TCGA Pan-Cancer Genomic Analysis of Alternative Lengthening of Telomeres (ALT) Related Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene Set
2.2. Genomic Alterations
2.3. Prevalence of the ALT Mechanism in Different Cancer Types
2.4. Protein-Protein Interaction Network
2.5. Gene Set Enrichment Analysis
3. Results
3.1. Role of the TM Gene Set in the ALT Mechanism
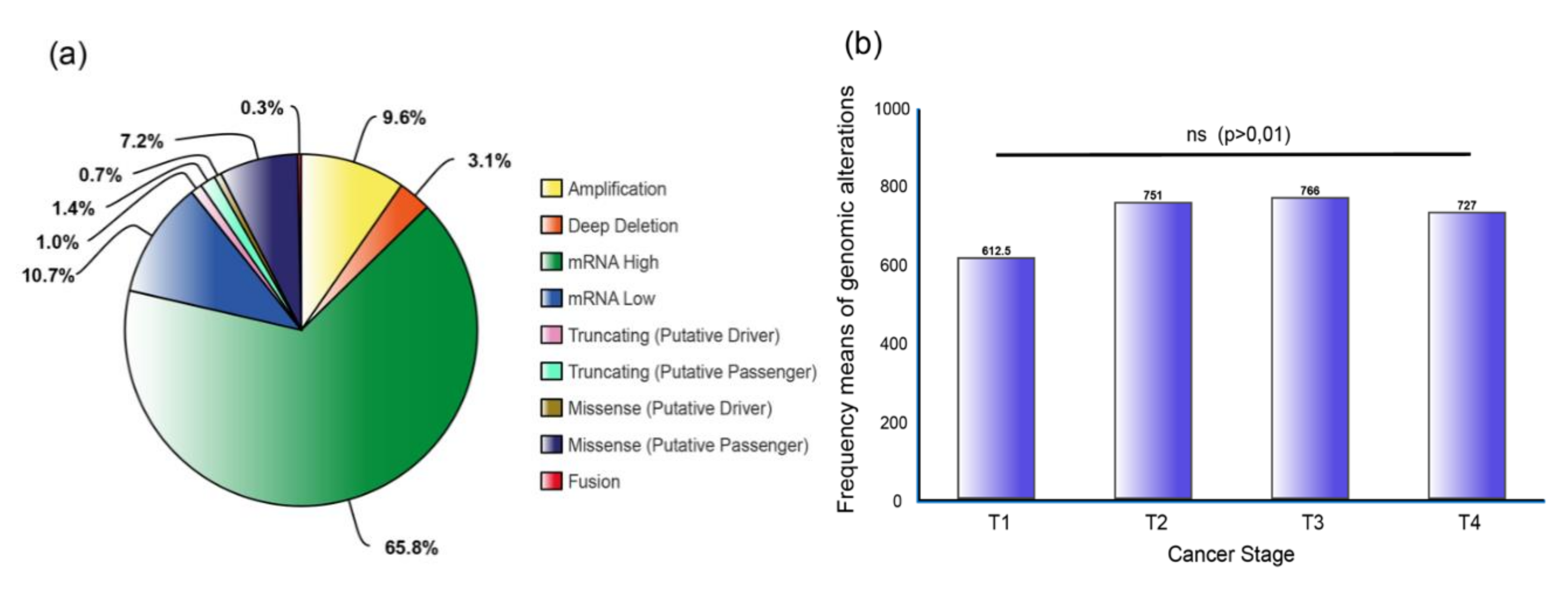
3.2. Genomic Alterations
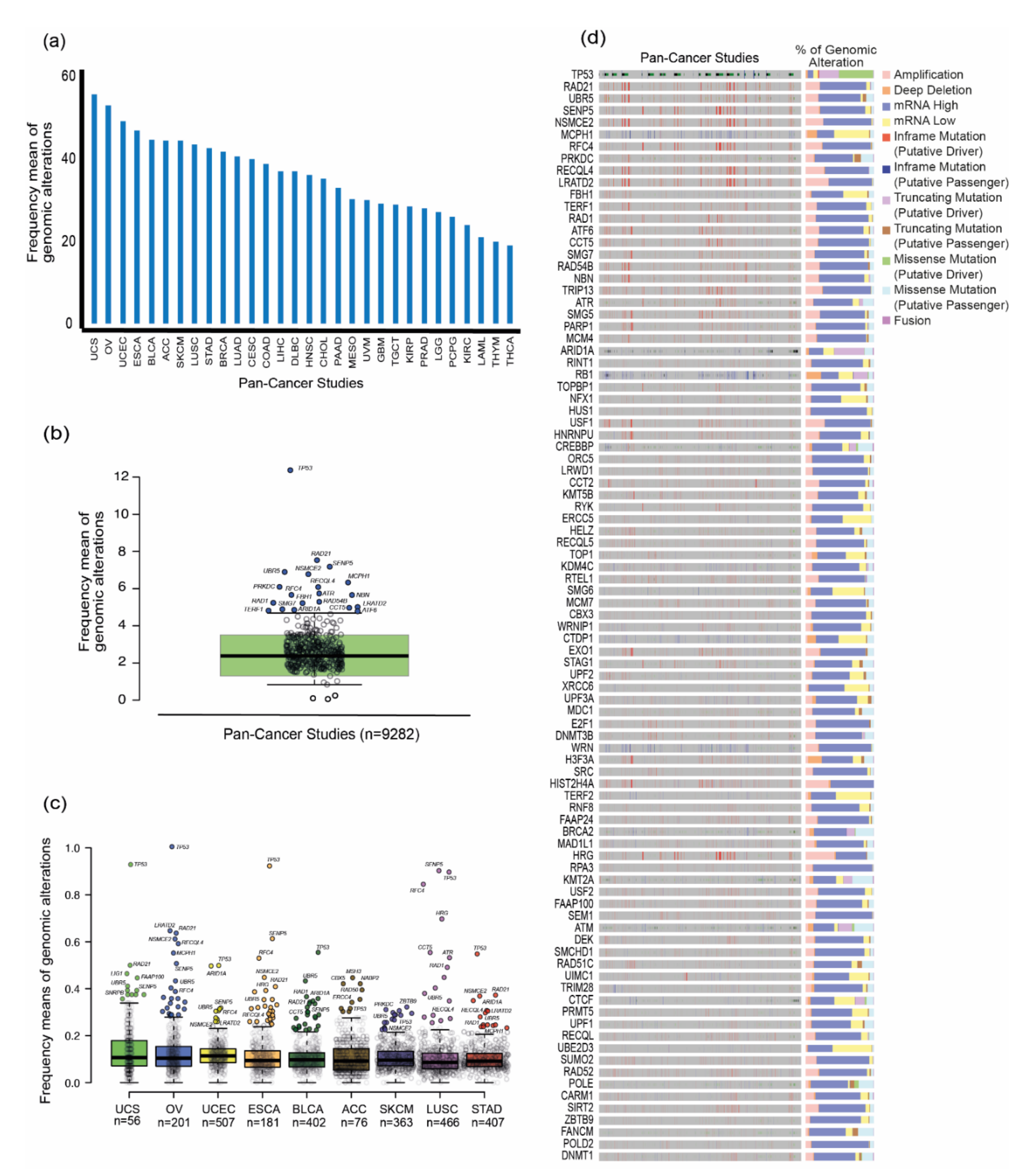
3.3. TM Genes Validation and TCGA Pan-Cancer Studies Frequencies
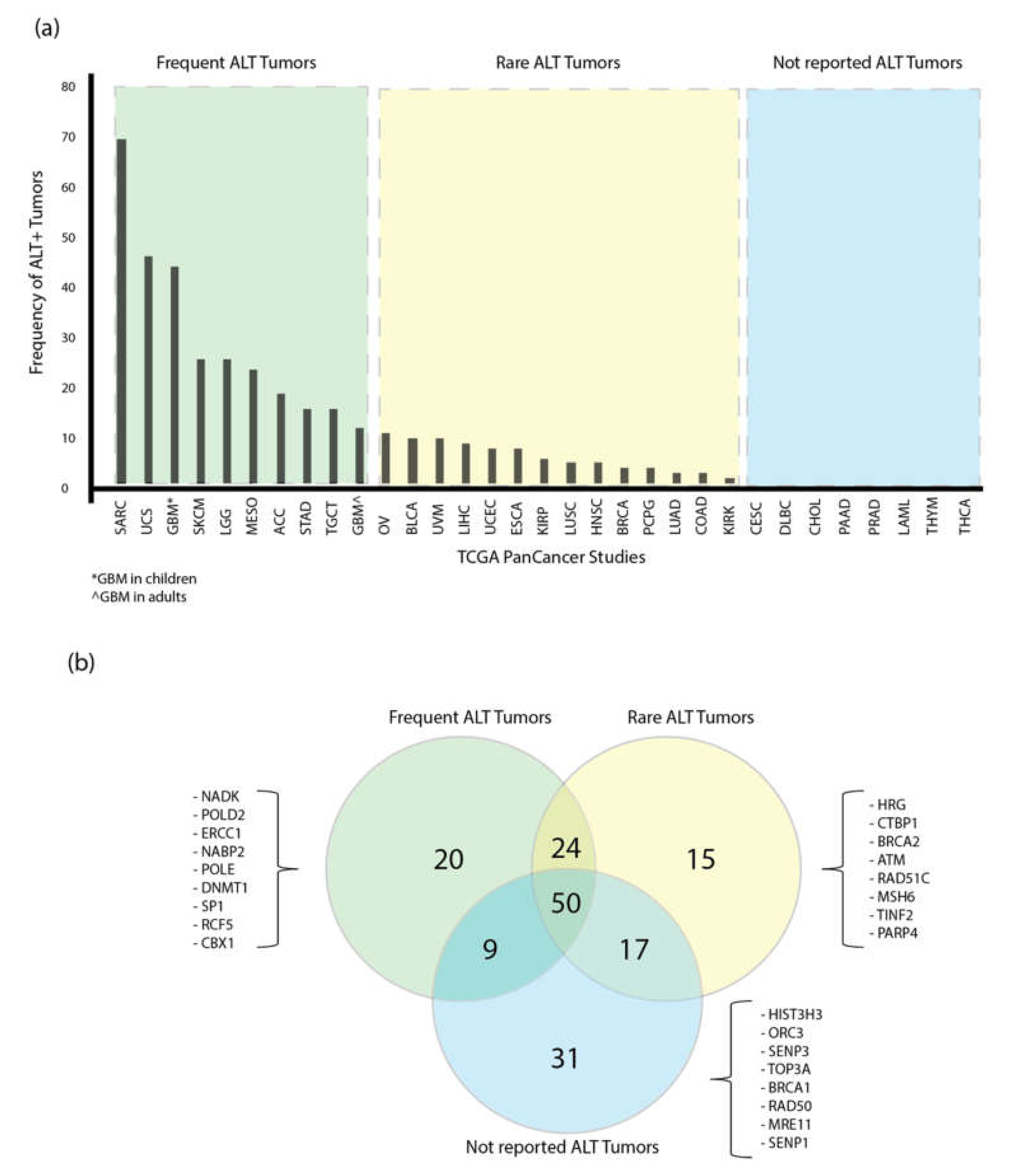
3.4. Prevalence of the ALT Mechanism in the TCGA PanCancer tumors
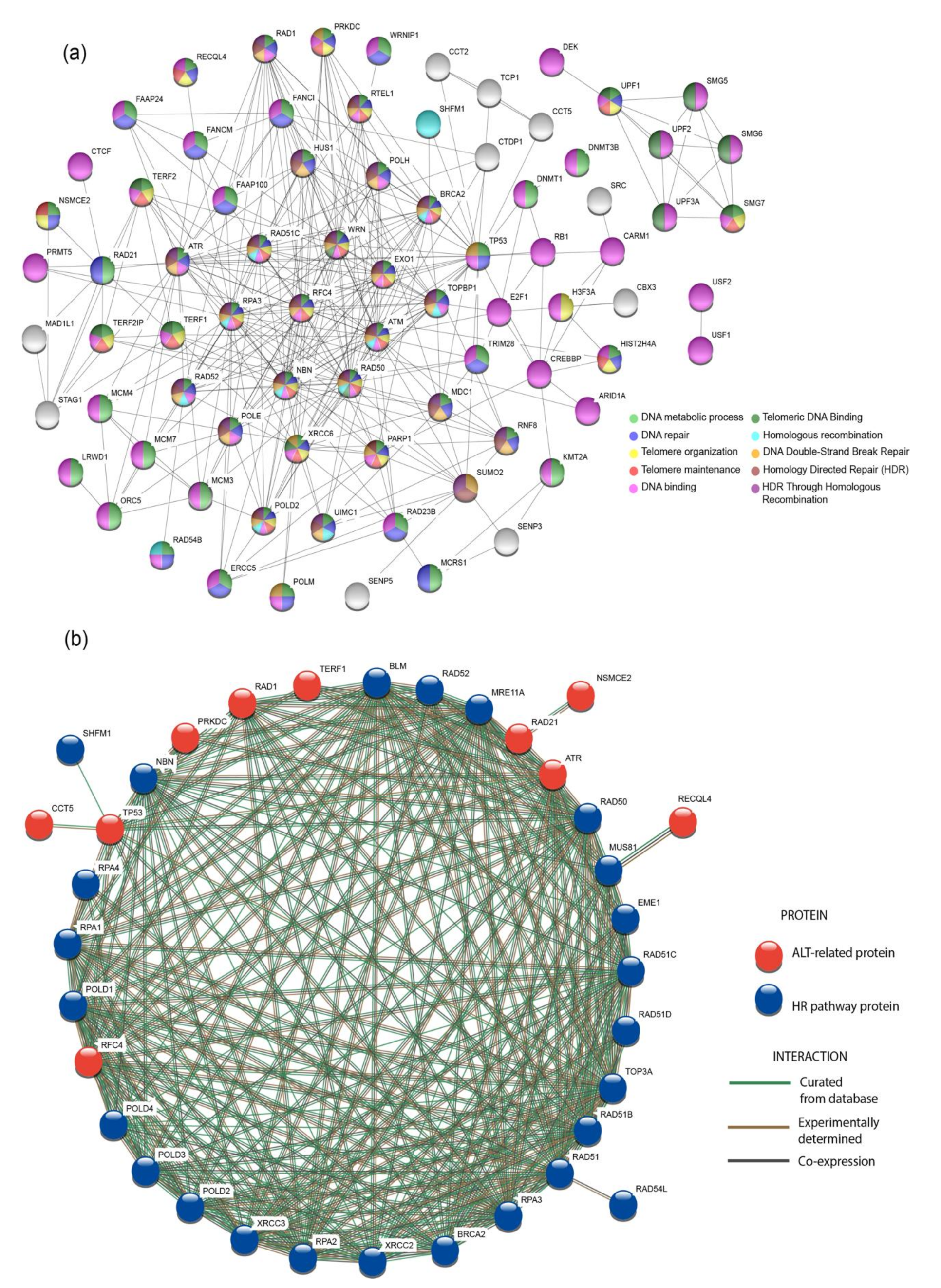
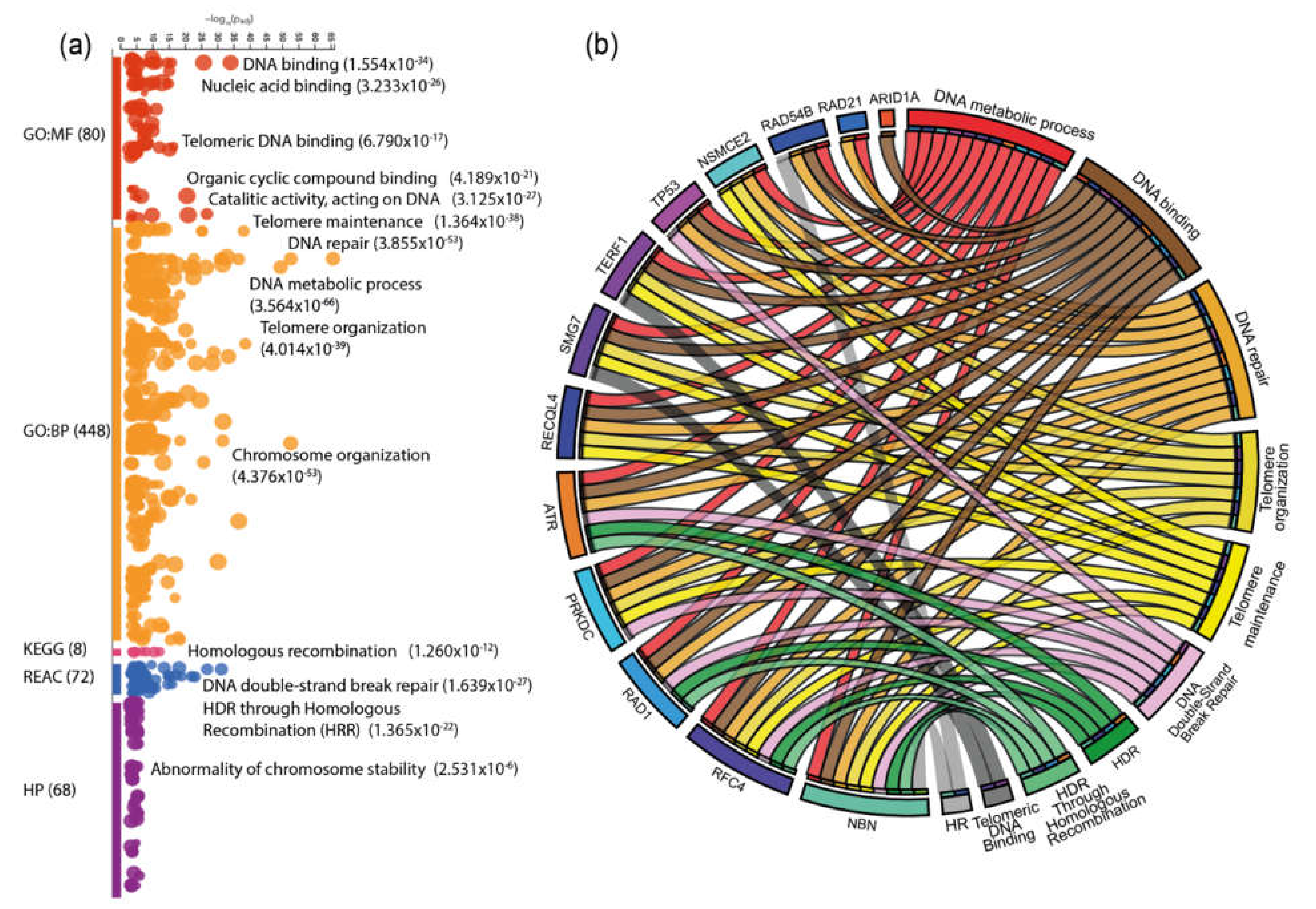
3.5. Protein-protein interaction (PPi) Network and Enrichment Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sommer, A.; Royle, N.J. ALT: A Multi-Faceted Phenomenon. Genes 2020, 11, 133. [Google Scholar] [CrossRef] [PubMed]
- Hoang, S.M.; O’Sullivan, R.J. Alternative Lengthening of Telomeres: Building Bridges to Connect Chromosome Ends. Trends Cancer 2020, 6, 247–260. [Google Scholar] [CrossRef]
- Recagni, M.; Bidzinska, J.; Zaffaroni, N.; Folini, M. The Role of Alternative Lengthening of Telomeres Mechanism in Cancer: Translational and Therapeutic Implications. Cancers 2020, 12, 949. [Google Scholar] [CrossRef]
- de Lange, T. How telomeres solve the end-protection problem. Science 2009, 326, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Turner, K.J.; Vasu, V.; Griffin, D.K. Telomere Biology and Human Phenotype. Cells 2019, 8, 73. [Google Scholar] [CrossRef]
- De Vitis, M.; Berardinelli, F.; Sgura, A. Telomere length maintenance in cancer: At the crossroad between telomerase and alternative lengthening of telomeres (ALT). Int. J. Mol. Sci. 2018, 19, 606. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.M.; Yadav, T.; Ouyang, J.; Lan, L.; Zou, L. Alternative Lengthening of Telomeres through Two Distinct Break-Induced Replication Pathways. Cell Rep. 2019, 26, 955–968.e3. [Google Scholar] [CrossRef] [PubMed]
- Heaphy, C.M.; Subhawong, A.P.; Hong, S.M.; Goggins, M.G.; Montgomery, E.A.; Gabrielson, E.; Netto, G.J.; Epstein, J.I.; Lotan, T.L.; Westra, W.H.; et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 2011, 179, 1608–1615. [Google Scholar] [CrossRef]
- Lee, M.; Teber, E.T.; Holmes, O.; Nones, K.; Patch, A.-M.; Dagg, R.A.; Lau, L.M.S.; Lee, J.H.; Napier, C.E.; Arthur, J.W.; et al. Telomere sequence content can be used to determine ALT activity in tumours. Nucleic Acids Res. 2018, 46, 4903–4918. [Google Scholar] [CrossRef]
- Lovejoy, C.A.; Takai, K.; Huh, M.S.; Picketts, D.J.; de Lange, T. ATRX affects the repair of telomeric DSBs by promoting cohesion and a DAXX-dependent activity. PLoS Biol. 2020, 18, e3000594. [Google Scholar] [CrossRef]
- Cox, K.E.; Maréchal, A.; Flynn, R.L. SMARCAL1 Resolves Replication Stress at ALT Telomeres. Cell Rep. 2016, 14, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-M.; Zou, L. Alternative lengthening of telomeres: From molecular mechanisms to therapeutic outlooks. Cell Biosci. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cesare, A.J.; Reddel, R.R. Alternative lengthening of telomeres: Models, mechanisms and implications. Nat. Rev. Genet. 2010, 11, 319–330. [Google Scholar] [CrossRef]
- Lawlor, R.T.; Veronese, N.; Pea, A.; Nottegar, A.; Smith, L.; Pilati, C.; Demurtas, J.; Fassan, M.; Cheng, L.; Luchini, C.; et al. Alternative lengthening of telomeres (ALT) influences survival in soft tissue sarcomas: A systematic review with meta-analysis. BMC Cancer 2019, 19, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Shi, G.; Zhang, L.; Li, F.; Jiang, Y.; Jiang, S.; Ma, W.; Zhao, Y.; Songyang, Z.; Huang, J.; et al. Switch telomerase to ALT mechanism by inducing telomeric DNA damages and dysfunction of ATRX and DAXX. Sci. Rep. 2016, 6, 32280. [Google Scholar] [CrossRef]
- Kumar, M.; Lechel, A.; Gunes, C. Telomerase: The Devil Inside. Genes 2016, 7, 43. [Google Scholar] [CrossRef]
- Brosnan-Cashman, J.A.; Yuan, M.; Graham, M.K.; Rizzo, A.J.; Myers, K.M.; Davis, C.; Zhang, R.; Esopi, D.M.; Raabe, E.H.; Eberhart, C.G.; et al. ATRX loss induces multiple hallmarks of the alternative lengthening of telomeres (ALT) phenotype in human glioma cell lines in a cell line-specific manner. PLoS ONE 2018, 13, e0204159. [Google Scholar] [CrossRef]
- Singhi, A.D.; Liu, T.-C.; Roncaioli, J.L.; Cao, D.; Zeh, H.J.; Zureikat, A.H.; Tsung, A.; Marsh, J.W.; Lee, K.K.; Hogg, M.E.; et al. Alternative Lengthening of Telomeres and Loss of DAXX/ATRX Expression Predicts Metastatic Disease and Poor Survival in Patients with Pancreatic Neuroendocrine Tumors. Clin. Cancer Res. 2017, 23, 600–609. [Google Scholar] [CrossRef]
- Braun, D.M.; Chung, I.; Kepper, N.; Deeg, K.I.; Rippe, K. TelNet—A database for human and yeast genes involved in telomere maintenance. BMC Genet. 2018, 19, 32. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304.e6. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.C.; Korkut, A.; Kanchi, R.S.; Hegde, A.M.; Lenoir, W.; Liu, W.; Liu, Y.; Fan, H.; Shen, H.; Ravikumar, V.; et al. A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell 2018, 33, 690–705.e9. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sethi, N.S.; Hinoue, T.; Schneider, B.G.; Cherniack, A.D.; Sanchez-Vega, F.; Seoane, J.A.; Farshidfar, F.; Bowlby, R.; Islam, M.; et al. Comparative Molecular Analysis of Gastrointestinal Adenocarcinomas. Cancer Cell 2018, 33, 721–735.e8. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.D.; Yau, C.; Bowlby, R.; Liu, Y.; Brennan, K.; Fan, H.; Taylor, A.M.; Wang, C.; Walter, V.; Akbani, R.; et al. Genomic, Pathway Network, and Immunologic Features Distinguishing Squamous Carcinomas. Cell Rep. 2018, 23, 194–212.e6. [Google Scholar] [CrossRef]
- Ricketts, C.J.; De Cubas, A.A.; Fan, H.; Smith, C.C.; Lang, M.; Reznik, E.; Bowlby, R.; Gibb, E.A.; Akbani, R.; Beroukhim, R.; et al. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep. 2018, 23, 313–326.e5. [Google Scholar] [CrossRef]
- Huang, K.L.; Mashl, R.J.; Wu, Y.; Ritter, D.I.; Wang, J.; Oh, C.; Paczkowska, M.; Reynolds, S.; Wyczalkowski, M.A.; Oak, N.; et al. Pathogenic Germline Variants in 10,389 Adult Cancers. Cell 2018, 173, 355–370.e14. [Google Scholar] [PubMed]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018, 173, 371–385.e18. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Liang, W.-W.; Foltz, S.M.; Mutharasu, G.; Jayasinghe, R.G.; Cao, S.; Liao, W.-W.; Reynolds, S.M.; Wyczalkowski, M.A.; Yao, L.; et al. Driver Fusions and Their Implications in the Development and Treatment of Human Cancers. Cell Rep. 2018, 23, 227–238.e3. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e11. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e10. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, S.; López-Cortés, A.; García-Cárdenas, J.M.; Armendáriz-Castillo, I.; Zambrano, A.K.; Indacochea, A.; Pérez-Villa, A.; Yumiceba, V.; Guevara-Ramírez, P.; Jacome-Alvarado, A.; et al. In silico analyses reveal new putative Breast Cancer RNA-binding proteins. BioRxiv 2020. [Google Scholar] [CrossRef]
- Malkova, A.; Ira, G. Break-induced replication: Functions and molecular mechanism. Curr. Opin. Genet. Dev. 2013, 23, 271–279. [Google Scholar] [CrossRef] [PubMed]
- KEGG Gene Set: KEGG_HOMOLOGOUS_RECOMBINATION. Available online: http://www.genome.jp/kegg/pathway/hsa/hsa03440.html (accessed on 30 June 2020).
- López-Cortés, A.; Paz-y-Miño, C.; Guerrero, S.; Cabrera-Andrade, A.; Barigye, S.J.; Munteanu, C.R.; González-Díaz, H.; Pazos, A.; Pérez-Castillo, Y.; Tejera, E.; et al. OncoOmics approaches to reveal essential genes in breast cancer: A panoramic view from pathogenesis to precision medicine. Sci. Rep. 2020, 10, 1–21. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g: Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- López-Cortés, A.; Guevara-Ramírez, P.; Guerrero, S.; Ortiz-Prado, E.; García-Cárdenas, J.M.; Zambrano, A.K.; Armendáriz-Castillo, I.; Pérez-Villa, A.; Yumiceba, V.; Varela, N.; et al. Metastatic signaling of hypoxia-related genes across TCGA Pan-Cancer types. BioRxiv 2020. [Google Scholar] [CrossRef]
- Pompili, L.; Leonetti, C.; Biroccio, A.; Salvati, E. Diagnosis and treatment of ALT tumors: Is Trabectedin a new therapeutic option? J. Exp. Clin. Cancer Res. 2017, 36, 1–10. [Google Scholar] [CrossRef]
- Ramamoorthy, M.; Smith, S. Loss of ATRX Suppresses Resolution of Telomere Cohesion to Control Recombination in ALT Cancer Cells. Cancer Cell 2015, 28, 357–369. [Google Scholar] [CrossRef]
- Dilley, R.L.; Greenberg, R.A. ALTernative Telomere Maintenance and Cancer. Trends Cancer 2015, 1, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Lovejoy, C.A.; Li, W.; Reisenweber, S.; Thongthip, S.; Bruno, J.; de Lange, T.; De, S.; Petrini, J.H.J.; Sung, P.A.; Jasin, M.; et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of Telomeres pathway. PLoS Genet. 2012, 8, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Nersisyan, L.; Hopp, L.; Loeffler-Wirth, H.; Galle, J.; Loeffler, M.; Arakelyan, A.; Binder, H. Telomere Length Maintenance and Its Transcriptional Regulation in Lynch Syndrome and Sporadic Colorectal Carcinoma. Front. Oncol. 2019, 9, 1172. [Google Scholar] [CrossRef]
- Lafferty-Whyte, K.; Cairney, C.J.; Will, M.B.; Serakinci, N.; Daidone, M.-G.; Zaffaroni, N.; Bilsland, A.; Keith, W.N. A gene expression signature classifying telomerase and ALT immortalization reveals an hTERT regulatory network and suggests a mesenchymal stem cell origin for ALT. Oncogene 2009, 28, 3765–3774. [Google Scholar] [CrossRef] [PubMed]
- Sobinoff, A.P.; Pickett, H.A. Mechanisms that drive telomere maintenance and recombination in human cancers. Curr. Opin. Genet. Dev. 2020, 60, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Lee, O.-H.; Kim, H.; He, Q.; Baek, H.J.; Yang, D.; Chen, L.-Y.; Liang, J.; Chae, H.K.; Safari, A.; Liu, D.; et al. Genome-wide YFP fluorescence complementation screen identifies new regulators for telomere signaling in human cells. Mol. Cell. Proteom. 2011, 10. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Englezou, A.; Dalla-Pozza, L.; Dunham, M.A.; Reddel, R.R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997, 3, 1271–1274. [Google Scholar] [CrossRef]
- Omori, Y.; Nakayama, F.; Li, D.; Kanemitsu, K.; Semba, S.; Ito, A.; Yokozaki, H. Alternative lengthening of telomeres frequently occurs in mismatch repair system-deficient gastric carcinoma. Cancer Sci. 2009, 100, 413–418. [Google Scholar] [CrossRef]
- Henson, J.D.; Lau, L.M.; Koch, S.; Martin La Rotta, N.; Dagg, R.A.; Reddel, R.R. The C-Circle Assay for alternative-lengthening-of-telomeres activity. Methods 2017, 114, 74–84. [Google Scholar] [CrossRef]
- Fogli, A.; Demattei, M.-V.; Corset, L.; Vaurs-Barriere, C.; Chautard, E.; Biau, J.; Kemeny, J.-L.; Godfraind, C.; Pereira, B.; Khalil, T.; et al. Detection of the alternative lengthening of telomeres pathway in malignant gliomas for improved molecular diagnosis. J. Neuro-Oncol. 2017, 135, 381–390. [Google Scholar] [CrossRef]
- Schwartzentruber, J.; Korshunov, A.; Liu, X.-Y.; Jones, D.T.W.; Pfaff, E.; Jacob, K.; Sturm, D.; Fontebasso, A.M.; Quang, D.-A.K.; Tonjes, M.; et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012, 482, 226–231. [Google Scholar] [CrossRef]
- Guièze, R.; Pages, M.; Véronèse, L.; Combes, P.; Lemal, R.; Gay-Bellile, M.; Chauvet, M.; Callanan, M.; Kwiatkowski, F.; Pereira, B.; et al. Telomere status in chronic lymphocytic leukemia with TP53 disruption. Oncotarget 2016, 7, 56976–56985. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chung, I.; Leonhardt, H.; Rippe, K. De novo assembly of a PML nuclear subcompartment occurs through multiple pathways and induces telomere elongation. J. Cell Sci. 2011, 124, 3603–3618. [Google Scholar] [CrossRef] [PubMed]
- Osterwald, S.; Deeg, K.I.; Chung, I.; Parisotto, D.; Worz, S.; Rohr, K.; Erfle, H.; Rippe, K. PML induces compaction, TRF2 depletion and DNA damage signaling at telomeres and promotes their alternative lengthening. J. Cell Sci. 2015, 128, 1887–1900. [Google Scholar] [CrossRef]
- Gaspar, T.B.; Sá, A.; Lopes, J.M.; Sobrinho-Simões, M.; Soares, P.; Vinagre, J. Telomere maintenance mechanisms in cancer. Genes 2018, 9, 241. [Google Scholar] [CrossRef]
- Dejardin, J.; Kingston, R.E. Purification of proteins associated with specific genomic Loci. Cell 2009, 136, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Potts, P.R.; Yu, H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 2007, 14, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Gomez, D.E.; Armando, R.G.; Farina, H.G.; Menna, P.L.; Cerrudo, C.S.; Ghiringhelli, P.D.; Alonso, D.F. Telomere structure and telomerase in health and disease. Int. J. Oncol. 2012, 41, 1561–1569. [Google Scholar] [CrossRef]
- Jiang, W.-Q.; Zhong, Z.-H.; Henson, J.D.; Neumann, A.A.; Chang, A.C.-M.; Reddel, R.R. Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1 complex. Mol. Cell. Biol. 2005, 25, 2708–2721. [Google Scholar] [CrossRef]
- Ramlee, M.K.; Wang, J.; Toh, W.X.; Li, S. Transcription Regulation of the Human Telomerase Reverse Transcriptase (hTERT) Gene. Genes 2016, 7, 50. [Google Scholar] [CrossRef]
- Uziel, O.; Yosef, N.; Sharan, R.; Ruppin, E.; Kupiec, M.; Kushnir, M.; Beery, E.; Cohen-Diker, T.; Nordenberg, J.; Lahav, M.; et al. The effects of telomere shortening on cancer cells: A network model of proteomic and microRNA analysis. Genomics 2015, 105, 5–16. [Google Scholar] [CrossRef]
- Tong, A.S.; Stern, J.L.; Sfeir, A.; Kartawinata, M.; de Lange, T.; Zhu, X.-D.; Bryan, T.M. ATM and ATR Signaling Regulate the Recruitment of Human Telomerase to Telomeres. Cell Rep. 2015, 13, 1633–1646. [Google Scholar] [CrossRef] [PubMed]
- Grolimund, L.; Aeby, E.; Hamelin, R.; Armand, F.; Chiappe, D.; Moniatte, M.; Lingner, J. A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nat. Commun. 2013, 4, 1–12. [Google Scholar]
- Lee, Y.-K.; Park, N.-H.; Lee, H. Prognostic value of alternative lengthening of telomeres-associated biomarkers in uterine sarcoma and uterine carcinosarcoma. Int. J. Gynecol. Cancer 2012, 22, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Rossi, M.L.; Singh, D.K.; Dunn, C.; Ramamoorthy, M.; Croteau, D.L.; Liu, Y.; Bohr, V.A. RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J. Biol. Chem. 2012, 287, 196–209. [Google Scholar] [CrossRef]
- Cerone, M.A.; Burgess, D.J.; Naceur-Lombardelli, C.; Lord, C.J.; Ashworth, A. High-throughput RNAi screening reveals novel regulators of telomerase. Cancer Res. 2011, 71, 3328–3340. [Google Scholar] [CrossRef]
- Zaboikin, M.; Zaboikina, T.; Freter, C.; Srinivasakumar, N. Non-Homologous End Joining and Homology Directed DNA Repair Frequency of Double-Stranded Breaks Introduced by Genome Editing Reagents. PLoS ONE 2017, 12, e0169931. [Google Scholar] [CrossRef]
- Kent, T.; Gracias, D.; Shepherd, S.; Clynes, D. Alternative Lengthening of Telomeres in Pediatric Cancer: Mechanisms to Therapies. Front. Oncol. 2020, 9, 1518. [Google Scholar] [CrossRef]
- Hyman, E.; Kauraniemi, P.; Hautaniemi, S.; Wolf, M.; Mousses, S.; Rozenblum, E.; Ringnér, M.; Sauter, G.; Monni, O.; Elkahloun, A.; et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002, 62, 6240–6245. [Google Scholar]
- Albertson, D.G. Gene amplification in cancer. Trends Genet. 2006, 22, 447–455. [Google Scholar] [CrossRef]
- García-Cárdenas, J.M.; Guerrero, S.; López-Cortés, A.; Armendáriz-Castillo, I.; Guevara-Ramírez, P.; Pérez-Villa, A.; Yumiceba, V.; Zambrano, A.K.; Leone, P.E.; Paz-y-Miño, C.; et al. Post-transcriptional Regulation of Colorectal Cancer: A Focus on RNA-Binding Proteins. Front. Mol. Biosci. 2019, 6, 65. [Google Scholar] [CrossRef]

| TCGA Study | n | TCGA Study | n |
|---|---|---|---|
| Acute Myeloid Leukemia (LAML) | 165 | Lung Squamous Cell Carcinoma (LUSC) | 466 |
| Adrenocortical Carcinoma (ACC) | 76 | Mesothelioma (MESO) | 82 |
| Bladder Urothelial Carcinoma (BLCA) | 402 | Ovarian Serous Cystadenocarcinoma (OV) | 201 |
| Brain Lower Grade Glioma (LGG) | 507 | Pancreatic Adenocarcinoma (PAAD) | 168 |
| Breast Invasive Carcinoma (BRCA) | 994 | Pheocromocytoma and Paraganlioma (PCPG) | 161 |
| Cervical Squamous Cell Carcinoma (CESC) | 275 | Prostate Adenocarcinoma (PRAD) | 488 |
| Cholangiocarcinoma (CHOL) | 36 | Sarcoma (SARC) | 251 |
| Colorectal Adenocarcinoma (COAD) | 524 | Skin Cutaneous Melanoma (SKCM) | 363 |
| Diffuse Large B-cell Lymphoma (DLBC) | 39 | Stomach Adenocarcinoma (STAD) | 407 |
| Esophageal Adenocarcinoma (ESCA) | 181 | Testicular Germ Cell Tumors (TGCT) | 144 |
| Glioblastoma Multiforme (GBM) | 145 | Thymoma (THYM) | 119 |
| Head and Neck Squamous Cell Carcinoma (HNSC) | 488 | Thyroid Carcinoma (THCA) | 480 |
| Kidney Renal Clear Cell Carcinoma (KIRC) | 352 | Uterine Carcinosarcoma (UCS) | 56 |
| Kidney Renal Papillary Cell Carcinoma (KIRP) | 274 | Uterine Corpus Endometrial Carcinoma (UCEC) | 507 |
| Liver Hepatocellular Carcinoma (LIHC) | 348 | Uveal Melanoma (UVM) | 80 |
| Lung Adenocarcinoma (LUAD) | 503 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Armendáriz-Castillo, I.; López-Cortés, A.; García-Cárdenas, J.; Guevara-Ramírez, P.; Leone, P.E.; Pérez-Villa, A.; Yumiceba, V.; Zambrano, A.K.; Guerrero, S.; Paz-y-Miño, C. TCGA Pan-Cancer Genomic Analysis of Alternative Lengthening of Telomeres (ALT) Related Genes. Genes 2020, 11, 834. https://doi.org/10.3390/genes11070834
Armendáriz-Castillo I, López-Cortés A, García-Cárdenas J, Guevara-Ramírez P, Leone PE, Pérez-Villa A, Yumiceba V, Zambrano AK, Guerrero S, Paz-y-Miño C. TCGA Pan-Cancer Genomic Analysis of Alternative Lengthening of Telomeres (ALT) Related Genes. Genes. 2020; 11(7):834. https://doi.org/10.3390/genes11070834
Chicago/Turabian StyleArmendáriz-Castillo, Isaac, Andrés López-Cortés, Jennyfer García-Cárdenas, Patricia Guevara-Ramírez, Paola E. Leone, Andy Pérez-Villa, Verónica Yumiceba, Ana K. Zambrano, Santiago Guerrero, and César Paz-y-Miño. 2020. "TCGA Pan-Cancer Genomic Analysis of Alternative Lengthening of Telomeres (ALT) Related Genes" Genes 11, no. 7: 834. https://doi.org/10.3390/genes11070834
APA StyleArmendáriz-Castillo, I., López-Cortés, A., García-Cárdenas, J., Guevara-Ramírez, P., Leone, P. E., Pérez-Villa, A., Yumiceba, V., Zambrano, A. K., Guerrero, S., & Paz-y-Miño, C. (2020). TCGA Pan-Cancer Genomic Analysis of Alternative Lengthening of Telomeres (ALT) Related Genes. Genes, 11(7), 834. https://doi.org/10.3390/genes11070834







