Salmonella enterica’s “Choice”: Itaconic Acid Degradation or Bacteriocin Immunity Genes
Abstract
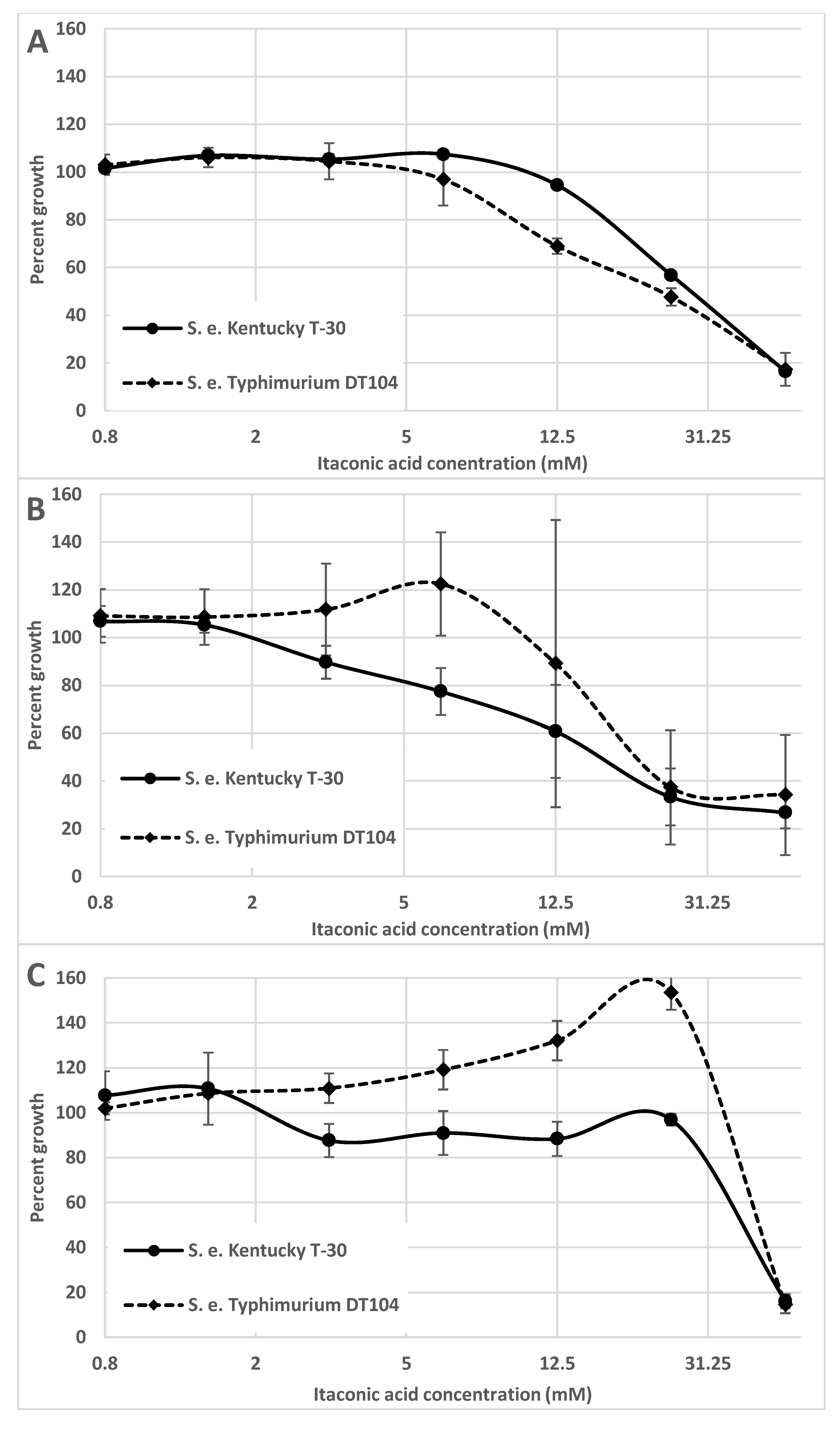
1. Salmonella and Itaconic Acid
2. Itaconic Acid Degradation Genes in Salmonella and Other Bacteria
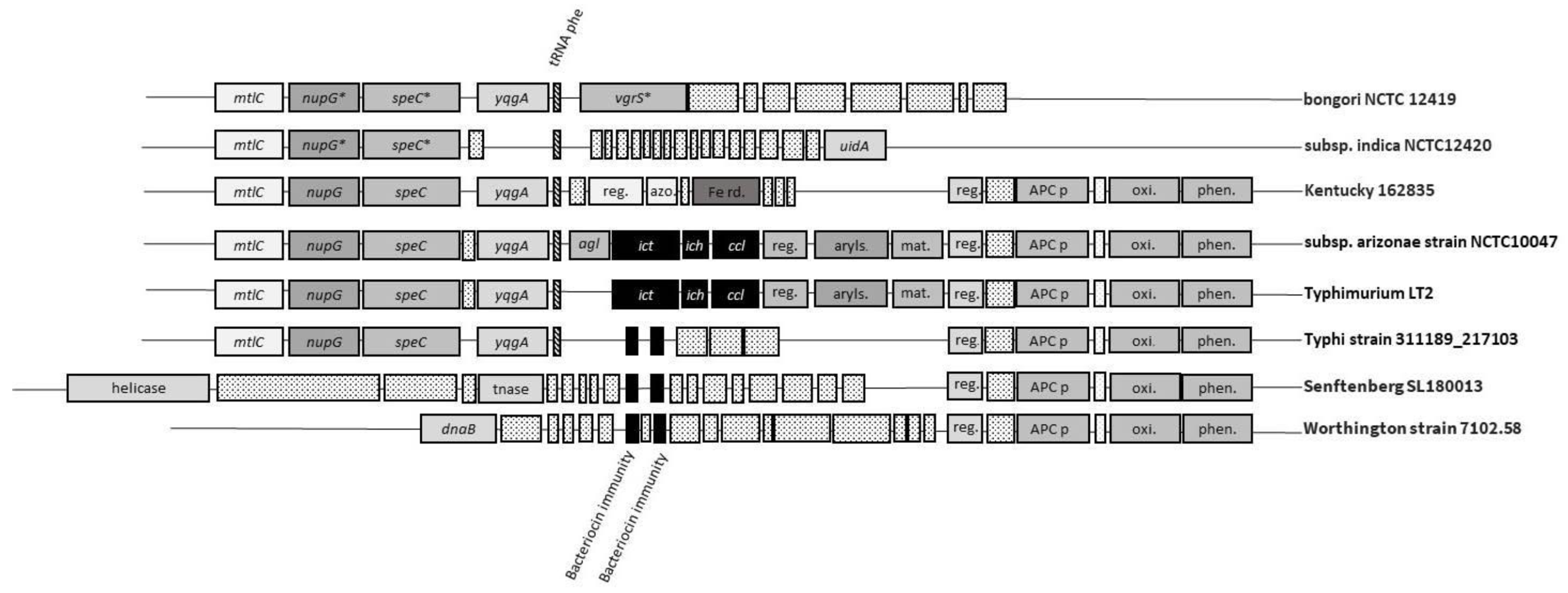
3. Distribution of Itaconic Acid Degradation Genes within the Genus Salmonella
4. Salmonella and Bacteriocins
5. Why Itaconic Acid Degradation or Bacteriocin Immunity Genes?
Funding
Acknowledgments
Conflicts of Interest
References
- Cordes, T.; Michelucci, A.; Hiller, K. Itaconic acid: The surprising role of an industrial compound as a mammalian antimicrobial metabolite. Annu. Rev. Nutr. 2015, 35, 451–473. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.V.; Alfaro, A.C.; Young, T.; Ravi, S.; Merien, F. Metabolomics of immune responses of New Zealand Greenshell™ mussels (Perna canaliculus) infected with pathogenic Vibrio sp. infection. Mar. Biotechnol. 2018, 20, 396–409. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.V.; Alfaro, A.C.; Merien, F.; Young, T.; Grandiosa, R. Metabolic and immunological response of male and female New Zealand Greenshell™ mussels (Perna canaliculus) infected with Vibrio sp. J. Invertebr. Pathol. 2018, 157, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Young, T.; Kesarcodi-Watson, A.; Alfaro, A.C.; Merien, F.; Nguyen, T.V.; Mae, H.; Le, D.V. Villas-Boas, S. Differential expression of novel metabolic and immunological biomarkers in oysters challenged with a virulent strain of OsHV-1. Dev. Comp. Immunol. 2017, 73, 229–245. [Google Scholar] [CrossRef] [PubMed]
- Strelko, C.L.; Lu, W.; Dufort, F.; Seyfried, T.N.; Chiles, T.C.; Rabinowitz, J.D.; Roberts, M.F. Itaconic acid is a mammalian metabolite induced during macrophage activation. J. Am. Chem. Soc. 2011, 133, 16386–16389. [Google Scholar] [CrossRef] [PubMed]
- Michelucci, A.; Cordes, T.; Ghelfi, J.; Pailot, A.; Reiling, N.; Goldmann, O.; Binz, T.; Wegner, A.; Tallam, A.; Rausell, A.; et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. USA 2013, 110, 7820–7825. [Google Scholar] [CrossRef]
- Rao, G.R.; McFadden, B.A. Isocitrate lyase from Pseudomonas indigofera IV. Specificity and inhibition. Arch. Biochem. Biophys. 1965, 112, 294–303. [Google Scholar] [CrossRef]
- Williams, J.O.; Roche, T.E.; McFadden, B.A. Mechanism of action of isocitrate lyase from Pseudomonas indigofera. Biochemistry 1971, 10, 1384–1390. [Google Scholar]
- Rittenhouse, J.W.; McFadden, B.A. Inhibition of isocitrate lyase from Pseudomonas indigofera by itaconate. Arch. Biochem. Biophys. 1974, 163, 79–86. [Google Scholar] [CrossRef]
- McFadden, B.A.; Purohit, S. Itaconate, an isocitrate lyase-directed inhibitor in Pseudomonas indigofera. J. Bacteriol. 1977, 131, 136–144. [Google Scholar] [CrossRef]
- Berg, I.A.; Filatova, L.V.; Ivanovsky, R.N. Inhibition of acetate and propionate assimilation by itaconate via propionyl-CoA carboxylase in isocitrate lyase-negative purple bacterium Rhodospirillum rubrum. FEMS Microbiol. Lett. 2002, 216, 49–54. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.J.; Artyomov, M.N. Itaconate: The poster child of metabolic reprogramming in macrophage function. Nat. Rev. Immunol. 2019, 19, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.B.; Maloy, S.R. Isolation and characterization of Salmonella typhimurium glyoxylate shunt mutants. J. Bacteriol. 1987, 169, 3029–3034. [Google Scholar] [CrossRef] [PubMed]
- Martin, W.R.; Frigan, F.; Bergman, E.H. Noninductive metabolism of itaconic acid by Pseudomonas and Salmonella species. J. Bacteriol. 1961, 82, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.C.; Libby, S.J.; Castor, M.E.; Fung, A.M. Isocitrate lyase (AceA) is required for Salmonella persistence but not acute lethal infection in mice. Infect. Immun. 2005, 73, 2547–2549. [Google Scholar] [CrossRef] [PubMed]
- Crousilles, A.; Dolan, S.K.; Brear, P.; Chirgadze, D.Y.; Welch, M. Gluconeogenic precursor availability regulates flux through the glyoxylate shunt in Pseudomonas aeruginosa. J. Biol. Chem. 2018, 293, 14260–14269. [Google Scholar] [CrossRef]
- Sasikaran, J.; Ziemski, M.; Zadora, P.K.; Fleig, A.; Berg, I.A. Bacterial itaconate degradation promotes pathogenicity. Nat. Chem. Biol. 2014, 10, 371–379. [Google Scholar] [CrossRef]
- Zhao, Y.; Jansen, R.; Gaastra, W.; Arkesteijn, G.; van der Zeijst, B.A.M.; van Putten, J.P.M. Identification of genes affecting Salmonella enterica serovar Enteritidis infection of chicken macrophages. Infect. Immun. 2002, 70, 5319–5321. [Google Scholar] [CrossRef]
- Shah, D.H.; Lee, M.J.; Park, J.H.; Lee, J.H.; Eo, S.K.; Kwon, J.T.; Chae, J.S. Identification of Salmonella gallinarum virulence genes in a chicken infection model using PCR-based signature-tagged mutagenesis. Microbiology 2005, 151, 3957–3968. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Adkins, J.N.; Coleman, J.R.; Schepmoes, A.A.; Dohnkova, A.; Mottaz, H.M.; Norbeck, A.D.; Purvine, S.O.; Manes, N.P.; Smallwood, H.S.; et al. Proteomic analysis of Salmonella enterica serovar Typhimurium isolates from RAW 264.7 macrophages. J. Biol. Chem. 2006, 281, 29131–29140. [Google Scholar] [CrossRef]
- Eriksson, S.; Lucchini, S.; Thompson, A.; Rhen, M.; Hinton, J.C.D. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 2003, 47, 103–118. [Google Scholar] [CrossRef] [PubMed]
- Hautefort, I.; Thompson, A.; Eriksson-Yberg, S.; Parker, M.L.; Lucchini, S.; Danino, V.; Bongaerts, R.J.M.; Ahmad, N.; Rhen, M.; Hinton, J.C.D. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell. Microbiol. 2008, 10, 958–984. [Google Scholar] [CrossRef] [PubMed]
- Santiviago, C.A.; Reynolds, M.M.; Porwollik, S.; Choi, S.-H.; Long, F.; Andrews-Polymenis, H.L.; McClelland, M. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 2009, 5, e100047. [Google Scholar] [CrossRef] [PubMed]
- Haneda, T.; Ishii, Y.; Danbara, H.; Okada, N. Genome-wide identification of novel genomic islands that contribute to Salmonella virulence in mouse systemic infection. FEMS Microbiol. Lett. 2009, 297, 241–249. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chaudhuri, R.R.; Morgan, E.; Peters, S.E.; Pleasance, S.J.; Hudson, D.L.; Davies, H.M.; Wang, J.; van Diemen, P.M.; Buckley, A.M.; Bowen, A.J.; et al. Comprehensive assignment of roles for Salmonella typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet. 2013, 9, e1003456. [Google Scholar] [CrossRef] [PubMed]
- Elder, J.R.; Chiok, K.L.; Paul, N.C.; Haldorson, G.; Guard, J.; Shah, D.H. The Salmonella pathogenicity island 13 contributes to pathogenesis in streptomycin pre-treated mice but not in day-old chickens. Gut Pathog. 2016, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Gogoi, M.; Chakravorty, D. Lactoylglutathione lyase, a critical enzyme in methylglyoxal detoxification, contributes to survival of Salmonella in the nutrient rich environment. Virulence 2014, 6, 50–65. [Google Scholar] [CrossRef]
- Baskaran, S.; Rajan, D.P.; Balasubraminian, K.A. Formation of methylglyoxal by bacteria isolated from human feces. J. Med. Microbiol. 1989, 28, 211–215. [Google Scholar] [CrossRef]
- Hammerer, F.; Chung, J.H.; Duncan, D.; Ruiz, A.C.; Auclair, K. Small molecule restores itaconate sensitivity in Salmonella enterica: A potential new approach to treating bacterial infections. ChemBioChem 2016, 17, 1513–1517. [Google Scholar] [CrossRef]
- Espinoza, R.A.; Silva-Valenzuela, C.A.; Amaya, F.A.; Urrutia, I.M.; Contreras, I.; Santiviago, C.A. Differential roles for pathogenicity islands SPI-13 and SPi-8 in the interaction of Salmonella Enteritidis and Salmonella Typhi with murine and human macrophages. Biol. Res. 2017, 50, 5. [Google Scholar] [CrossRef]
- Joerger, R.D.; Sartori, C.A.; Kniel, K.E. Comparison of genetic and physiological properties of Salmonella enterica isolates from chickens reveals one major difference between serovar Kentucky and other serovars: Response to acid. Foodborne Pathog. Dis. 2009, 6, 503–512. [Google Scholar] [CrossRef]
- CDC. Available online: https://www.cdc.gov/nationalsurveillance/pdfs/2016-Salmonella-report-508.pdf (accessed on 30 May 2020).
- CDC. Available online: https://www.cdc.gov/nationalsurveillance/pdfs/salmonella-serotypes-isolated-animals-and-related-sources-508.pdf (accessed on 30 May 2020).
- Parkhill, J.; Dougan, G.; James, K.D.; Thomson, N.R.; Pickard, D.; Wain, J.; Churcher, C.; Mungall, K.L.; Bentley, S.D.; Holden, M.T.G.; et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 2001, 431, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gutierrez, E.; Mayer, M.J.; Cotter, P.D.; Narbad, A. Gut microbiota as a source of novel antimicrobials. Gut Microbes 2019, 10, 1–21. [Google Scholar] [CrossRef]
- Kim, J.Y.; Young, J.A.; Gunther, N.W., IV; Lee, J.-L. Inhibition of Salmonella by bacteriocin-producing lactic acid bacteria derived from U.S. kimchi and broiler chickens. J. Food Saf. 2014, 35, 1–12. [Google Scholar] [CrossRef]
- Lei, S.; Zhao, R.; Sun, J.; Ran, J.; Ruan, X.; Zhu, Y. Partial purification and characterization of a broad-spectrun bacteriocin produced by a Lactobacillus plantarum zrx03 isolated from infant’s feces. Food Sci. Nutr. 2020, 8, 2214–2222. [Google Scholar] [CrossRef] [PubMed]
- Fredericq, P. Recherche des propriétés lysogène et antibiotiques chez les Salmonella. Comptes Rendus Séances Soc. Biol. 1952, 146, 298–300. [Google Scholar]
- Fredericq, P. Colicins. Annu. Rev. Microbiol. 1957, 11, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Hamon, Y.; Péron, Y. Contribution a l’étude de la propriété bacteriocinogène dans la tribu des Salmonelleae. 1. Les bacteroicines des Salmonella. Ann. Inst. Pasteur 1966, 110, 389–402. [Google Scholar]
- Atkinson, N. Colicin-like antibiotics and bacteriophages of Salmonellas. Aust. J. Exp. Biol. Med. Sci. 1970, 48, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Barker, R.M. Colicinogeny in Salmonella typhimurium. J. Gen. Microbiol. 1980, 120, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, N. Salmonellin—A new colicin-like antibiotic. Nature 1967, 213, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, N. Colicin-like antibiotics of 100 strains of Salmonella. Aust. J. Exp. Biol. Med. Sci. 1973, 51, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.; Hahn-Loebmann, S.; Stephan, A.; Schulz, S.; Giritch, A.; Naumann, M.; Kleinschmidt, M.; Tusé, D.; Gleba, Y. Plant-made Salmonella bacteriocins salmocins for control of Salmonella pathovars. Sci. Rep. 2018, 8, 4078. [Google Scholar] [CrossRef] [PubMed]
- Kaldhone, P.R.; Han, J.; Deck, J.; Khajanchi, B.; Nayak, R.; Foley, S.L.; Ricke, S.C. Evaluation of the genetics and functionality of plasmids in incompatibility group I1-positive Salmonella enterica. Foodborne Pathog. Dis. 2018, 15, 168–176. [Google Scholar] [CrossRef]
- Kalthone, P.R.; Carlton, A.; Aijahdali, N.; Khajanchi, B.K.; Sanad, Y.M.; Han, J.; Deck, J.; Ricke, S.C.; Foley, S.L. Evaluation of incompatibility group I1 (IncI1) plasmid-containing Salmonella enterica and assessment of the plasmids in bacteriocin production and biofilm development. Front. Vet. Sci. 2019, 6, 298. [Google Scholar] [CrossRef]
- Lehrbach, P.R.; Broda, P. Molecular comparisons of plasmids isolated from colicinogenic strains of Escherichia coli. J. Gen. Microbiol. 1984, 130, 401–410. [Google Scholar] [CrossRef]
- Fricke, W.F.; McDermott, P.F.; Mammel, M.K.; Zhao, S.H.; Johnson, T.J.; Rasko, D.A.; Fedorka-Cray, P.J.; Pedroso, A.; Whichard, J.M.; Eugene LeClerc, J.; et al. Antimicrobial resistance-conferring plasmids with similarity to virulence plasmids from avian pathogenic Escherichia coli Strains in Salmonella enterica serovar Kentucky isolates from poultry. Appl. Environ. Microbiol. 2009, 75, 5963–5971. [Google Scholar] [CrossRef]
- Johnson, T.J.; Thorsness, J.L.; Cole, P. Horizontal gene transfer of a ColV plasmid has resulted in a dominant avian clonal type of Salmonella enterica serovar Kentucky. PLoS ONE 2010, 5, e15524. [Google Scholar] [CrossRef]
- Cascales, E.; Buchanan, S.K.; Duché, D.; Kleanthous, C.; Lloubès, R.; Postle, K.; Riley, M.; Slatin, S.; Cavard, D. Colicin biology. Microbiol. Mol. Biol. Rev. 2007, 71, 158–229. [Google Scholar] [CrossRef]
- Azpiroz, M.F.; Bascuas, T.; Laviňa, M. Microcin H47 system: An Escherichia coli small genomic island with novel features. PLoS ONE 2011, 6, e26179. [Google Scholar] [CrossRef]
- Hacker, J.; Kaper, J.B. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 2000, 54, 641–679. [Google Scholar] [CrossRef] [PubMed]
- Wooley, R.E.; Gibbs, P.S.; Shotts, E.B. Inhibition of Salmonella typhimurium in the chicken intestinal tract by a transformed avirulent avian Escherichia coli. Avian Dis. 1999, 43, 245–250. [Google Scholar] [CrossRef]
- Wooley, R.E.; Ritchie, B.W.; Currin, F.P.; Chitwood, S.W.; Sanchez, S.; Crane, M.M.; Lamberski, N. In vitro inhibition of Salmonella organisms isolated from reptiles by an inactivated culture of microcin-producing Escherichia coli. Am. J. Vet. Res. 2001, 62, 1399–1401. [Google Scholar] [CrossRef] [PubMed]
- Zihler, A.; Le Blay, G.; de Wouten, T.; Lacroix, C.; Braegger, C.P.; Lehner, A.; Tischler, P.; Rattei, T.; Haechler, H.; Stephan, R. In vitro inhibition activity of different bacteriocin-producing Escherichia coli against Salmonella strains isolated from clinical cases. Lett. Appl. Microbiol. 2009, 49, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Quereda, J.J.; Dussurget, O.; Nahori, M.-A.; Ghozlane, A.; Volant, S.; Dillies, M.-A.; Regnault, B.; Kennedy, S.; Mondot, S.; Villoing, B.; et al. Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc. Natl. Acad. Sci. USA 2016, 113, 5706–5711. [Google Scholar] [CrossRef]
- Kommineni, S.; Bretl, D.J.; Lam, V.; Chakraborty, R.; Hayward, M.; Simpson, P.; Cao, Y.; Bousounis, P.; Kristich, C.J.; Salzman, N.H. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 2015, 526, 719–724. [Google Scholar] [CrossRef]
- Stecher, B.; Denzler, R.; Maier, L.; Bernet, F.; Sanders, M.J.; Pickard, D.J.; Barthel, M.; Westendorf, A.M.; Krogfelt, K.A.; Walker, A.W.; et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 2012, 109, 1269–1274. [Google Scholar] [CrossRef]
- Nedialkova, L.P.; Denzler, R.; Koeppel, M.B.; Diehl, M.; Ring, D.; Wille, T.; Gerlach, R.G.; Stecher, B. Inflammation fuels colicin Ib-dependent competition of Salmonella serovar Typhimurium and E. coli in enterobacterial Blooms. PLoS Pathog. 2014, 10, e100384. [Google Scholar] [CrossRef]
- Sassone-Corsi, M.; Nuccio, S.; Liu, H.; Hernandez, D.; Vu, C.T.; Takahashi, A.A.; Edwards, R.A.; Raffatellu, M. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 2016, 540, 280–283. [Google Scholar] [CrossRef]
- He, H.; Genovese, K.J.; Swaggerty, C.L.; Nisbet, D.J.; Kogut, M.H. A comparative study on invasion, modulation of oxidative burst, and nitric oxide responses of macrophages (HD11), and systemic infection in chickens by prevalent poultry Salmonella serovars. Foodborne Pathog. Dis. 2012, 9, 1104–1110. [Google Scholar] [CrossRef]
- Salehi, S.; Howe, K.; Lawrence, M.L.; Brooks, J.P.; Bailey, R.H.; Karsi, A. Salmonella enterica serovar Kentucky flagella are required for broiler skin adhesion and Caco-2 cell invasion. Appl. Environ. Microbiol. 2017, 83, e02115-16. [Google Scholar] [CrossRef] [PubMed]
- Parry, C.; Dougan, C. Typhoid fever. N. Engl. J. Med. 2002, 347, 1770–1782. [Google Scholar] [CrossRef] [PubMed]
- Gunn, J.S.; Marshall, J.M.; Baker, S.; Dongol, S.; Charles, R.C.; Ryan, E.T. Salmonella chronic carriage: Epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol. 2014, 22, 648–655. [Google Scholar] [CrossRef]
| Species | Subspecies | Serovar | A * | B * | C * | Species | Subspecies | Serovar | A * | B * | C * |
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. bongori | 0 | 0 | 12 | S. enterica | enterica | Hillingdon | 0 | 1 | 0 | ||
| S. enterica | houtenae | 4 | 0 | 0 | Hvittingfoss | 2 | 0 | 0 | |||
| diarizonae | 10 | 0 | 0 | Indiana | 0 | 0 | 6 | ||||
| arizonae | 8 | 0 | 0 | Infantis | 10 | 3 | 0 | ||||
| salamae | 0 | 0 | 11 | Iverness | 1 | 0 | 0 | ||||
| indica | 0 | 0 | 9 | Java | 1 | 0 | 0 | ||||
| enterica | unkown | 93 | 21 | 9 | Javiana | 2 | 0 | 0 | |||
| 1,4,[5],12:i:- | 29 | 0 | 0 | Johannesburg | 1 | 0 | 0 | ||||
| 4,[5],12:i:- | 8 | 0 | 0 | Kentucky | 0 | 4 | 1 | ||||
| Abaetetuba | 1 | 0 | 0 | Koessen | 1 | 0 | 0 | ||||
| Aberdeen | 1 | 0 | 0 | Krefeld | 0 | 1 | 0 | ||||
| Abony | 1 | 0 | 0 | Macclesfield | 1 | 0 | 1 | ||||
| Adjame | 0 | 0 | 11 | Manchester | 0 | 0 | 1 | ||||
| Agona | 0 | 14 | 0 | Manhattan | 1 | 0 | 0 | ||||
| Albany | 0 | 2 | 0 | Mbandaka | 1 | 0 | 2 | ||||
| Anatum | 29 | 0 | 0 | Mikawasima | 2 | 0 | 0 | ||||
| Antsalova | 1 | 0 | 0 | Milwaukee | 0 | 1 | 0 | ||||
| Apapa | 0 | 1 | 0 | Minnesota | 2 | 0 | 0 | ||||
| Bardo | 1 | 0 | 0 | Moscow | 1 | 0 | 0 | ||||
| Bareilly | 29 | 0 | 0 | Muenchen | 3 | 0 | 0 | ||||
| Bergen | 0 | 1 | 0 | Muenster | 3 | 0 | 0 | ||||
| Berta | 1 | 0 | 0 | Montevideo | 18 | 0 | 1 | ||||
| Birkenhead | 1 | 0 | 0 | Newport | 32 | 0 | 0 | ||||
| Blegdam | 1 | 0 | 0 | Ohio | 0 | 0 | 1 | ||||
| Blockly | 1 | 0 | 0 | Onderstepoort | 1 | 0 | 0 | ||||
| Borreze | 0 | 1 | 0 | Oranienburg | 2 | 0 | 0 | ||||
| Bovismorbificans | 1 | 0 | 0 | Ouakam | 0 | 0 | 1 | ||||
| Brancaster | 0 | 1 | 0 | Panama | 1 | 0 | 0 | ||||
| Brandenburg | 3 | 0 | 0 | Pomona | 2 | 0 | 0 | ||||
| Braenderup | 3 | 0 | 0 | Poona | 2 | 0 | 0 | ||||
| Bredeney | 3 | 0 | 0 | Paratyphi A | 0 | 6 | 0 | ||||
| California | 1 | 0 | 0 | Paratyphi B | 1 | 0 | 0 | ||||
| Carmel | 1 | 0 | 0 | Paratyphi C | 1 | 0 | 0 | ||||
| Cerro | 2 | 0 | 0 | Pullorum | 4 | 0 | 0 | ||||
| Chester | 1 | 0 | 0 | Quebec | 1 | 0 | 0 | ||||
| Cholerasuis | 4 | 0 | 0 | Rissen | 0 | 1 | 3 | ||||
| Concord | 1 | 0 | 0 | Rough C:-:- | 2 | 0 | 0 | ||||
| Corvallis | 0 | 2 | 0 | Saintpaul | 8 | 1 | 0 | ||||
| Crossness | 1 | 0 | 0 | Sanjuan | 0 | 0 | 1 | ||||
| Cubana | 0 | 1 | 0 | Senftenberg | 0 | 14 | 1 | ||||
| Daytona | 1 | 0 | 0 | Schwarzengrund | 3 | 0 | 0 | ||||
| Derby | 0 | 2 | 3 | Sloterdijk | 1 | 0 | 0 | ||||
| Djakarta | 1 | 0 | 0 | Stanley | 2 | 0 | 0 | ||||
| Dublin | 12 | 0 | 0 | Stanleyville | 4 | 1 | 0 | ||||
| Enteritidis | 241 | 0 | 0 | Sundsvall | 1 | 0 | 0 | ||||
| Florida | 1 | 0 | 0 | Tennessee | 0 | 7 | 0 | ||||
| Fresno | 0 | 1 | 0 | Thompson | 9 | 0 | 0 | ||||
| Gallinarum | 5 | 0 | 0 | Typhi | 0 | 122 | 0 | ||||
| Gaminara | 2 | 0 | 0 | Typhimurium | 96 | 1 | 1 | ||||
| Give | 2 | 0 | 0 | Virchow | 2 | 0 | 0 | ||||
| Goaldcoast | 3 | 0 | 0 | Wandsworth | 1 | 0 | 0 | ||||
| Hadar | 1 | 0 | 0 | Weltevreden | 5 | 0 | 0 | ||||
| Havana | 0 | 1 | 0 | Worthington | 0 | 2 | 0 | ||||
| Hayindogo | 1 | 0 | 0 | Yovokome | 1 | 0 | 0 | ||||
| Heidelberg | 33 | 0 | 0 |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joerger, R.D. Salmonella enterica’s “Choice”: Itaconic Acid Degradation or Bacteriocin Immunity Genes. Genes 2020, 11, 797. https://doi.org/10.3390/genes11070797
Joerger RD. Salmonella enterica’s “Choice”: Itaconic Acid Degradation or Bacteriocin Immunity Genes. Genes. 2020; 11(7):797. https://doi.org/10.3390/genes11070797
Chicago/Turabian StyleJoerger, Rolf D. 2020. "Salmonella enterica’s “Choice”: Itaconic Acid Degradation or Bacteriocin Immunity Genes" Genes 11, no. 7: 797. https://doi.org/10.3390/genes11070797
APA StyleJoerger, R. D. (2020). Salmonella enterica’s “Choice”: Itaconic Acid Degradation or Bacteriocin Immunity Genes. Genes, 11(7), 797. https://doi.org/10.3390/genes11070797




