Quantitative Proteomics of Urinary Bladder Cancer Cell Lines Identify UAP1 as a Potential Therapeutic Target
Abstract
1. Introduction
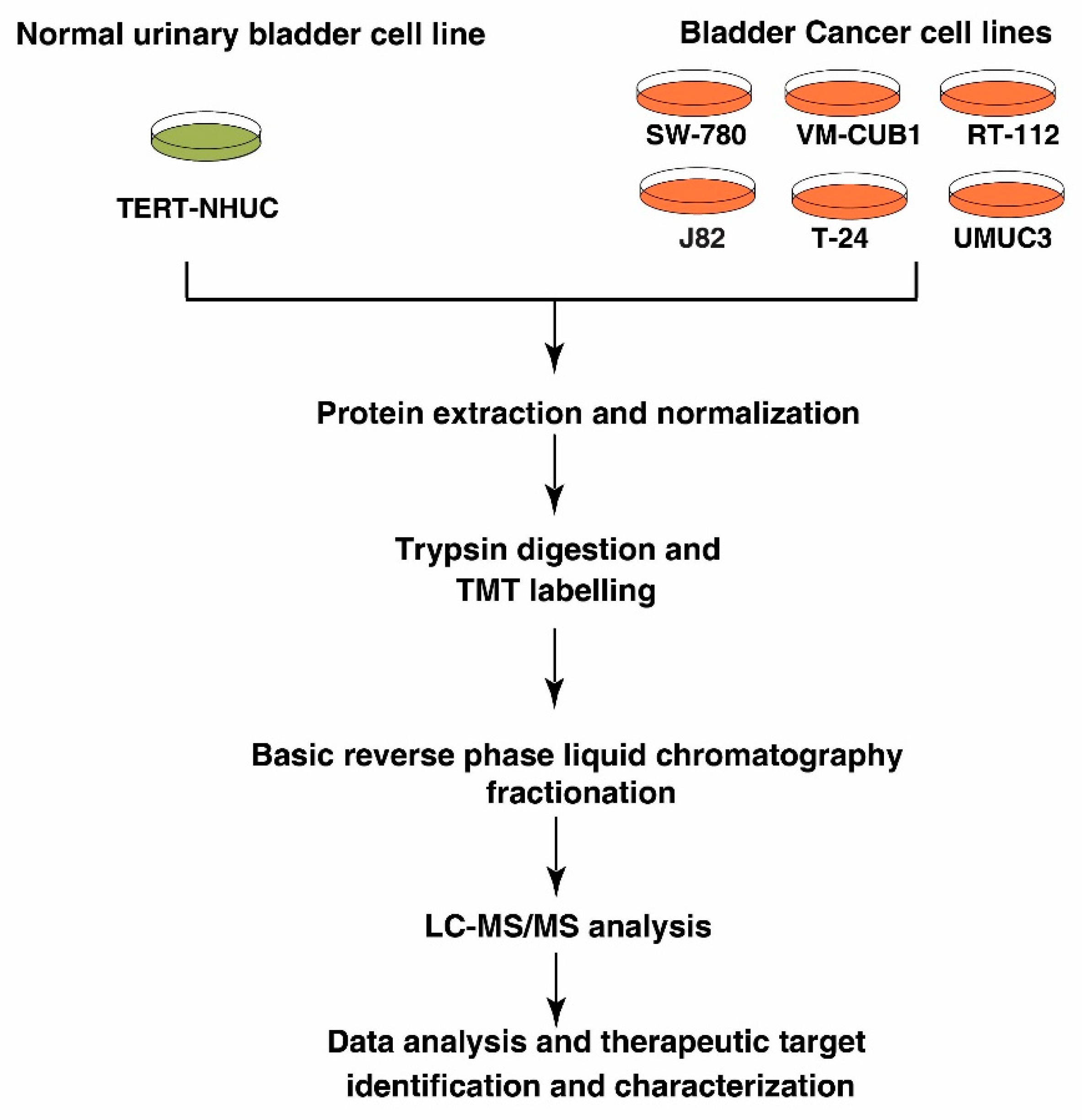
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
2.2. Cell lysis and Protein Digestion
2.3. TMT Labeling and Basic pH Reverse Phase Liquid Chromatography (bRPLC)
2.4. LC-MS/MS Analysis
2.5. Data Analysis
2.6. Protein-Protein Interaction Networks
2.7. Western Blotting
2.8. siRNA Transfection
2.9. Cell Proliferation Assay
2.10. Colony Formation Assay
2.11. Wound Healing Assay
2.12. Invasion Assay
2.13. Statistical Analysis
3. Results
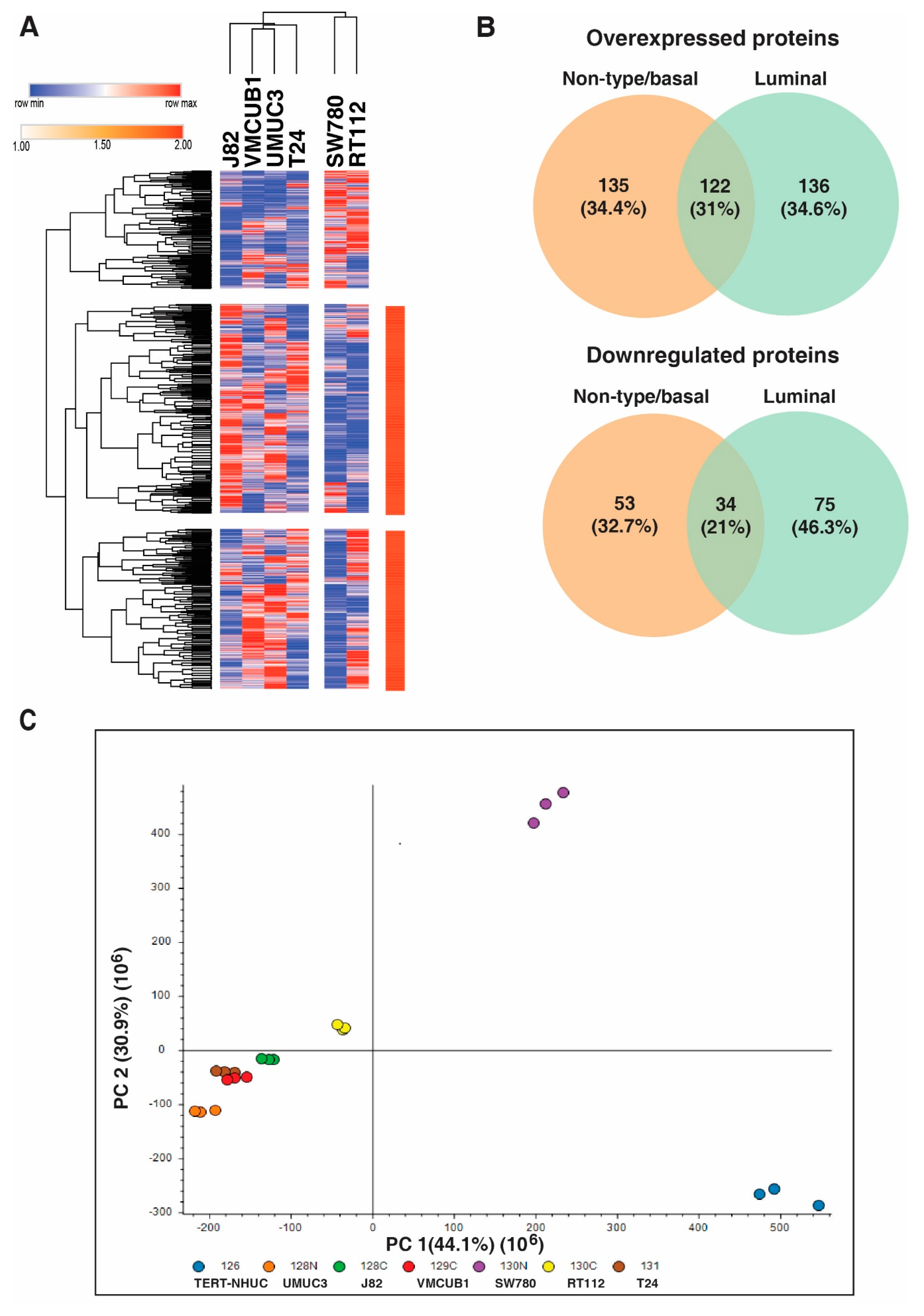
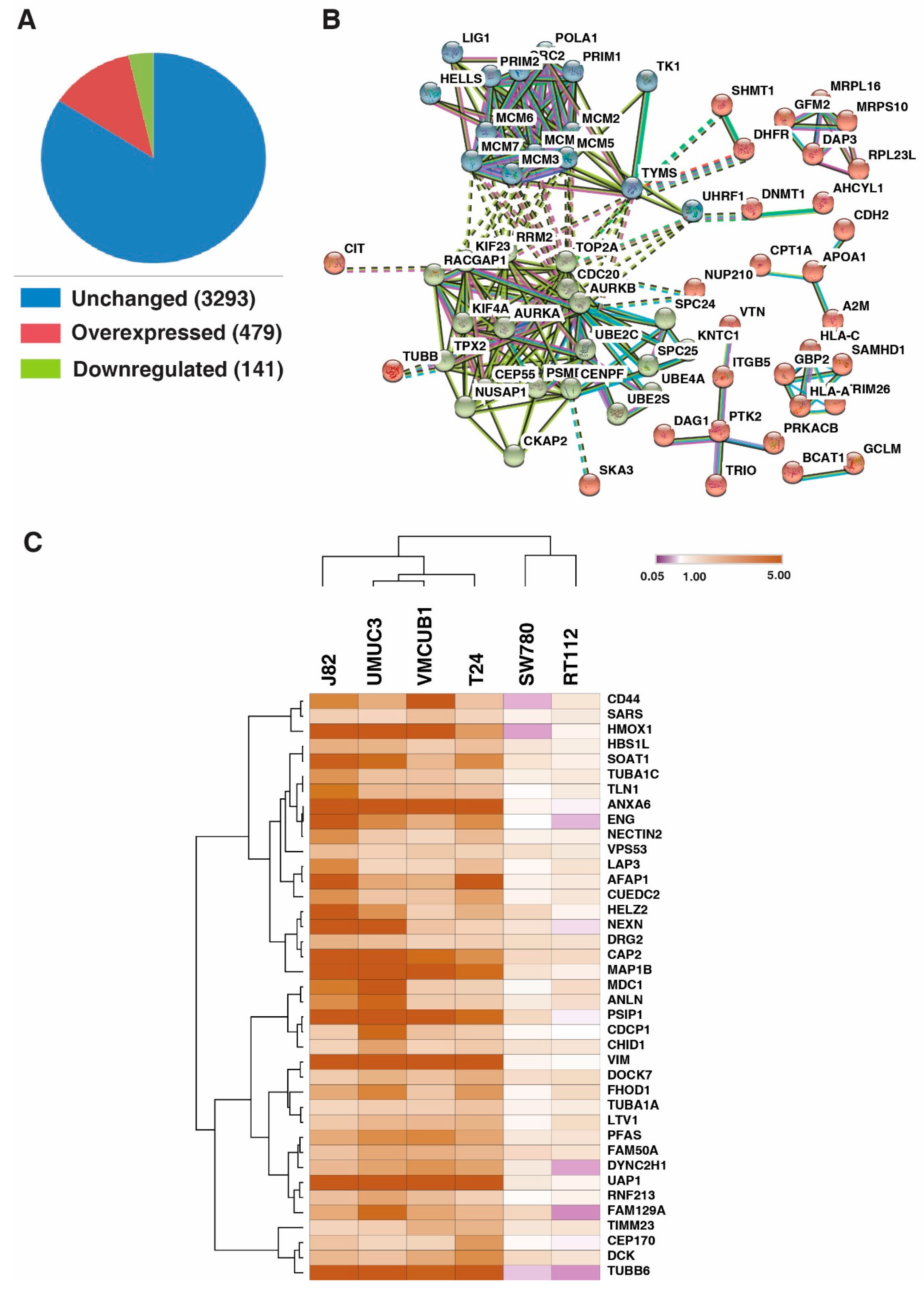
3.1. Identification of Proteins with Altered Expression in Urinary Bladder Cancer Cell Lines through Quantitative Proteomics
3.2. Distinct Protein Expression Pattern of Non-Type/Basal as Compared to Luminal Bladder Carcinoma Cell Lines
3.3. DNA Replication Process and Cell Cycle Regulation and Progression Were Dysregulated across All Bladder Carcinoma Cell Lines
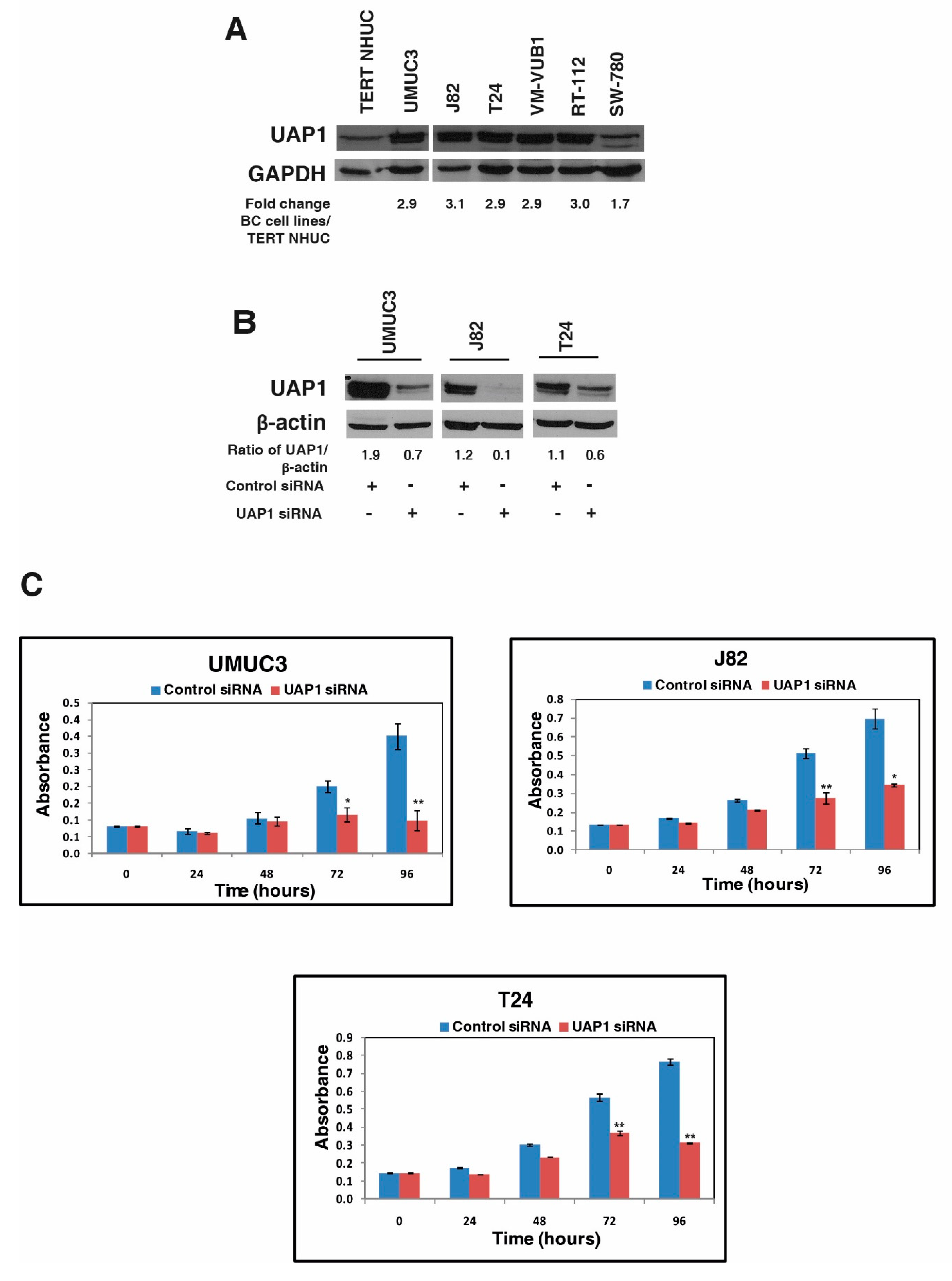
3.4. UAP1 Was Overexpressed in Bladder Carcinoma
3.5. Silencing of UAP1 Decreases Cellular Proliferation in Urinary Bladder Cancer Cells
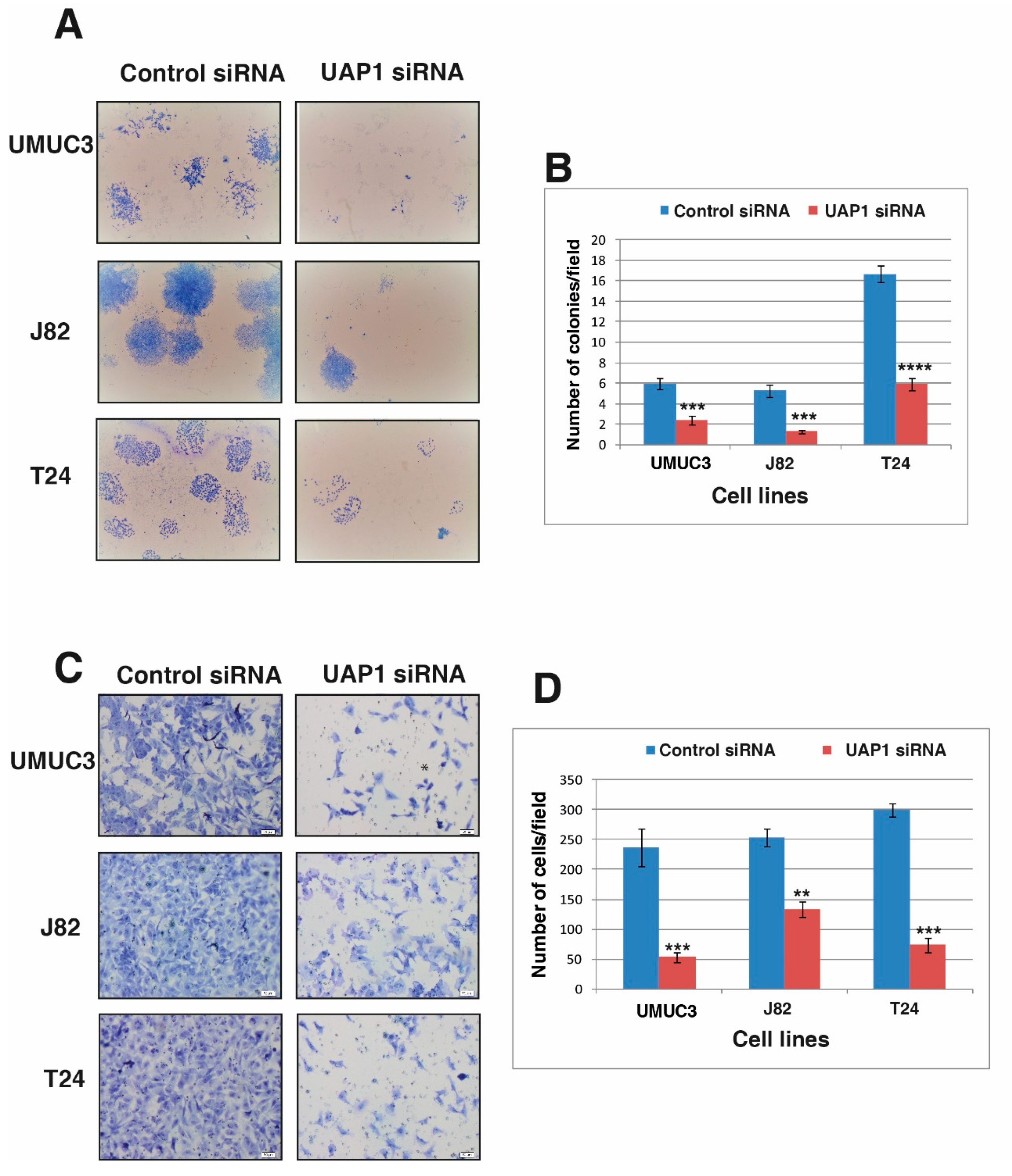
3.6. Silencing of UAP1 Decreases Colony Forming Ability in Urinary Bladder Cancer Cells
3.7. Silencing of UAP1 Decreases Invasive Property in Urinary Bladder Cancer Cells
3.8. Silencing of UAP1 Decreases Cell Motility in Urinary Bladder Cancer Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Knowles, M.A.; Hurst, C.D. Molecular biology of bladder cancer: New insights into pathogenesis and clinical diversity. Nat. Rev. Cancer 2014, 15, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Sylvester, R.J.; Van Der Meijden, A.P.; Oosterlinck, W.; Witjes, J.A.; Bouffioux, C.; Denis, L.; Newling, D.W.; Kurth, K. Predicting Recurrence and Progression in Individual Patients with Stage Ta T1 Bladder Cancer Using EORTC Risk Tables: A Combined Analysis of 2596 Patients from Seven EORTC Trials. Eur. Urol. 2006, 49, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Mandhani, A.; Mitash, N.; Agnihotri, S.; Mittal, B.; Tiwari, S. Molecular cystoscopy: Micro-RNAs could be a marker for identifying genotypic changes for transitional cell carcinoma of the urinary bladder. Indian J. Urol. 2016, 32, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Lobo, N.; Mount, C.; Omar, K.; Nair, R.; Thurairaja, R.; Khan, M.S. Landmarks in the treatment of muscle-invasive bladder cancer. Nat. Rev. Urol. 2017, 14, 565–574. [Google Scholar] [CrossRef]
- Zehnder, P.; Studer, U.E.; Skinner, E.C.; Thalmann, G.; Miranda, G.; Roth, B.; Cai, J.; Birkhäuser, F.D.; Mitra, A.P.; Burkhard, F.C.; et al. Unaltered oncological outcomes of radical cystectomy with extended lymphadenectomy over three decades. BJU Int. 2013, 112, E51–E58. [Google Scholar] [CrossRef]
- Gakis, G. Management of Muscle-invasive Bladder Cancer in the 2020s: Challenges and Perspectives. Eur. Urol. Focus 2020. [Google Scholar] [CrossRef]
- Bellmunt, J.; Powles, T.; Vogelzang, N.J. A review on the evolution of PD-1/PD-L1 immunotherapy for bladder cancer: The future is now. Cancer Treat. Rev. 2017, 54, 58–67. [Google Scholar] [CrossRef]
- Gopalakrishnan, D.; Koshkin, V.S.; Ornstein, M.C.; Papatsoris, A.; Grivas, P. Immune checkpoint inhibitors in urothelial cancer: Recent updates and future outlook. Ther. Clin. Risk Manag. 2018, 14, 1019–1040. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, X.; Yang, C.; Li, P.; Yuan, W.; Deng, X.; Cheng, Y.; Li, P.; Yang, H.; Tao, J.; et al. Targeting protein kinase CK2 suppresses bladder cancer cell survival via the glucose metabolic pathway. Oncotarget 2016, 7, 87361–87372. [Google Scholar] [CrossRef]
- Kawanishi, H.; Matsui, Y.; Ito, M.; Watanabe, J.; Takahashi, T.; Nishizawa, K.; Nishiyama, H.; Kamoto, T.; Mikami, Y.; Tanaka, Y.; et al. Secreted CXCL1 Is a Potential Mediator and Marker of the Tumor Invasion of Bladder Cancer. Clin. Cancer Res. 2008, 14, 2579–2587. [Google Scholar] [CrossRef]
- Latosinska, A.; Mokou, M.; Makridakis, M.; Mullen, W.; Zoidakis, J.; Lygirou, V.; Frantzi, M.; Katafigiotis, I.; Stravodimos, K.; Hupe, M.C.; et al. Proteomics analysis of bladder cancer invasion: Targeting EIF3D for therapeutic intervention. Oncotarget 2017, 8, 69435–69455. [Google Scholar] [CrossRef] [PubMed]
- Gershoni-Baruch, R.; Nachlieli, T.; Guilburd, J.N. Apert’s syndrome with occipital encephalocele and absence of corpus callosum. Child’s nervous system: ChNS: Official journal of the International Society for Pediatric. Neurosurgery 1991, 7, 231–232. [Google Scholar]
- Guo, M.; Lu, S.; Huang, H.; Wang, Y.; Yang, M.Q.; Yang, Y.; Fan, Z.; Jiang, B.; Deng, Y. Increased AURKA promotes cell proliferation and predicts poor prognosis in bladder cancer. BMC Syst. Boil. 2018, 12, 118. [Google Scholar] [CrossRef] [PubMed]
- Rebouissou, S.; Bernard-Pierrot, I.; De Reyniès, A.; Lepage, M.-L.; Krucker, C.; Chapeaublanc, E.; Herault, A.; Kamoun, A.; Caillault, A.; Letouzé, E.; et al. EGFR as a potential therapeutic target for a subset of muscle-invasive bladder cancers presenting a basal-like phenotype. Sci. Transl. Med. 2014, 6, 244ra91. [Google Scholar] [CrossRef]
- Baldia, P.H.; Maurer, A.; Heide, T.; Rose, M.; Stoehr, R.; Hartmann, A.; Williams, S.V.; Knowles, M.A.; Knuechel, R.; Gaisa, N.T. Fibroblast growth factor receptor (FGFR) alterations in squamous differentiated bladder cancer: A putative therapeutic target for a small subgroup. Oncotarget 2016, 7, 71429–71439. [Google Scholar] [CrossRef]
- Zou, J.; Huang, R.; Li, H.; Wang, B.; Chen, Y.; Chen, S.; Ou, K.; Wang, X. Secreted TGF-β-induced protein promotes aggressive progression in bladder cancer cells. Cancer Manag. Res. 2019, 11, 6995–7006. [Google Scholar] [CrossRef]
- Warrick, J.I.; Walter, V.; Yamashita, H.; Chung, E.; Shuman, L.; Amponsa, V.O.; Zheng, Z.; Chan, W.; Whitcomb, T.L.; Yue, F.; et al. FOXA1, GATA3 and PPARɣ Cooperate to Drive Luminal Subtype in Bladder Cancer: A Molecular Analysis of Established Human Cell Lines. Sci. Rep. 2016, 6, 38531. [Google Scholar] [CrossRef]
- Deb, B.; Puttamallesh, V.N.; Gondkar, K.; Thiery, J.; Gowda, H.; Kumar, P. Phosphoproteomic Profiling Identifies Aberrant Activation of Integrin Signaling in Aggressive Non-Type Bladder Carcinoma. J. Clin. Med. 2019, 8, 703. [Google Scholar] [CrossRef]
- Itkonen, H.M.; Engedal, N.; Babaie, E.; Lühr, M.; Guldvik, I.J.; Minner, S.; Hohloch, J.; Tsourlakis, M.C.; Schlomm, T.; Mills, I.G. UAP1 is overexpressed in prostate cancer and is protective against inhibitors of N-linked glycosylation. Oncogene 2014, 34, 3744–3750. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; Von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012, 41, D808–D815. [Google Scholar] [CrossRef]
- Subbannayya, Y.; Syed, N.; Barbhuiya, M.A.; Raja, R.; Marimuthu, A.; Sahasrabuddhe, N.; Pinto, S.M.; Manda, S.S.; Renuse, S.; Manju, H.; et al. Calcium calmodulin dependent kinase kinase 2—A novel therapeutic target for gastric adenocarcinoma. Cancer Boil. Ther. 2015, 16, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Syed, N.; Chavan, S.; Sahasrabuddhe, N.A.; Renuse, S.; Sathe, G.; Nanjappa, V.; Radhakrishnan, A.; Raja, R.; Pinto, S.M.; Srinivasan, A.; et al. Silencing of high-mobility group box 2 (HMGB2) modulates cisplatin and 5-fluorouracil sensitivity in head and neck squamous cell carcinoma. Proteomics 2015, 15, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Raja, R.; Sahasrabuddhe, N.A.; Radhakrishnan, A.; Syed, N.; Solanki, H.S.; Puttamallesh, V.N.; Balaji, S.A.; Nanjappa, V.; Datta, K.K.; Babu, N.; et al. Chronic exposure to cigarette smoke leads to activation of p21 (RAC1)-activated kinase 6 (PAK6) in non-small cell lung cancer cells. Oncotarget 2016, 7, 61229–61245. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, A.; Nanjappa, V.; Raja, R.; Sathe, G.; Puttamallesh, V.N.; Jain, A.; Pinto, S.M.; Balaji, S.A.; Chavan, S.; Sahasrabuddhe, N.A.; et al. A dual specificity kinase, DYRK1A, as a potential therapeutic target for head and neck squamous cell carcinoma. Sci. Rep. 2016, 6, 36132. [Google Scholar] [CrossRef]
- Ma, G.; Yang, X.; Liang, Y.; Wang, L.; Li, D.; Chen, Y.; Liang, Z.; Wang, Y.; Niu, H. Precision medicine and bladder cancer heterogeneity. Bull Cancer 2018, 105, 925–931. [Google Scholar] [CrossRef]
- Burger, M.; Denzinger, S.; Hartmann, A.; Wieland, W.-F.; Stoehr, R.; Obermann, E.C. Mcm2 predicts recurrence hazard in stage Ta/T1 bladder cancer more accurately than CK20, Ki67 and histological grade. Br. J. Cancer 2007, 96, 1711–1715. [Google Scholar] [CrossRef]
- Choy, B.; LaLonde, A.; Que, J.; Wu, T.; Zhou, Z. MCM4 and MCM7, potential novel proliferation markers, significantly correlated with Ki-67, Bmi1, and cyclin E expression in esophageal adenocarcinoma, squamous cell carcinoma, and precancerous lesions. Hum. Pathol. 2016, 57, 126–135. [Google Scholar] [CrossRef]
- Gou, K.; Liu, J.; Feng, X.; Li, H.; Yuan, Y.; Xing, C. Expression of Minichromosome Maintenance Proteins (MCM) and Cancer Prognosis: A meta-analysis. J. Cancer 2018, 9, 1518–1526. [Google Scholar] [CrossRef]
- Hyrien, O. How MCM loading and spreading specify eukaryotic DNA replication initiation sites. F1000 Res. 2016, 5, 2063. [Google Scholar] [CrossRef]
- Lau, K.-M.; Chan, Q.K.Y.; Pang, J.; Li, K.K.W.; Yeung, W.W.; Chung, N.Y.F.; Lui, P.C.; Tam, Y.-S.; Li, H.-M.; Zhou, L.; et al. Minichromosome maintenance proteins 2, 3 and 7 in medulloblastoma: Overexpression and involvement in regulation of cell migration and invasion. Oncogene 2010, 29, 5475–5489. [Google Scholar] [CrossRef]
- DaFonseca, C.J.; Shu, F.; Zhang, J.J. Identification of two residues in MCM5 critical for the assembly of MCM complexes and Stat1-mediated transcription activation in response to IFN-. Proc. Natl. Acad. Sci. USA 2001, 98, 3034–3039. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Chen, M.; Han, S.; Zhang, L.; Bai, Y.; Fang, X.; Ding, S.-Z.; Yang, Y. Plasma minichromosome maintenance complex component 6 is a novel biomarker for hepatocellular carcinoma patients. Hepatol. Res. 2014, 44, 1347–1356. [Google Scholar] [CrossRef] [PubMed]
- Korkolopoulou, P.; Givalos, N.; Saetta, A.A.; Goudopoulou, A.; Gakiopoulou, H.; Thymara, I.; Thomas-Tsagli, E.; Patsouris, E. Minichromosome maintenance proteins 2 and 5 expression in muscle-invasive urothelial cancer: A multivariate survival study including proliferation markers and cell cycle regulators. Hum. Pathol. 2005, 36, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Rausch, S.; Hennenlotter, J.; Teepe, K.; Kuehs, U.; Aufderklamm, S.; Bier, S.; Mischinger, J.; Gakis, G.; Stenzl, A.; Schwentner, C.; et al. Muscle-invasive bladder cancer is characterized by overexpression of thymidine kinase 1. Urol. Oncol. Semin. Orig. Investig. 2015, 33, 426.e21–426.e29. [Google Scholar] [CrossRef]
- Simon, R.; Atefy, R.; Wagner, U.; Forster, T.; Bruderer, J.; Wilber, K.; Mihatsch, M.J.; Gasser, T.; Sauter, G. HER-2 andTOP2A coamplification in urinary bladder cancer. Int. J. Cancer 2003, 107, 764–772. [Google Scholar] [CrossRef]
- Choi, J.-W.; Kim, Y.; Lee, J.H.; Kim, Y.-S. High expression of spindle assembly checkpoint proteins CDC20 and MAD2 is associated with poor prognosis in urothelial bladder cancer. Virchows Arch. A Pathol. Anat. Histopathol. 2013, 463, 681–687. [Google Scholar] [CrossRef]
- Mobley, A.; Zhang, S.; Bondaruk, J.; Wang, Y.; Majewski, T.; Caraway, N.P.; Huang, L.; Shoshan, E.; Velazquez-Torres, G.; Nitti, G.; et al. Aurora Kinase A is a Biomarker for Bladder Cancer Detection and Contributes to its Aggressive Behavior. Sci. Rep. 2017, 7, 40714. [Google Scholar] [CrossRef]
- Otto, T.; Sicinski, P. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer 2017, 17, 93–115. [Google Scholar] [CrossRef]
- Jagarlamudi, K.K.; Shaw, M. Thymidine kinase 1 as a tumor biomarker: Technical advances offer new potential to an old biomarker. Biomark. Med. 2018, 12, 1035–1048. [Google Scholar] [CrossRef]
- Langbein, S.; Lehmann, J.; Harder, A.; Steidler, A.; Michel, M.S.; Alken, P.; Badawi, J.K. Protein profiling of bladder cancer using the 2D-PAGE and SELDI-TOF-MS technique. Technol. Cancer Res. Treat. 2006, 5, 67–71. [Google Scholar] [CrossRef]
- Rahmani, A.H.; Babiker, A.Y.; AlWanian, W.M.; Elsiddig, S.A.; Faragalla, H.E.; Aly, S.M. Association of Cytokeratin and Vimentin Protein in the Genesis of Transitional Cell Carcinoma of Urinary Bladder Patients. Dis. Markers 2015, 2015, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Nitschke, K.; Heinkele, J.; Weis, C.-A.; Worst, T.S.; Eckstein, M.; Porubsky, S.; Erben, P. ANLN and TLE2 in Muscle Invasive Bladder Cancer: A Functional and Clinical Evaluation Based on In Silico and In Vitro Data. Cancers 2019, 11, 1840. [Google Scholar] [CrossRef] [PubMed]
- Akella, N.M.; Ciraku, L.; Reginato, M.J. Fueling the fire: Emerging role of the hexosamine biosynthetic pathway in cancer. BMC Boil. 2019, 17, 52. [Google Scholar] [CrossRef]
- Chiaradonna, F.; Ricciardiello, F.; Palorini, R. The Nutrient-Sensing Hexosamine Biosynthetic Pathway as the Hub of Cancer Metabolic Rewiring. Cells 2018, 7, 53. [Google Scholar] [CrossRef]
- De Queiroz, R.M.; Oliveira, I.A.; Piva, B.; Catão, F.B.; Rodrigues, B.D.C.; Pascoal, A.D.C.; Diaz, B.; Todeschini, A.R.; Caarls, M.B.; Dias, W. Hexosamine Biosynthetic Pathway and Glycosylation Regulate Cell Migration in Melanoma Cells. Front. Oncol. 2019, 9, 116. [Google Scholar] [CrossRef]
- Deb, B.; Patel, K.; Sathe, G.; Kumar, P. N-Glycoproteomic Profiling Reveals Alteration In Extracellular Matrix Organization In Non-Type Bladder Carcinoma. J. Clin. Med. 2019, 8, 1303. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puttamallesh, V.N.; Deb, B.; Gondkar, K.; Jain, A.; Nair, B.; Pandey, A.; Chatterjee, A.; Gowda, H.; Kumar, P. Quantitative Proteomics of Urinary Bladder Cancer Cell Lines Identify UAP1 as a Potential Therapeutic Target. Genes 2020, 11, 763. https://doi.org/10.3390/genes11070763
Puttamallesh VN, Deb B, Gondkar K, Jain A, Nair B, Pandey A, Chatterjee A, Gowda H, Kumar P. Quantitative Proteomics of Urinary Bladder Cancer Cell Lines Identify UAP1 as a Potential Therapeutic Target. Genes. 2020; 11(7):763. https://doi.org/10.3390/genes11070763
Chicago/Turabian StylePuttamallesh, Vinuth N., Barnali Deb, Kirti Gondkar, Ankit Jain, Bipin Nair, Akhilesh Pandey, Aditi Chatterjee, Harsha Gowda, and Prashant Kumar. 2020. "Quantitative Proteomics of Urinary Bladder Cancer Cell Lines Identify UAP1 as a Potential Therapeutic Target" Genes 11, no. 7: 763. https://doi.org/10.3390/genes11070763
APA StylePuttamallesh, V. N., Deb, B., Gondkar, K., Jain, A., Nair, B., Pandey, A., Chatterjee, A., Gowda, H., & Kumar, P. (2020). Quantitative Proteomics of Urinary Bladder Cancer Cell Lines Identify UAP1 as a Potential Therapeutic Target. Genes, 11(7), 763. https://doi.org/10.3390/genes11070763




