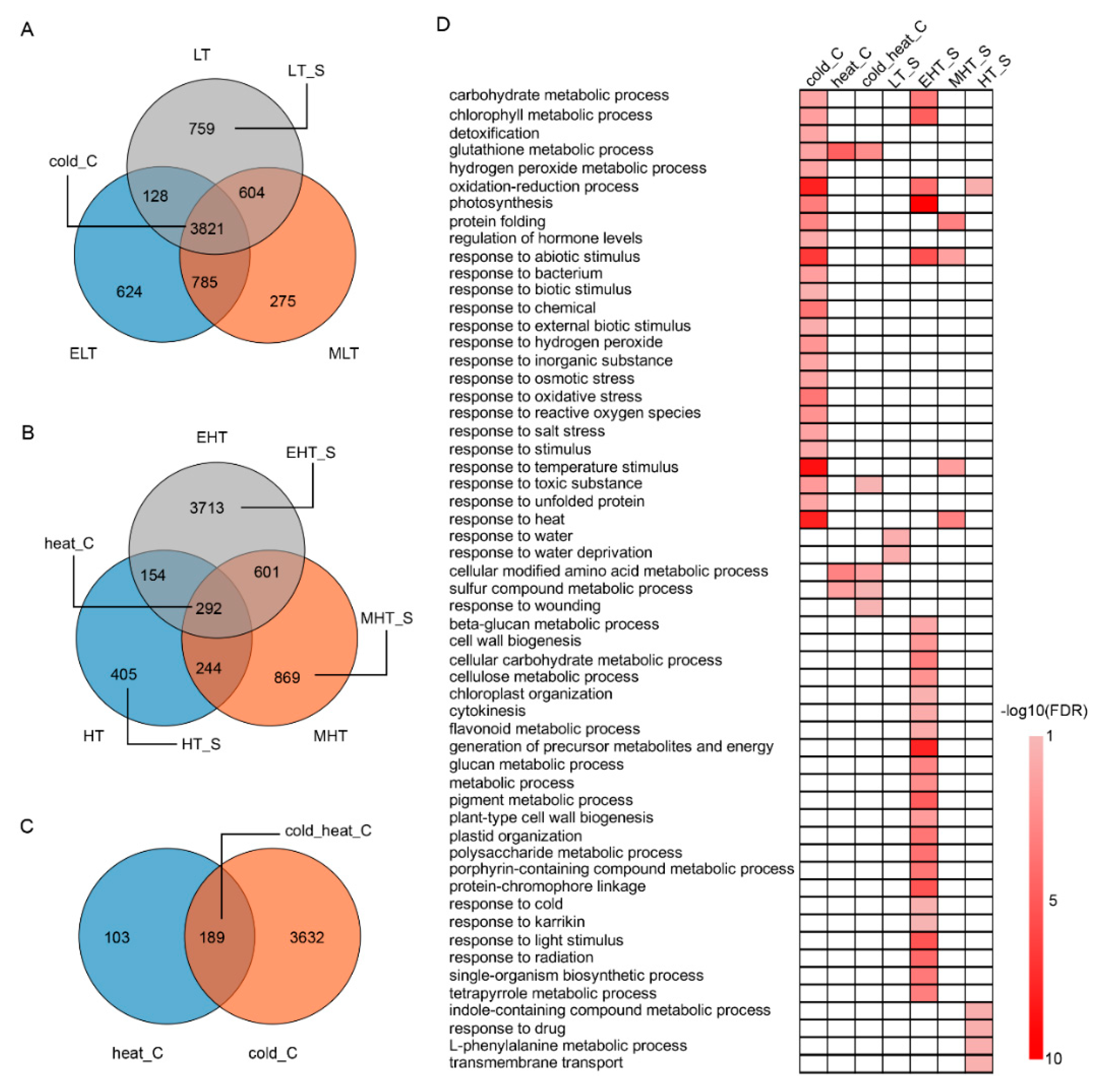
3.2. Common and Specific DEGs Responded to TS
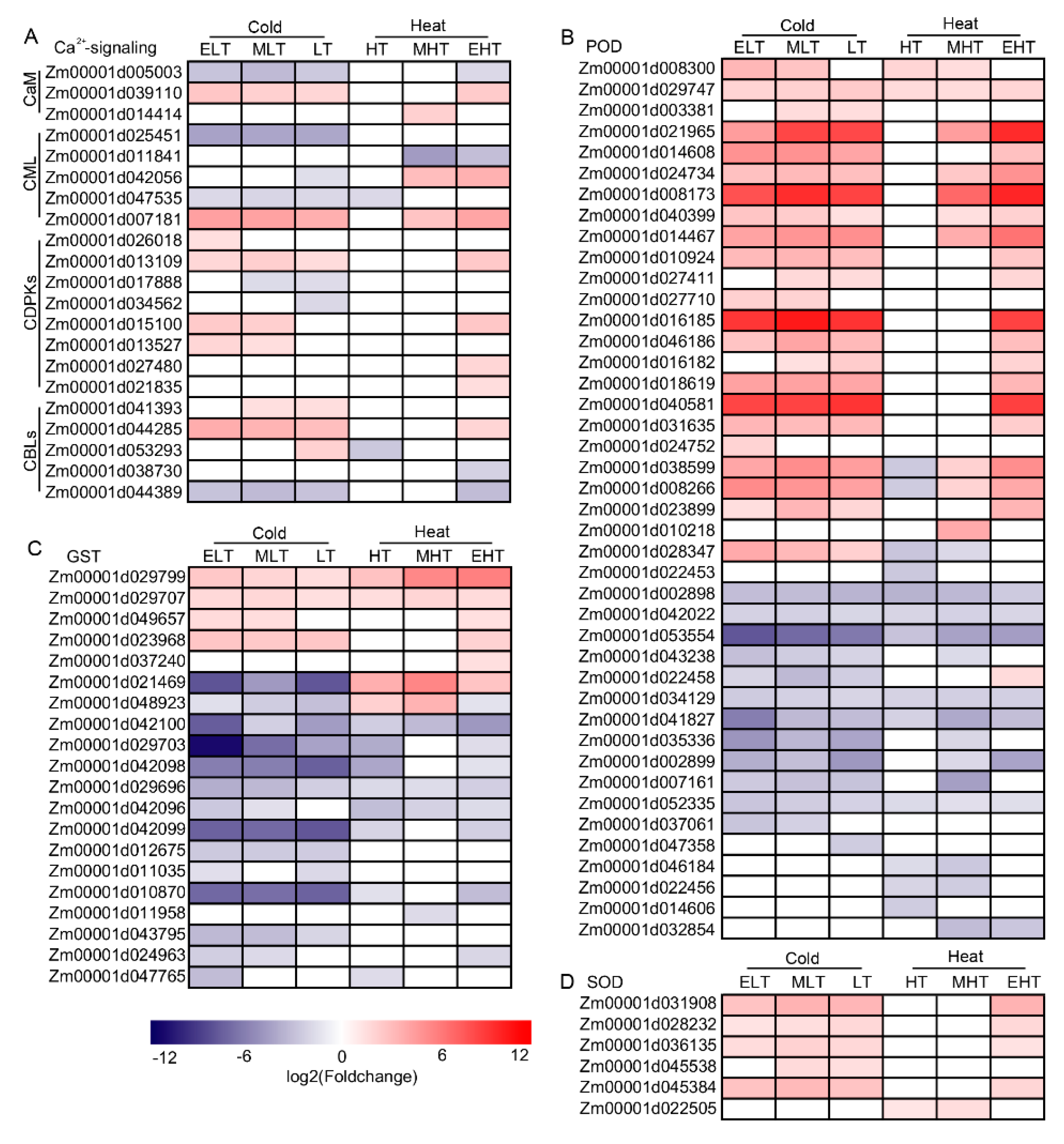
3.3. Ca2+ and ROS Signaling Were Induced under TS
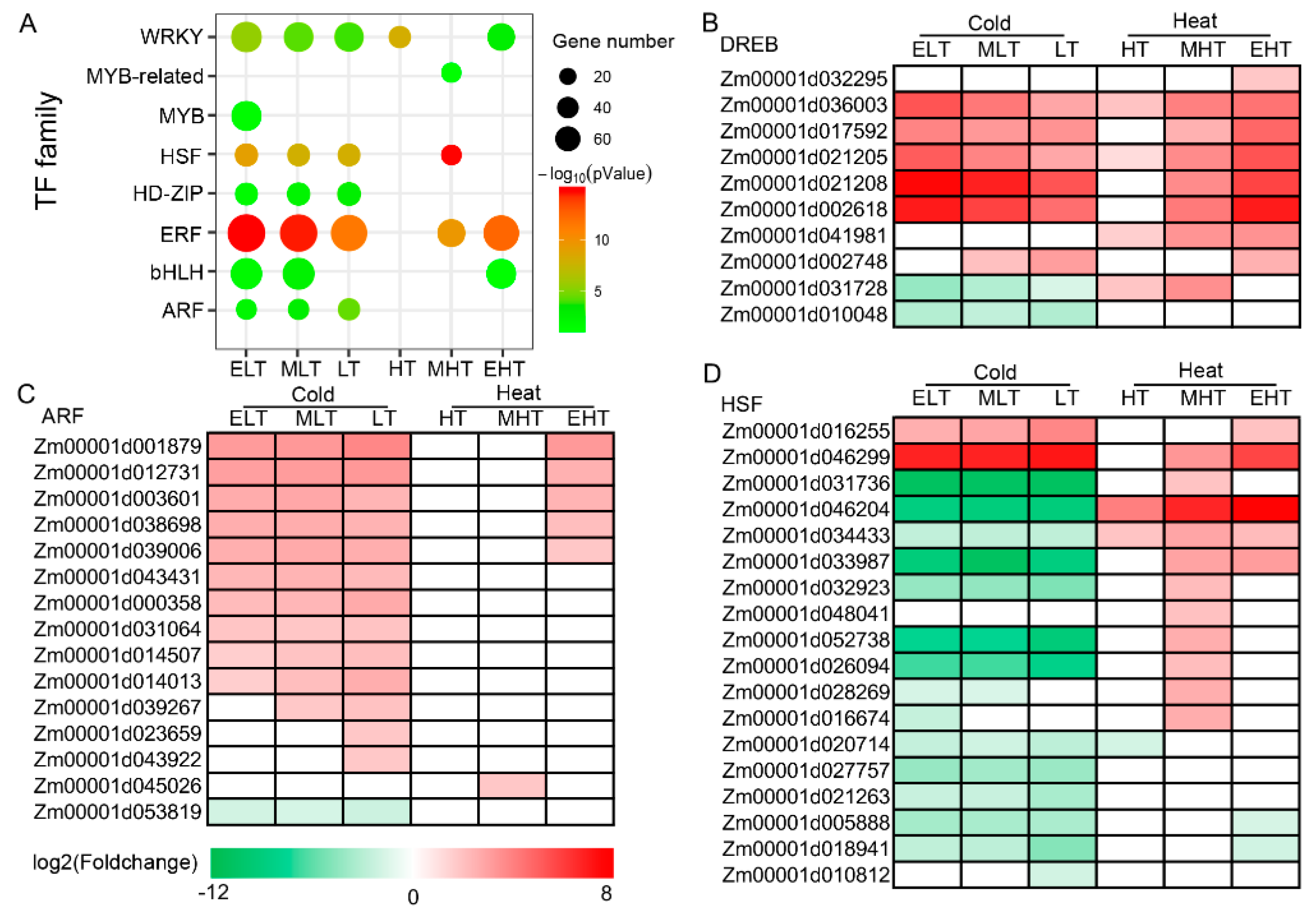
3.4. Dynamic Expression of Transcription Factors in Response to TS
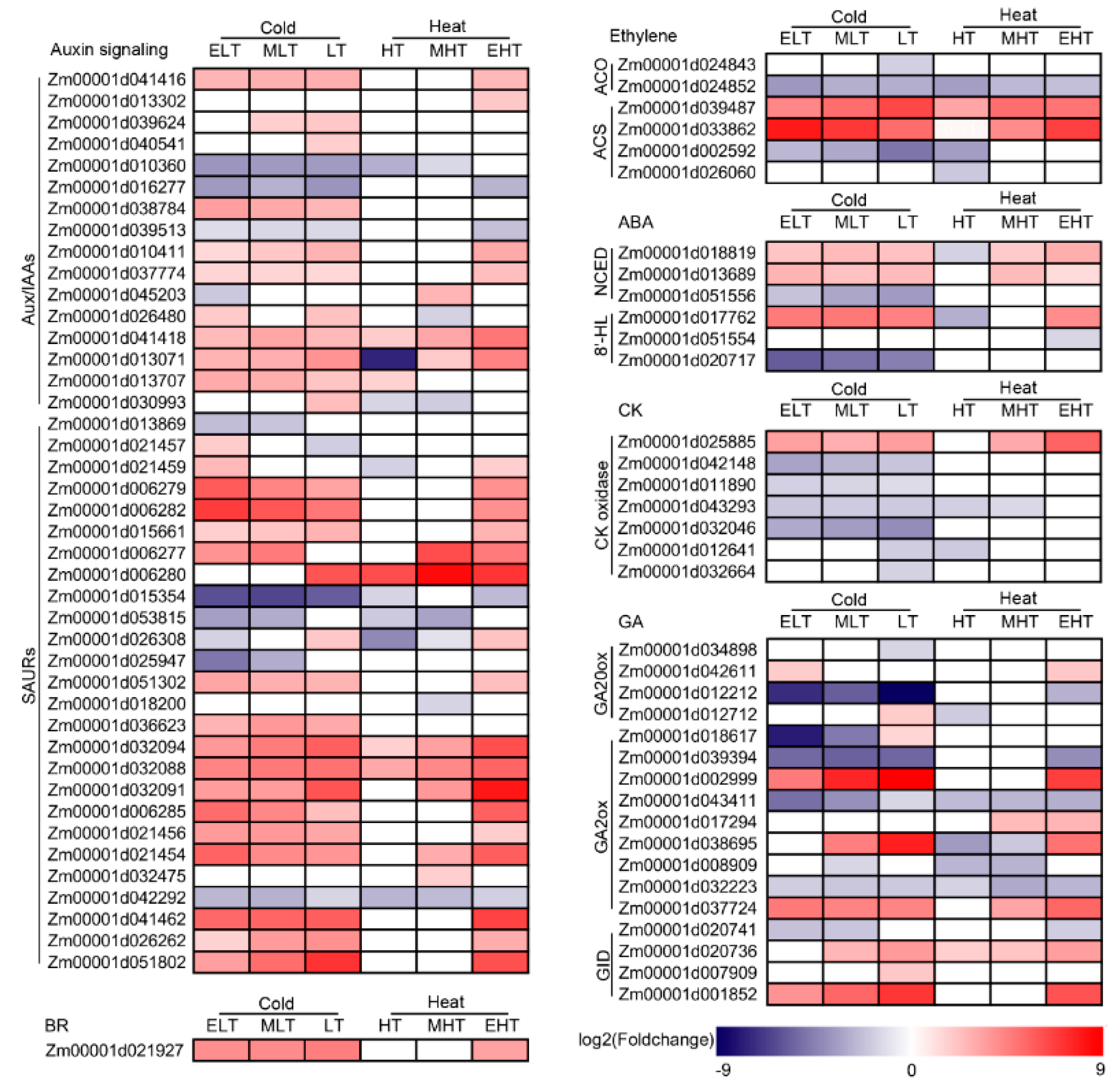
3.5. Hormone Metabolism Was Significantly Induced
3.6. Heat Shock Proteins (HSPs) Were Mainly Up-Regulated under Heat Conditions
3.7. The Hub Genes Associated with Cold and Heat Stress
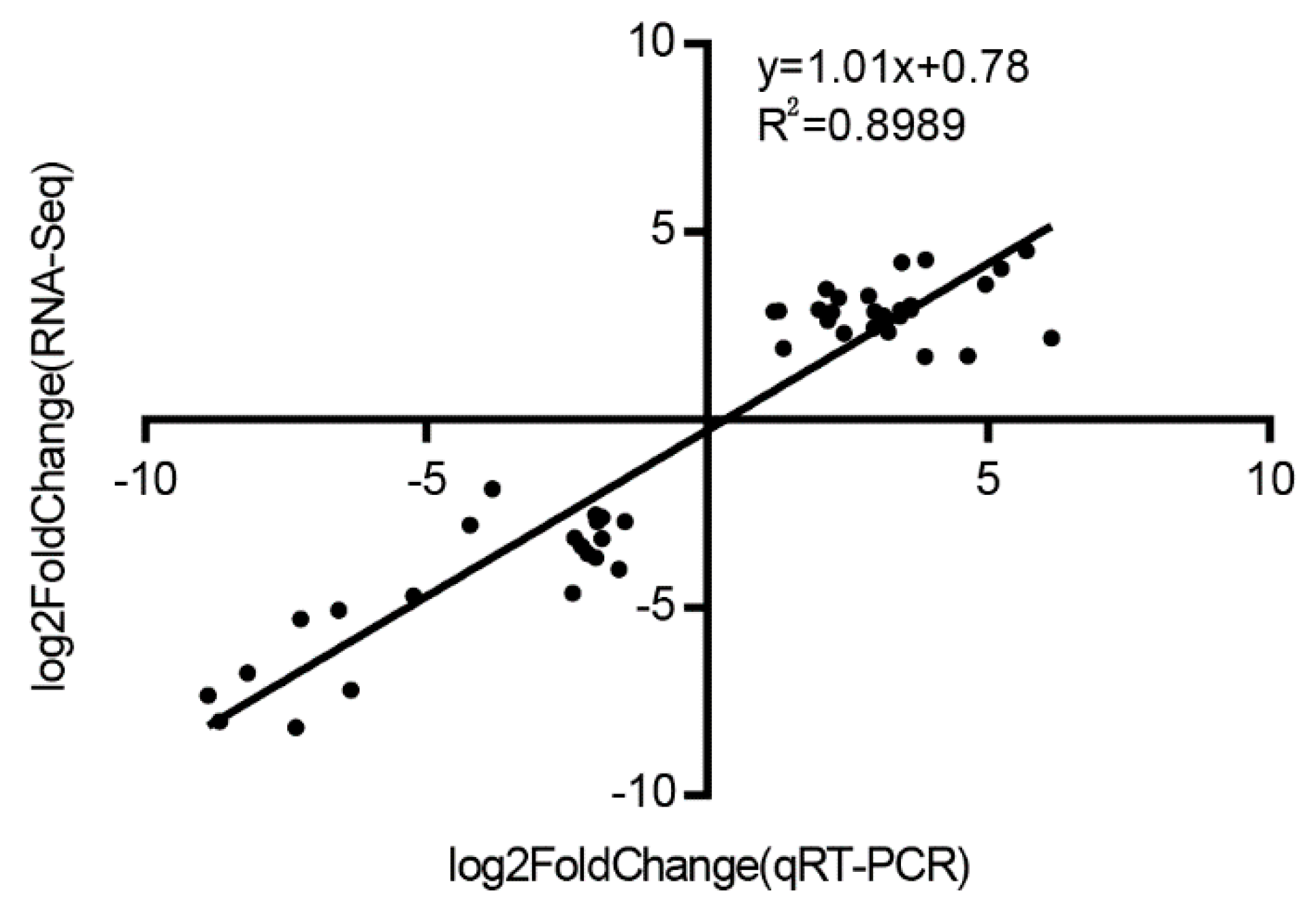
3.8. Validation of RNA-Seq Analysis by qRT-PCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tenaillon, M.I.; Charcosset, A. A European perspective on maize history. C. R. Biol. 2011, 334, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Karim, M.A.; Fracheboud, Y.; Stamp, P. Effect of high temperature on seedling growth and photosynthesis of tropical maize genotypes. J. Agron. Crop Sci. 2000, 184, 217–223. [Google Scholar] [CrossRef]
- Gong, F.; Wu, X.; Zhang, H.; Chen, Y.; Wang, W. Making better maize plants for sustainable grain production in a changing climate. Front. Plant Sci. 2015, 6, 835. [Google Scholar] [CrossRef] [PubMed]
- Presterl, T.; Ouzunova, M.; Schmidt, W.; Möller, E.M.; Röber, F.K.; Knaak, C.; Ernst, K.; Westhoff, P.; Geiger, H.H. Quantitative trait loci for early plant vigour of maize grown in chilly environments. Theor. Appl. Genet. 2007, 114, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Vanacker, H.; Gomez, L.D.; Harbinson, J. Regulation of photosynthesis and antioxidant metabolism in maize leaves at optimal and chilling temperatures. Plant Physiol. Biochem. 2002, 40, 659–668. [Google Scholar] [CrossRef]
- Leipner, J.; Stamp, P. Chilling stress in maize seedlings. In Handbook of Maize: Its Biology; Bennetzen, J.L., Hake, S.C., Eds.; Springer: New York, NY, USA, 2009; pp. 291–310. [Google Scholar]
- Sowiński, P.; Dalbiak, A.; Tadeusiak, J.; Ochodzki, P. Relations between carbohydrate accumulation in leaves, sucrose phosphate synthase activity and photoassimilate transport in chilling treated maize seedlings. Acta Physiol. Plant. 1999, 21, 375–381. [Google Scholar] [CrossRef]
- Marocco, A.; Lorenzoni, C.; Francheboud, Y. Chilling stress in maize. Maydica 2005, 50, 571–580. [Google Scholar]
- Bilska-Kos, A.; Solecka, D.; Dziewulska, A.; Ochodzki, P.; Jończyk, M.; Bilski, H.; Sowiński, P. Low temperature caused modifications in the arrangement of cell wall pectins due to changes of osmotic potential of cells of maize leaves (Zea mays L.). Protoplasma 2017, 254, 713–724. [Google Scholar] [CrossRef]
- Janowiak, F.; Markowski, A. Changes in leaf water relations and injuries in maize seedlings induced by different chilling conditions. J. Agron. Crop Sci. 1994, 172, 19–28. [Google Scholar] [CrossRef]
- Guo, X.; Liu, D.; Chong, K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [CrossRef]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.K.; Sunkar, R. Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 2010, 639, 39–55. [Google Scholar] [PubMed]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef]
- Kasuga, M.; Liu, Q.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress—Inducible transcription factor. Nat. Biotechnol. 1999, 17, 287–291. [Google Scholar] [CrossRef]
- Gilmour, S.J.; Fowler, S.G.; Thomashow, M.F. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004, 54, 767–781. [Google Scholar] [CrossRef]
- Dong, C.H.; Agarwal, M.; Zhang, Y.Y.; Xie, Q.; Zhu, J.K. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 2006, 103, 8281–8286. [Google Scholar] [CrossRef]
- Park, H.J.; Park, H.C.; Lee, S.Y.; Bohnert, H.J.; Yun, D.J. Ubiquitin and ubiquitin-like modifiers in plants. J. Plant Biol. 2011, 54, 275–285. [Google Scholar] [CrossRef]
- Agarwal, M.; Hao, Y.J.; Kapoor, A.; Dong, C.H.; Fujii, H.; Zheng, X.W.; Zhu, J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef]
- Shi, Y.; Tian, S.; Hou, L.; Huang, X.; Zhang, X.; Guo, H.; Yang, S. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 2012, 24, 2578–2595. [Google Scholar] [CrossRef]
- Ji, H.; Wang, Y.; Cloix, C.; Li, K.; Jenkins, G.I.; Wang, S.; Shang, Z.; Shi, Y.; Yang, S.; Li, X. The Arabidopsis RCC1 family protein TCF1 regulates freezing tolerance and cold acclimation through modulating lignin biosynthesis. PLoS Genet. 2015, 11, e1005471. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sun, D.; Zhou, R. Ca2+ and AtCaM3 are involved in the expression of heat shock protein gene in Arabidopsis. Plant Cell Environ. 2005, 28, 1276–1284. [Google Scholar] [CrossRef]
- Sangwan, V.; Orvar, B.L.; Beyerly, J.; Hirt, H.; Dhindsa, R.S. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 2002, 31, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Suri, S.S.; Dhindsa, R.S. A heat-activated MAP kinase (HAMK) as a mediator of heat shock response in tobacco cells. Plant Cell Environ. 2008, 31, 218–226. [Google Scholar] [CrossRef]
- Saidi, Y.; Finka, A.; Muriset, M.; Bromberg, Z.; Weiss, Y.G.; Maathuis, F.J.M.; Goloubinoff, P. The heat shock response in moss plants is regulated by specific calcium-permeable channels in the plasma membrane. Plant Cell 2009, 21, 2829–2843. [Google Scholar] [CrossRef]
- Liu, H.C.; Liao, H.T.; Charng, Y.Y. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011, 34, 738–751. [Google Scholar] [CrossRef]
- Mishra, S.K.; Tripp, J.; Winkelhaus, S.; Tschiersch, B.; Theres, K.; Nover, L.; Scharf, K.D. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 2002, 16, 1555–1567. [Google Scholar] [CrossRef]
- Yoshida, T.; Ohama, N.; Nakajima, J.; Kidokoro, S.; Mizoi, J.; Nakashima, K.; Maruyama, K.; Kim, J.M.; Seki, M.; Todaka, D.; et al. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genom. 2011, 286, 321–332. [Google Scholar] [CrossRef]
- Schramm, F.; Larkindale, J.; Kiehlmann, E.; Ganguli, A.; Englich, G.; Vierling, E.; Koskull-Döring, P. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 2008, 53, 264–274. [Google Scholar] [CrossRef]
- Guan, Q.; Yue, X.; Zeng, H.; Zhu, J. The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 2014, 26, 438–453. [Google Scholar] [CrossRef]
- Srivastava, R.; Deng, Y.; Howell, S.H. Stress sensing in plants by an ER stress sensor/transducer, bZIP28. Front. Plant Sci. 2014, 5, 59. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yan, B.; Lou, X.; Ma, H.; Ruan, S. Comparative transcriptome analysis reveals the transcriptional alterations in heat-resistant and heat-sensitive sweet maize (Zea mays L.) varieties under heat stress. BMC Plant Biol. 2017, 17, 26. [Google Scholar] [CrossRef] [PubMed]
- Sobkowiak, A.; Jończyk, M.; Jarochowska, E.; Biecek, P.; Trzcinska-Danielewicz, J.; Leipner, J.; Fronk, J.; Sowiński, P. Genome-wide transcriptomic analysis of response to low temperature reveals candidate genes determining divergent cold-sensitivity of maize inbred lines. Plant Mol. Biol. 2014, 85, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hu, G.; Liu, X.; Zhou, Y.; Li, Y.; Zhang, X.; Yuan, X.; Zhang, Q.; Yang, D.; Wang, T.; et al. Transcriptome sequencing identified genes and gene ontologies associated with early Freezing tolerance in maize. Front. Plant Sci. 2016, 7, 1477. [Google Scholar] [CrossRef]
- Qian, Y.; Ren, Q.; Zhang, J.; Chen, L. Transcriptomic analysis of the maize (Zea mays L.) inbred line B73 response to heat stress at the seedling stage. Gene 2019, 692, 68–78. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Jiang, Z.; Gao, L.; You, C.; Ma, X.; Wang, X.; Xu, X.; Mo, B.; Chen, X.; Liu, L. Genome-Wide Transcript and Small RNA Profiling Reveals Transcriptomic Responses to Heat Stress. Plant Physiol. 2019, 181, 609–629. [Google Scholar] [CrossRef]
- McGettigan, P.A. Transcriptomics in the RNA-seq era. Curr. Opin. Chem. Biol. 2013, 17, 4–11. [Google Scholar] [CrossRef]
- Yu, F.; Liang, K.; Fang, T.; Zhao, H.; Han, X.; Cai, M.; Qiu, F. A group VII ethylene response factor gene, ZmEREB180, coordinates waterlogging tolerance in maize seedlings. Plant Biotechnol. J. 2019, 17, 2286–2298. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. BioRxiv 2018, 289660. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.; Han, Y.; He, Q. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package Version 1.0.12. 2019. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 1 November 2019).
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Yu, F.; Tan, Z.; Fang, T.; Tang, K.; Liang, K.; Qiu, F. A comprehensive transcriptomics analysis reveals long non-coding RNA to be involved in the key metabolic pathway in response to waterlogging stress in maize. Genes 2020, 11, 267. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yang, S.; Hoffman, N. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. Mol. Biol. 1984, 35, 155–189. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Cutler, A.J.; Krochko, J.E. Formation and breakdown of ABA. Trends Plant Sci. 1999, 4, 472–478. [Google Scholar] [CrossRef]
- Grigorova, B.; Vaseva, I.I.; Demirevska, K.; Feller, U. Expression of selected heat shock proteins after individually applied and combined drought and heat stress. Acta Physiol. Plant. 2011, 33, 2041–2049. [Google Scholar] [CrossRef]
- Hu, X.; Li, Y.; Li, C.; Yang, H.; Wang, W.; Lu, M. Characterization of small heat shock proteins associated with maize tolerance to combined drought and heat stress. J. Plant Growth Regul. 2010, 29, 455–464. [Google Scholar] [CrossRef]
- Jaya, N.; Garcia, V.; Vierling, E. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc. Natl. Acad. Sci. USA 2009, 106, 15604–15609. [Google Scholar] [CrossRef]
- Timperio, A.M.; Egidi, M.G.; Zolla, L. Proteomics applied on plant abiotic stresses: Role of heat shock protein (HSP). J. Proteome Res. 2008, 71, 391–411. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.; Li, J.; Wang, P.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Ruelland, E.; Vaultier, M.N.; Zachowski, A.; Hurry, V. Cold signalling and cold acclimation in plants. Adv. Bot. Res. 2009, 49, 35–150. [Google Scholar]
- Siddiqui, K.S.; Cavicchioli, R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006, 75, 403–433. [Google Scholar] [CrossRef]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Kudla, J.; Becker, D.; Grill, E.; Hedrich, R.; Hippler, M.; Kummer, U.; Parniske, M.; Romeis, T.; Schumacher, K. Advances and current challenges in calcium signaling. New Phytol. 2018, 218, 414–431. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ding, Y.; Yang, S. Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol. 2015, 56, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Gong, F.; Cheminant, S.; Alioua, M.; Hedden, P.; Genschik, P. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 2008, 20, 2117–2129. [Google Scholar] [CrossRef] [PubMed]
- Eremina, M.; Unterholzner, S.J.; Rathnayake, A.I.; Castellanos, M.; Khan, M.; Kugler, K.G.; May, S.T.; Mayer, K.F.X.; Rozhon, W.; Poppenberger, B. Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc. Natl. Acad. Sci. USA 2016, 113, E5982–E5991. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.H.; Savina, M.; Du, J.; Devendran, A.; Kannivadi Ramakanth, K.; Tian, X.; Sim, W.S.; Mironova, V.V.; Xu, J. A sacrifice—For survival mechanism protects root stem cell niche from chilling stress. Cell 2017, 170, 102–113. [Google Scholar] [CrossRef]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Döring, P.; Vierling, E.; Scharf, K.D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K.D. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef]
- Yamada, K.; Fukao, Y.; Hayashi, M.; Fukazawa, M.; Suzuki, I.; Nishimura, M. Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J. Biol. Chem. 2007, 282, 37794–37804. [Google Scholar] [CrossRef]
- Li, P.; Cao, W.; Fang, H.; Xu, S.; Yin, S.; Zhang, Y.; Lin, D.; Wang, J.; Chen, Y.; Xu, C.; et al. Transcriptomic profiling of the maize (Zea mays L.) leaf response to abiotic stresses at the seedling stage. Front. Plant Sci. 2017, 8, 290. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Wang, X.; Li, Y.; Zhang, Y.; Gou, Z.; Qi, X.; Zhang, J. Transcriptomic Analysis Revealed the Common and Divergent Responses of Maize Seedling Leaves to Cold and Heat Stresses. Genes 2020, 11, 881. https://doi.org/10.3390/genes11080881
Li Y, Wang X, Li Y, Zhang Y, Gou Z, Qi X, Zhang J. Transcriptomic Analysis Revealed the Common and Divergent Responses of Maize Seedling Leaves to Cold and Heat Stresses. Genes. 2020; 11(8):881. https://doi.org/10.3390/genes11080881
Chicago/Turabian StyleLi, Yongsheng, Xingrong Wang, Yue Li, Yanjun Zhang, Zuowang Gou, Xusheng Qi, and Jinlin Zhang. 2020. "Transcriptomic Analysis Revealed the Common and Divergent Responses of Maize Seedling Leaves to Cold and Heat Stresses" Genes 11, no. 8: 881. https://doi.org/10.3390/genes11080881
APA StyleLi, Y., Wang, X., Li, Y., Zhang, Y., Gou, Z., Qi, X., & Zhang, J. (2020). Transcriptomic Analysis Revealed the Common and Divergent Responses of Maize Seedling Leaves to Cold and Heat Stresses. Genes, 11(8), 881. https://doi.org/10.3390/genes11080881




