SpCas9- and LbCas12a-Mediated DNA Editing Produce Different Gene Knockout Outcomes in Zebrafish Embryos
Abstract
1. Introduction
2. Materials and Methods
3. Results
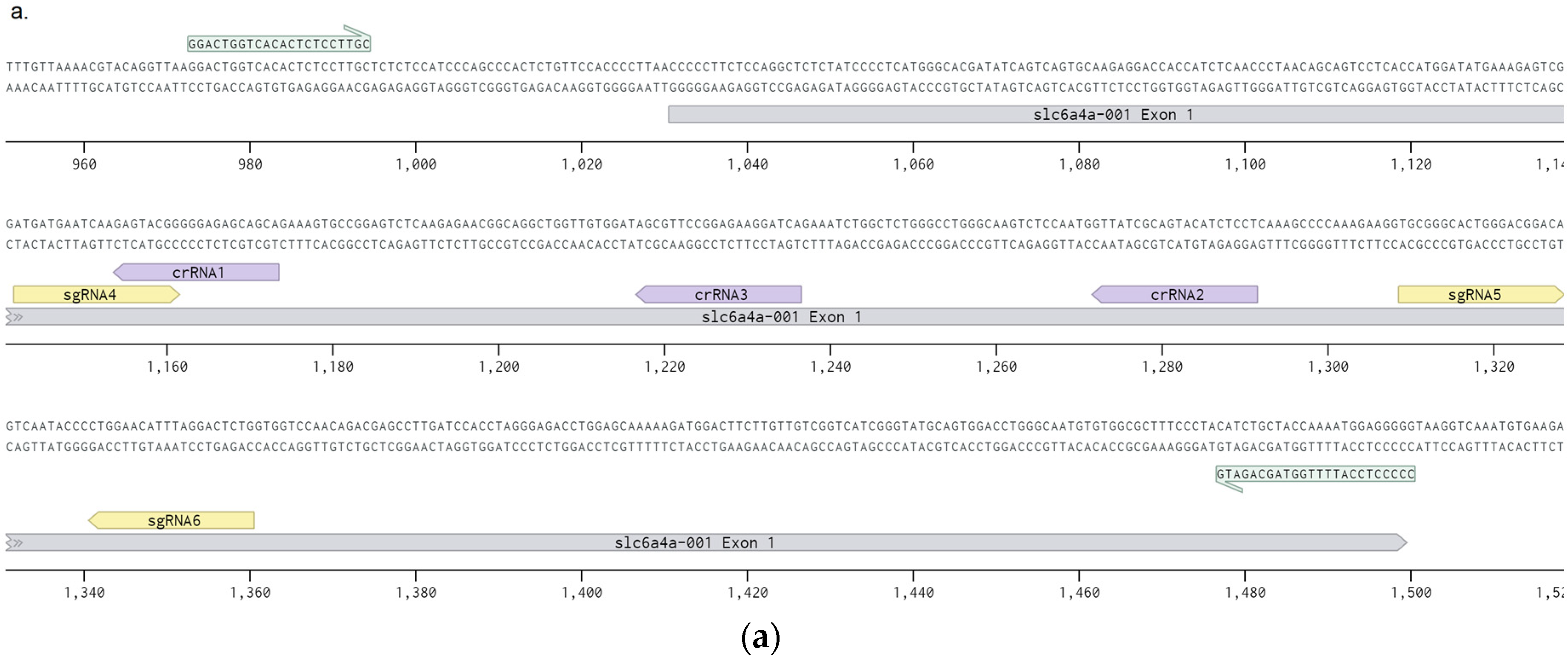
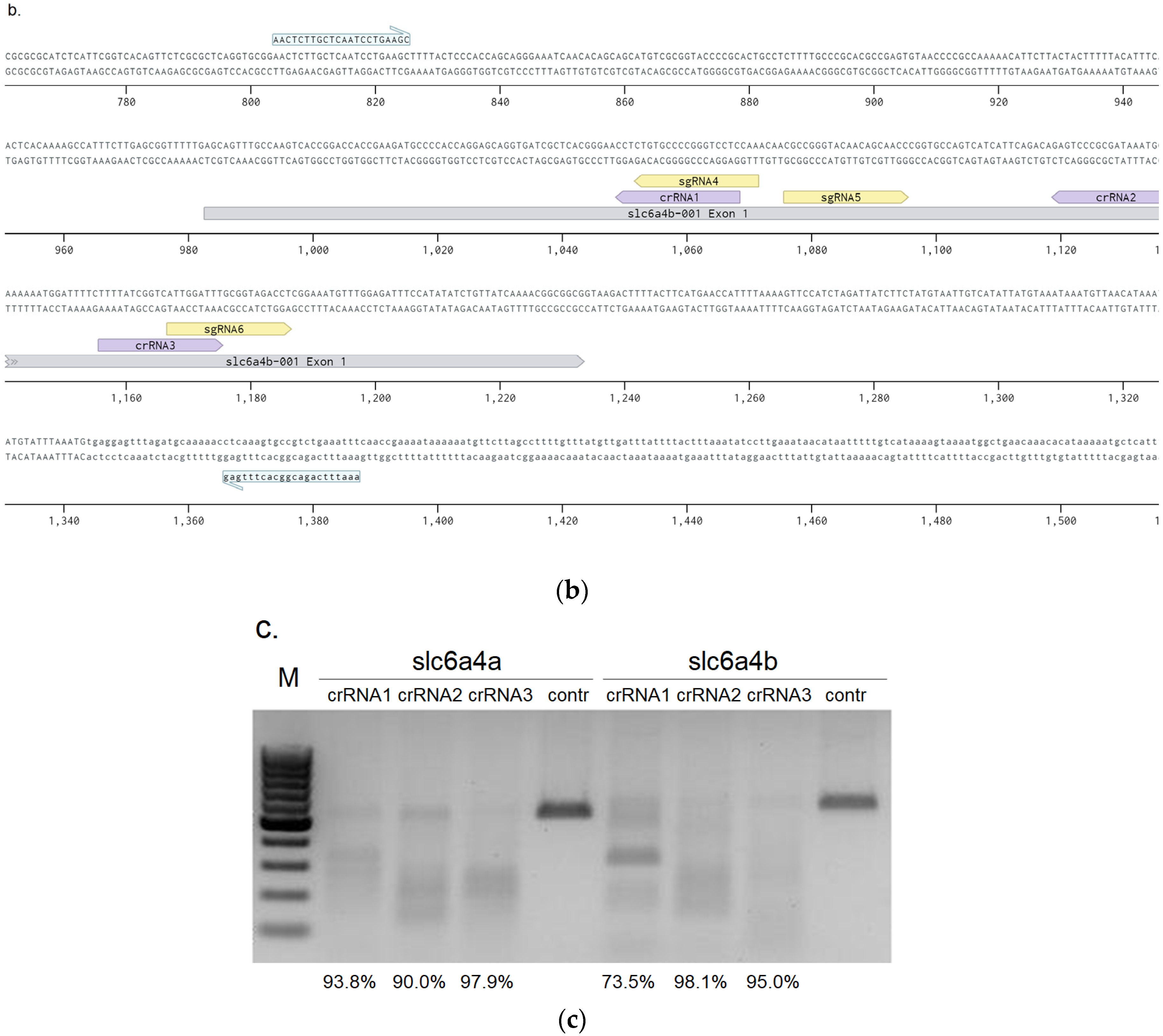
3.1. crRNA and sgRNA Selection and In Vitro Efficiency Assessment
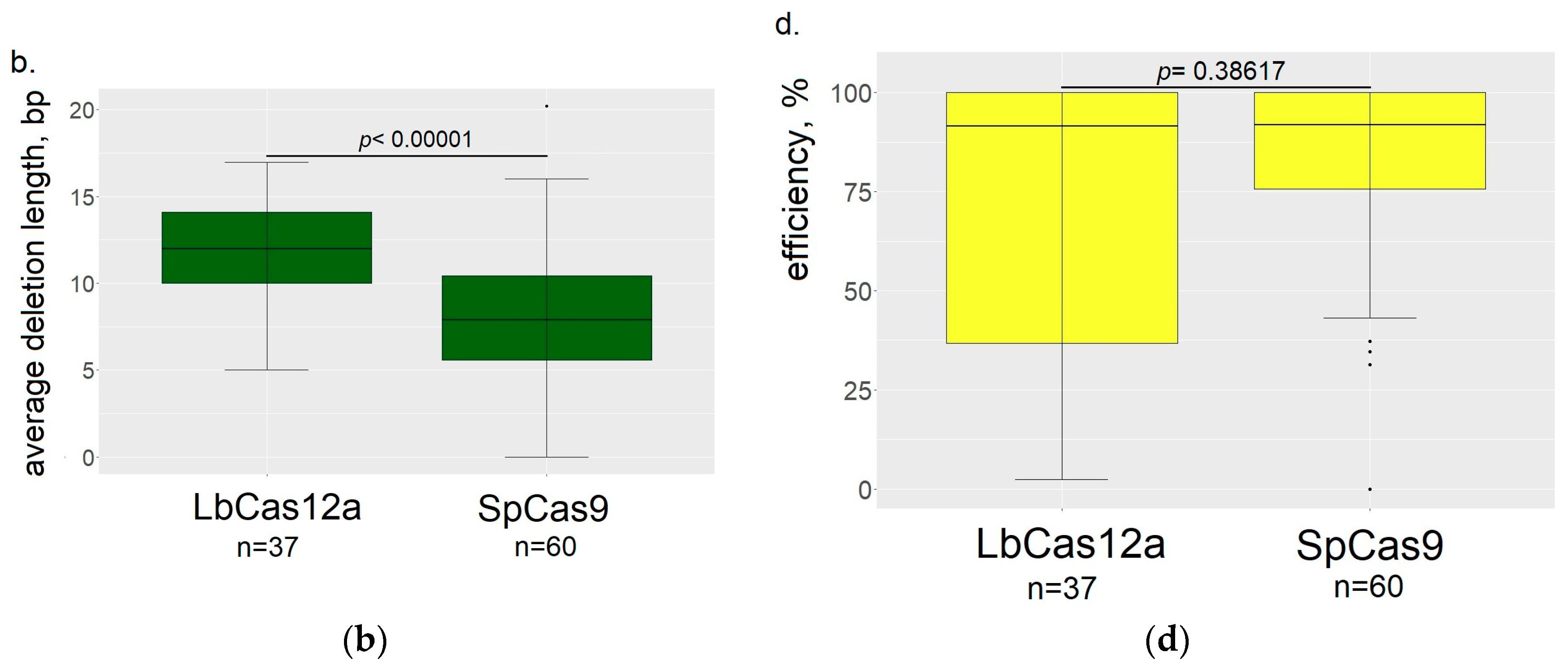
3.2. Comparison of the ResultingMutants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| RNP | ribonucleoprotein |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| CRISPR/Cas | Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associated protein |
| crRNA | crispr RNA |
| tracrRNA | trans-activating crispr RNA |
| TIDE | Tracking of Indels by DEcomposition |
| Cas | CRISPR associated protein |
| SpCas9 | Streptococcus pyogenes Cas9 |
| LbCas12a | Lachnospiraceae bacterium Cas12a |
| PAM | protospacer adjacent motive |
| NLS | nuclear localization signal |
References
- Barrangou, R.; Doudna, J.A. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 2016, 34, 933–941. [Google Scholar] [CrossRef]
- Blader, P.; Strähle, U. Zebrafish developmental genetics and central nervous system development. Hum. Mol. Genet. 2000, 9, 945–951. [Google Scholar] [CrossRef]
- Cornet, C.; Di Donato, V.; Terriente, J. Combining Zebrafish and CRISPR/Cas9: Toward a more efficient drug discovery pipeline. Front. Pharmacol. 2018, 9, 703. [Google Scholar] [CrossRef]
- Bengel, D.; Murphy, D.L.; Andrews, A.M.; Wichems, C.H.; Feltner, U.; Heils, A.; Mössner, R.; Westphal, H.; Lesch, K.-P. Altered brain serotonin homeostasis and Locomotor insensitivity to 3,4-Methylenedioxymethamphetamine (“Ecstasy”) in serotonin transporter-deficient mice. Mol. Pharmacol. 1998, 53, 649–655. [Google Scholar] [CrossRef]
- Homberg, J.; Olivier, J.D.; Smits, B.; Mul, J.D.; Mudde, J.; Verheul, M.; Nieuwenhuizen, O.; Cools, A.; Ronken, E.; Cremers, T.; et al. Characterization of the serotonin transporter knockout rat: A selective change in the functioning of the serotonergic system. Neuroscience 2007, 146, 1662–1676. [Google Scholar] [CrossRef]
- Wang, Y.; Takai, R.; Yoshioka, H.; Shirabe, K. Characterization and expression of serotonin transporter genes in zebrafish. Tohoku J. Exp. Med. 2006, 208, 267–274. [Google Scholar] [CrossRef]
- Terns, M.P. CRISPR-based technologies: Impact of RNA-targeting systems. Mol. Cell 2018, 72, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, J.; Hur, J.K.; Been, K.W.; Yoon, S.-H.; Kim, J.-S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016, 34, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Cheong, S.-A.; Lee, J.G.; Lee, S.-W.; Lee, M.S.; Baek, I.-J.; Sung, Y.H. Generation of knockout mice by Cpf1-mediated gene targeting. Nat. Biotechnol. 2016, 34, 808–810. [Google Scholar] [CrossRef] [PubMed]
- Strohkendl, I. Kinetic basis for DNA target specificity of CRISPR-Cas12a. Mol. Cell 2018, 7, 816–824. [Google Scholar] [CrossRef] [PubMed]
- Swarts, D.; Jínek, M. Cas9 versus Cas12a/Cpf1: Structure-function comparisons and implications for genome editing. Wiley Interdiscip. Rev. RNA 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.; Liu, J. Cas9-catalyzed DNA cleavage generates staggered ends: Evidence from molecular dynamics simulations. Sci. Rep. 2016, 6, 37584. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Biswal, A.; Dionora, J.; Perdigon, K.M.; Balahadia, C.P.; Mazumdar, S.; Chater, C.C.; Lin, H.-C.; Coe, R.A.; Kretzschmar, T.; et al. CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 2017, 30, 1473–1757. [Google Scholar] [CrossRef] [PubMed]
- Benchling [Biology Software]. 2020. Available online: https://benchling.com (accessed on 7 June 2020).
- Fedorova, I.; Arseniev, A.; Selkova, P.; Pobegalov, G.; Goryanin, I.; Vasileva, A.; Musharova, O.; Abramova, M.; Kazalov, M.; Zyubko, T.; et al. DNA targeting by clostridium cellulolyticum CRISPR-Cas9 type II-C system. Nucleic Acids Res. 2020, 48, 2026–2034. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (DanioRerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- Matthews, J.L.; Murphy, J.M.; Carmichael, C.; Yang, H.; Tiersch, T.; Westerfield, M.; Varga, Z.M. Changes to extender, Cryoprotective medium, andInVitroFertilization improve Zebrafish sperm cryopreservation. Zebrafish 2018, 15, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Rosen, J.N.; Sweeney, M.F.; Mably, J.D. Microinjection of Zebrafish embryos to analyze gene function. J. Vis. Exp. 2009. [Google Scholar] [CrossRef]
- Moreno-Mateos, M.A.; Fernandez, J.P.; Rouet, R.; Vejnar, C.E.; Lane, M.A.; Mis, E.; Khokha, M.K.; Doudna, J.A.; Giraldez, A.J. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat. Commun. 2017, 8, 2024. [Google Scholar] [CrossRef]
- Brinkman, E.K.; Chen, T.; Amendola, M.; Van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014, 42. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2015. [Google Scholar]
- Doench, J.G.; Fusi, N.; Sullender, M.E.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.B.; Orchard, R.; et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef]
- Hsu, P.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Nerys-Junior, A.; Braga-Dias, L.P.; Pezzuto, P.; Cotta-De-Almeida, V.; Tanuri, A. Comparison of the editing patterns and editing efficiencies of TALEN and CRISPR-Cas9 when targeting the human CCR5 gene. Genet. Mol. Biol. 2018, 41, 167–179. [Google Scholar] [CrossRef]
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal Transduct. Target. Ther. 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Sentmanat, M.F.; Peters, S.T.; Florian, C.P.; Connelly, J.P.; Pruett-Miller, S.M. A survey of validation strategies for CRISPR-Cas9 editing. Sci. Rep. 2018, 8, 888. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, T.; Liao, C.; Beisel, C.L. The Acidaminococcus sp. Cas12a nuclease recognizes GTTV and GCTV as non-canonical PAMs. FEMS Microbiol. Lett. 2019, 366. [Google Scholar] [CrossRef]
- Hendel, A.; Bak, R.O.; Clark, J.T.; Kennedy, A.B.; Ryan, D.E.; Roy, S.; Steinfeld, I.; Lunstad, B.D.; Kaiser, R.J.; Wilkens, A.B.; et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015, 33, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Bloh, K.M.; Bialk, P.A.; Gopalakrishnapillai, A.; Kolb, E.A.; Kmiec, E.B. CRISPR/Cas9-directed reassignment of the GATA1 initiation codon in K562 cells to recapitulate AML in down syndrome. Mol. Ther.-Nucleic Acids 2017, 7, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Luk, K.; Shin, M.; Idrizi, F.; Kwok, S.; Roscoe, B.; Mintzer, E.; Suresh, S.; Morrison, K.; Frazão, J.B.; et al. Enhanced Cas12a editing in mammalian cells and zebrafish. Nucleic Acids Res. 2019, 47, 4169–4180. [Google Scholar] [CrossRef]
- Hu, X.; Wang, C.; Liu, Q.; Fu, Y.; Wang, K. Targeted mutagenesis in rice using CRISPR-Cpf1 system. J. Genet. Genom. 2017, 44, 71–73. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; Watanabe, G.; Gerodimos, C.A.; Ochi, T.; Blundell, T.L.; Jackson, S.P.; Lieber, M.R. Different DNA end configurations dictate which NHEJ components are most important for joining efficiency. J. Biol. Chem. 2016, 291, 24377–24389. [Google Scholar] [CrossRef]
- Gao, Z.; Fan, M.; Das, A.T.; Herrera-Carrillo, E.; Berkhout, B. Extinction of all infectious HIV in cell culture by the CRISPR-Cas12a system with only a single crRNA. Nucleic Acids Res. 2020, 48, 5527–5539. [Google Scholar] [CrossRef]
- Van Overbeek, M.; Capurso, D.; Carter, M.M.; Thompson, M.S.; Frias, E.; Russ, C.; Reece-Hoyes, J.S.; Nye, C.; Gradia, S.; Vidal, B.; et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell 2016, 63, 633–646. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meshalkina, D.A.; Glushchenko, A.S.; Kysil, E.V.; Mizgirev, I.V.; Frolov, A. SpCas9- and LbCas12a-Mediated DNA Editing Produce Different Gene Knockout Outcomes in Zebrafish Embryos. Genes 2020, 11, 740. https://doi.org/10.3390/genes11070740
Meshalkina DA, Glushchenko AS, Kysil EV, Mizgirev IV, Frolov A. SpCas9- and LbCas12a-Mediated DNA Editing Produce Different Gene Knockout Outcomes in Zebrafish Embryos. Genes. 2020; 11(7):740. https://doi.org/10.3390/genes11070740
Chicago/Turabian StyleMeshalkina, Darya A., Aleksei S. Glushchenko, Elana V. Kysil, Igor V. Mizgirev, and Andrej Frolov. 2020. "SpCas9- and LbCas12a-Mediated DNA Editing Produce Different Gene Knockout Outcomes in Zebrafish Embryos" Genes 11, no. 7: 740. https://doi.org/10.3390/genes11070740
APA StyleMeshalkina, D. A., Glushchenko, A. S., Kysil, E. V., Mizgirev, I. V., & Frolov, A. (2020). SpCas9- and LbCas12a-Mediated DNA Editing Produce Different Gene Knockout Outcomes in Zebrafish Embryos. Genes, 11(7), 740. https://doi.org/10.3390/genes11070740





