Phenotypic Variability and Genetic Diversity in a Pinus koraiensis Clonal Trial in Northeastern China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Plant Materials
2.2. Growth, Wood, and Fruit Trait Measurements
2.3. DNA Extraction and Simple Sequence Repeats (SSR) Analysis
2.4. Statistical Analysis
3. Results
3.1. Phenotypic Variability
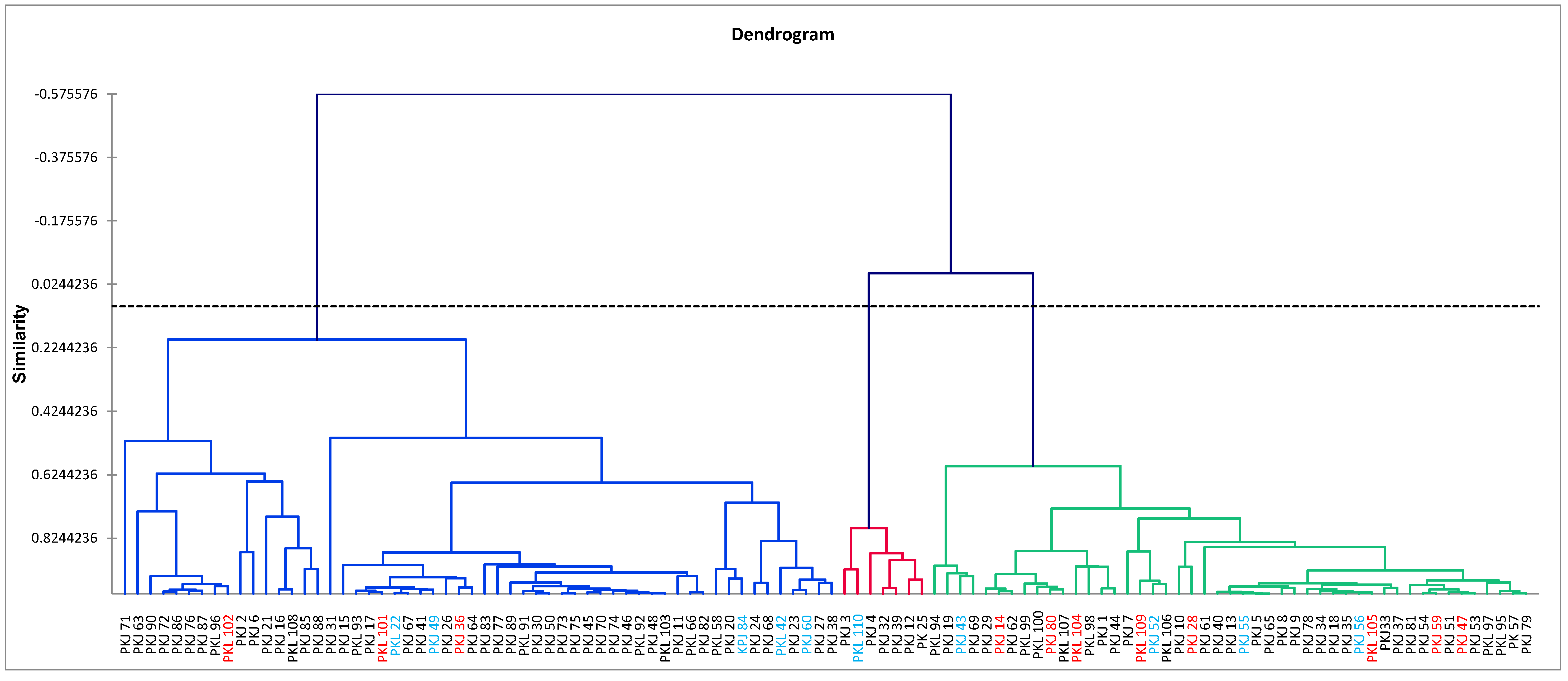
3.2. Classification Analysis of Phenotypic Characteristics
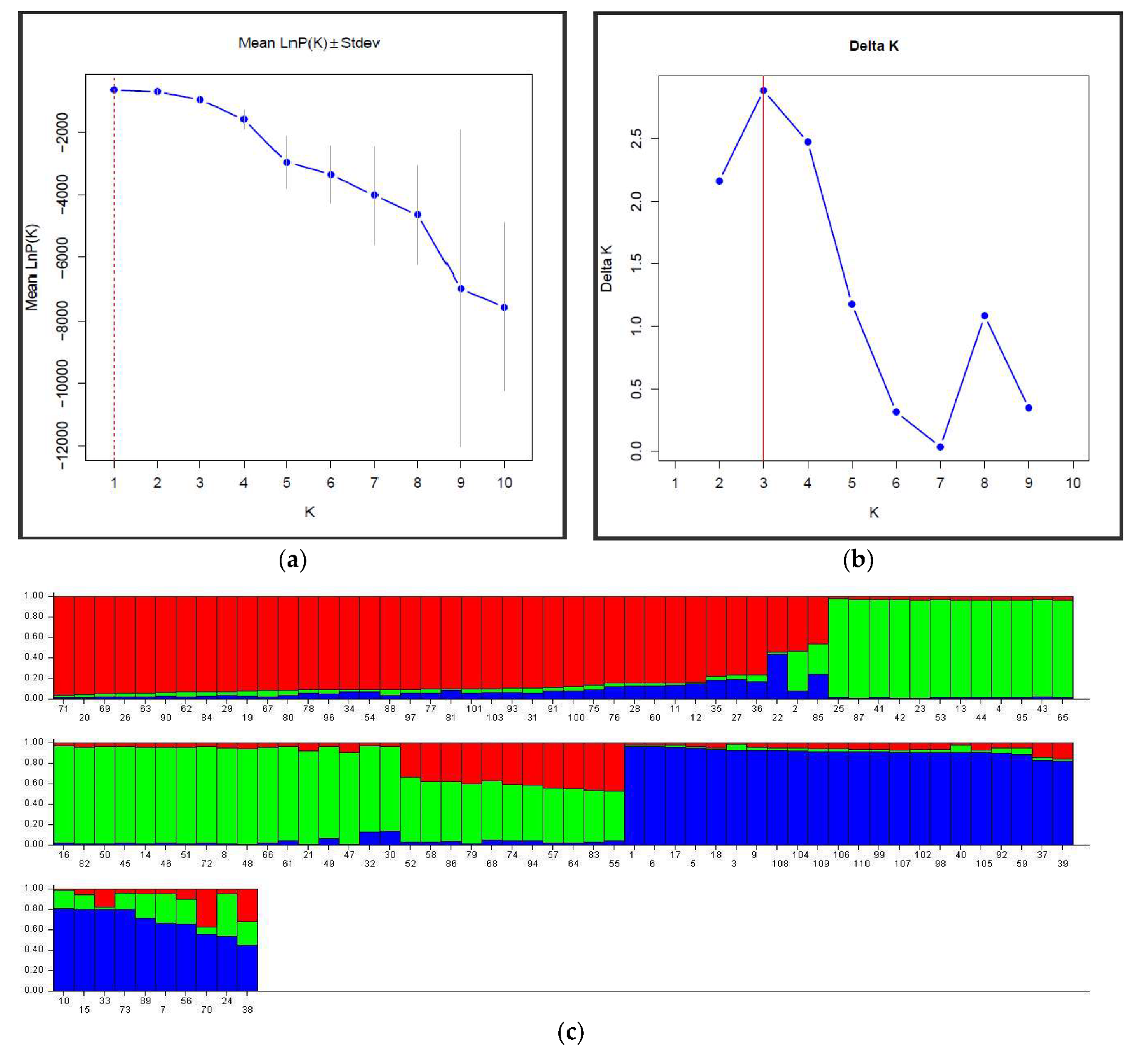
3.3. Clone Structure and Genetic Diversity
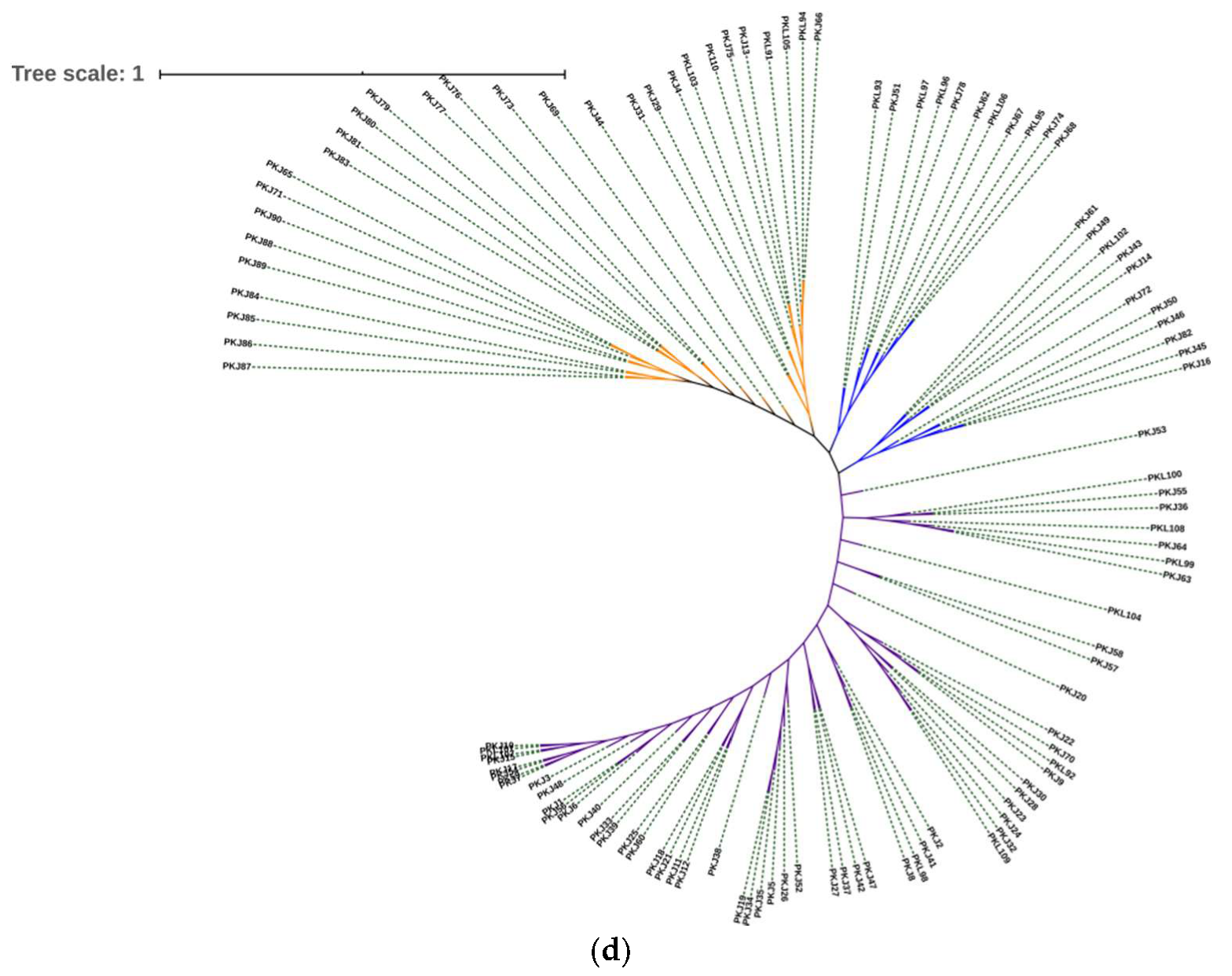
3.4. Cluster Analysis by Molecular Markers
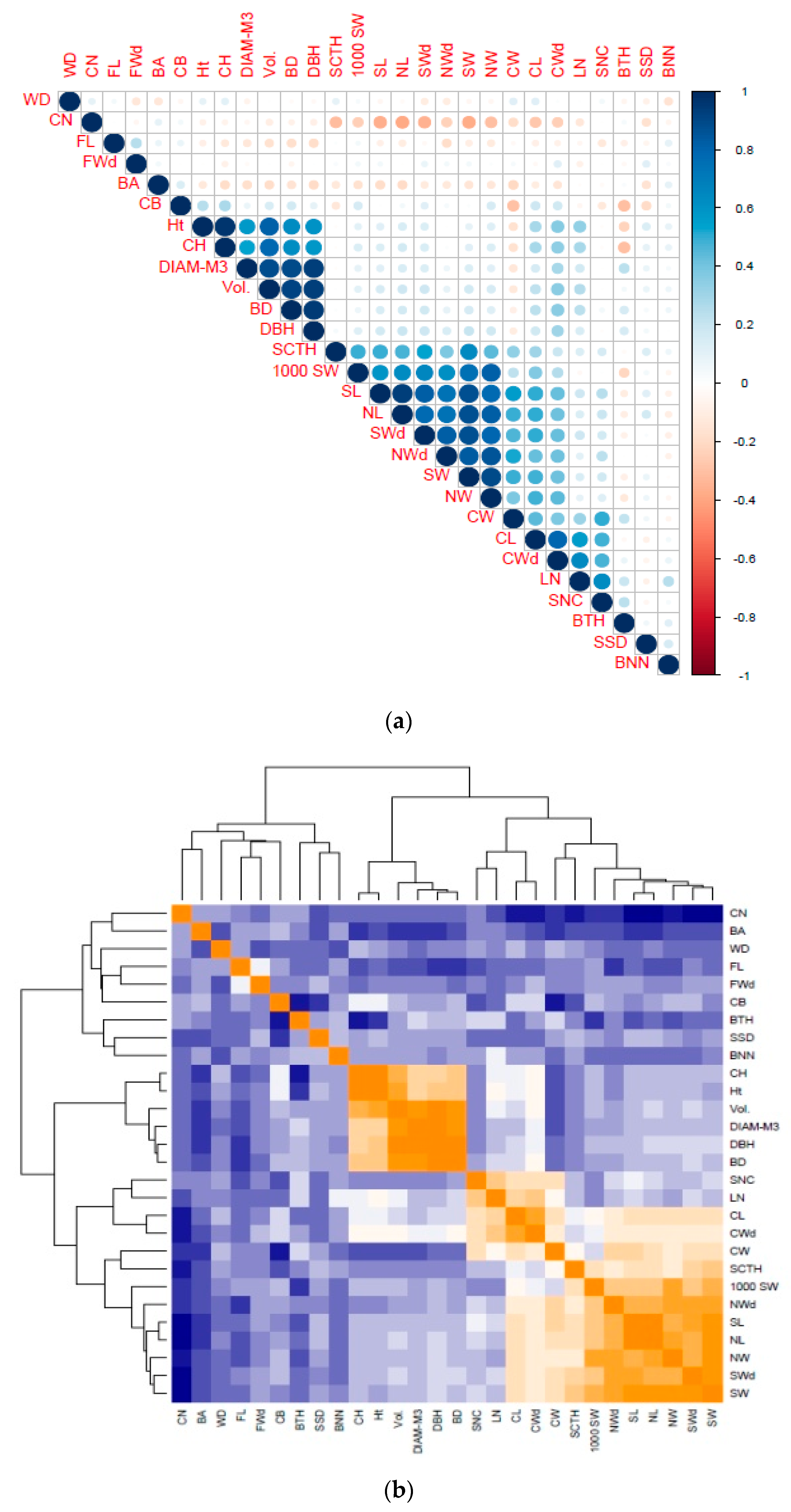
3.5. Correlation between Phenotypic and Molecular Markers
4. Discussion
4.1. Clone Variability Based on Phenotypic Characteristics
4.2. Cluster Analysis Based on Phenotypic Traits
4.3. SSR Polymorphism and Clone Genetic Diversity
4.4. Genetic Structure and Clone Genetic Relatedness
4.5. Correlations between Phenotypic and Molecular Markers
5. Final Considerations
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, J.; Zhou, Y.; Zhou, G.; Xiao, C. Composition and structure of Pinus koraiensis mixed forest respond to spatial climatic changes. PLoS ONE 2014, 9, e97192. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Xiao, X.; Dong, J.; Zhang, G.; Shimada, M.; Liu, J.; Li, C.; Kou, W.; Moore, B., III. Forest cover maps of China in 2010 from multiple approaches and data sources: PALSAR, Landsat, MODIS, FRA, and NFI. ISPRS J. Photogramm. 2015, 109, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Li, C.Z.; Löfgren, K.G. A theory of red pine (Pinus koraiensis) management for both timber and commercial seeds. For. Sci. 2000, 46, 284–290. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, H.; Yang, C.; Zhang, L.; Du, J.; Jiang, Y. Clonal variations in nutritional components of Pinus koreansis seeds collected from seed orchards in Northeastern China. J. For. Res. 2016, 27, 295–311. [Google Scholar] [CrossRef]
- Yu, D.; Zhou, L.; Zhou, W.; Ding, H.; Wang, Q.; Wang, Y.; Wu, X.; Dai, L. Forest management in Northeast China: History, problems, and challenges. J. Environ. Manag. 2011, 48, 1122–1135. [Google Scholar] [CrossRef]
- Owari, T.; Tatsumi, S.; Ning, L.; Yin, M. Height growth of Korean pine seedlings planted under strip-cut larch plantations in Northeast China. J. Res. 2015, 2015, 178681. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Dai, E.; Li, Y.; Xi, W. Climate change and sustainable forestry: A regional perspective from northeast China. For. Chron. 2018, 94, 201–207. [Google Scholar] [CrossRef]
- Wang, G.; Ning, Y.; Jin, J.; Zhang, S.; Yi, H. Study on the establishment technique of seed orchard for Korean pine improved generations. J. For. Res. 2000, 28, 68–69. [Google Scholar]
- Liang, D.; Ding, C.; Zhao, G.; Leng, W.; Zhang, M.; Zhao, X.; Qu, G. Variation and selection analysis of Pinus koraiensis clones in northeast China. J. For. Res. 2018, 29, 611–622. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, Q.; Tian, Y.; Yang, S.; Wang, H.; Wang, L.; Li, Y.; Zhang, P.; Zhao, X. Comprehensive assessment of growth traits and wood properties in half-sib Pinus koraiensis families. Euphytica 2018, 214, 202–217. [Google Scholar] [CrossRef]
- Liang, D.; Wang, B.; Song, S.; Wang, J.; Wang, L.; Wang, Q.; Ren, X.; Zhao, X. Analysis of genetic effects on a complete diallel cross test of Pinus koraiensis. Euphytica 2019, 215, 92. [Google Scholar] [CrossRef]
- Kaviriri, D.K.; Li, Y.; Zhang, D.; Li, H.; Fan, Z.; Wang, J.; Wang, L.; Wang, Q.; Wang, D.; Chiang, V.L. Clonal variations in cone, seed and nut traits in a Pinus koraiensis seed orchard in Northeast China. J. For. Res. 2020, 1–9. [Google Scholar] [CrossRef]
- Man, B.X.; Zhang, L.; Sun, J.Z.; Yuan, H.S.; Sun, J.J.; Wang, X.Y.; Sun, Y.B.; Qiu, L.; Wang, Y.L.; Wang, Y.Z. On plant growth regulator accelerate seed setting earlier of Pinus koraiensis. J. Beihua Univ. (Nat. Sci.) 2012, 13, 329–334. [Google Scholar]
- Li, Y.; Liang, L.; Ge, X.J. Development of microsatellite loci for Pinus koraiensis (Pinaceae). Am. J. Bot. 2010, 97, e39–e41. [Google Scholar] [CrossRef]
- Feng, F.J.; Zhao, D.; Sui, X.; Sun, X.Y. Study on mating system of Pinus koraiensis in natural population based on cpSSR technology. In Advanced Materials Research; Trans Tech Publications Ltd.: Bäch SZ, Switzerland, 2011; Volume 183, pp. 700–704. [Google Scholar] [CrossRef]
- Du, J.; Zhang, Z.; Zhang, H.; Junhong, T. EST–SSR marker development and transcriptome sequencing analysis of different tissues of Korean pine (Pinus koraiensis Sieb. et Zucc.). Biotechnol. Biotechnol. Equip. 2017, 31, 679–689. [Google Scholar] [CrossRef] [Green Version]
- Atangana, A.R.; Beaulieu, J.; Khasa, D.P. Wild genetic diversity preservation in a small-sized first generation breeding population of Allanblackia floribunda (Clusiaceae). Tree Genet. Genomes 2010, 6, 127–136. [Google Scholar] [CrossRef]
- Sui, X.; Feng, F.; Zhao, D.; Xing, M.; Sun, X.; Han, S.; Li, M. Mating system patterns of natural populations of Pinus koraiensis along its post-glacial colonization route in northeastern China. Genet. Mol. Res. 2015, 14, 4113–4124. [Google Scholar] [CrossRef]
- Porth, I.; El-kassaby, Y. Assessment of the genetic diversity in forest tree populations using molecular markers. Diversity 2014, 6, 283–295. [Google Scholar] [CrossRef] [Green Version]
- Lefort-Buson, M.; Hebert, Y.; Damerval, C. Tools for assessing genetic and phenotypic diversity. Agronomy 1988, 8, 173–178. (In French) [Google Scholar] [CrossRef]
- Rezende, G.D.S.P.; de Resende, M.D.V.; de Assis, T.F. Eucalyptus Breeding for Clonal Forestry. In Challenges and Opportunities for the World’s Forests in the 21st Century; Fenning, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2014; Forestry Sciences Vol. 81; pp. 393–424. [Google Scholar]
- Schaberg, P.G.; Dehayes, D.H.; Hawley, G.J.; Nijensohn, S.E. Anthropogenic alterations of genetic diversity within tree populations: Implications for forest ecosystem resilience. For. Ecol. Manag. 2008, 256, 855–862. [Google Scholar] [CrossRef]
- Chan, W.Y.; Hoffmann, A.A.; Van Oppen, M.J. Hybridization as a conservation management tool. Conserv. Lett. 2019, 12, e12652. [Google Scholar] [CrossRef]
- Namkoong, G.; Kang, H.C.; Brouard, J.S. Tree Breeding: Principles and Strategies, 1st ed.; Springer Science and Business Media: New York, NY, USA, 2012; Volume 11, pp. 37–111. [Google Scholar]
- Meyer, R.C.; Törjék, O.; Becher, M.; Altmann, T. Heterosis of biomass production in Arabidopsis. Establishment during early development. Plant. Physiol. 2004, 134, 1813–1823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birchler, J.A.; Auger, D.L.; Riddle, N.C. In search of the molecular basis of heterosis. Plant Cell 2003, 15, 2236–2239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šijačić-Nikolić, M.; Isajev, V. Assesment of heterotic effect in Serbian spruce hybrid combination. Genetika 2004, 36, 257–263. [Google Scholar] [CrossRef]
- Lynch, M.; Ritland, K. Estimation of pairwise relatedness with molecular markers. Genetics 1999, 152, 1753–1766. [Google Scholar] [PubMed]
- Hardy, O.J. Estimation of pairwise relatedness between individuals and characterization of isolation-by-distance processes using dominant genetic markers. Mol. Ecol. 2003, 12, 1577–1588. [Google Scholar] [CrossRef]
- Oliveira, E.J.; Pádua, J.G.; Zucchi, M.I.; Vencovsky, R.; Vieira, M.L.C. Origin, evolution and genome distribution of microsatellites. Genet. Mol. Biol. 2006, 29, 294–307. [Google Scholar] [CrossRef]
- Xu, Z.; Yu, G.; Zhang, X.; He, N.; Wang, Q.; Wang, S.; Xu, X.; Wang, R.; Zhao, N. Biogeographical patterns of soil microbial community as influenced by soil characteristics and climate across Chinese forest biomes. Appl. Soil Ecol. 2018, 124, 298–305. [Google Scholar] [CrossRef]
- Zhu, B.; Wang, X.; Fang, J.; Piao, S.; Shen, H.; Zhao, S.; Peng, C. Altitudinal changes in carbon storage of temperate forests on Mt Changbai, Northeast China. J. Plant Res. 2010, 123, 439–452. [Google Scholar] [CrossRef]
- Zhao, X.; Bian, X.; Liu, M.; Li, Z.; Li, Y.; Zheng, M.; Teng, W.; Jiang, J.; Liu, G. Analysis of genetic effects on a complete diallel cross test of Betula platyphylla. Euphytica 2014, 200, 221–229. [Google Scholar] [CrossRef]
- Louppe, D. Forest estimate. In Tropical Forester Memento; Editions Quae: Versailles Cedex, France, 2015. (In French) [Google Scholar]
- Yin, S.; Xiao, Z.; Zhao, G.; Zhao, X.; Sun, X.; Zhang, Y.; Wang, F.; Li, S.; Zhao, X.; Qu, G.Z. Variation analyses of growth and wood properties of Larix olgensis clones in China. J. For. Res. 2017, 28, 687–697. [Google Scholar] [CrossRef]
- Allen, G.; Flores-vergara, M.; Krasynanski, S.; Kumar, S.; Thompson, W. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006, 1, 2320–2325. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, H.; Mo, C.; Zhang, L. SSR analysis of Pinus koraiensis transcriptome and development of EST-SSR markers. For. Sci. 2015, 51, 114–120. (In Chinese) [Google Scholar]
- Fox, J.; Bouchet-Valat, M.; Andronic, L.; Ash, M.; Boye, T.; Calza, S.; Chang, A.; Grosjean, P.; Heiberger, R.; Pour, K.K. Package ‘Rcmdr’. 2018. Available online: http://cran.ma.imperial.ac.uk/web/packages/Rcmdr/Rcmdr.pdf (accessed on 14 April 2020).
- Li, Y.; Suontama, M.; Burdon, R.D.; Dungey, H.S. Genotype by environment interactions in forest tree breeding: Review of methodology and perspectives on research and application. Tree Genet. Genomes 2017, 13, 60–78. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Chen, Z.; Lundqvist, S.O.; Olsson, L.; Grahn, T.; Karlsson, B.; Wu, H.X.; García-GIL, M.R. Genetic analysis of wood quality traits in Norway spruce open-pollinated progenies and their parent plus trees at clonal archives and the evaluation of phenotypic selection of plus trees. Can. J. For. Res. 2019, 49, 810–818. [Google Scholar] [CrossRef]
- Zhao, S.; Guo, Y.; Sheng, Q.; Shyr, Y. Heatmap3: An improved heatmap package with more powerful and convenient features. BMC Bioinform. 2014, 15, 16. [Google Scholar] [CrossRef] [Green Version]
- Brommer, J. Whither PST? The approximation of QST by PST in evolutionary and conservation biology. J. Evol. Biol. 2011, 24, 1160–1168. [Google Scholar] [CrossRef]
- Junior, P.C.; Ikeda, A.C.; Schuhli, G.S.; Silva, L.D.; Higa, A.R. Repeatability and genetic dissimilarity using biometric characteristics of black wattle seeds. Adv. For. Sci. 2018, 5, 333–337. [Google Scholar]
- Vidal, N.P.; Manful, C.F.; Pham, T.H.; Stewart, P.; Keough, D.; Thomas, R. The use of XLSTAT in conducting principal component analysis (PCA) when evaluating the relationships between sensory and quality attributes in grilled foods. MethodsX 2020, 7, 100835. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Manter, D.K.; Delgado, J.A.; Holm, D.G.; Stong, R.A. Pyrosequencing reveals a highly diverse and cultivar-specific bacterial endophyte community in potato roots. Microb. Ecol. 2010, 60, 157–166. [Google Scholar] [CrossRef]
- Hosking, L.; Lumsden, S.; Lewis, K.; Yeo, A.; Mccarthy, L.; Bansal, A.; Riley, J.; Purvis, I.; Xu, C.F. Detection of genotyping errors by Hardy–Weinberg equilibrium testing. Eur. J. Hum. Genet. 2004, 12, 395–399. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.Q.; Shen, S.K.; Zhang, X.J.; Wang, Y.H.; Sun, W.B. Genetic diversity and population structure of an extremely endangered species: The world’s largest Rhododendron. Aob Plants 2015, 7, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peakall, R.; Smousse, P.E. GeneAlEx 6.5: Genetics analysis in Excel. Population genetics software for teaching and research- an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porras-Hurtado, L.; Ruiz, Y.; Santos, C.; Phillips, C.; Carracedo, Á.; Lareu, M. An overview of STRUCTURE: Applications, parameter settings, and supporting software. Front. Genet. 2013, 4, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.L.; Liu, J.X. StructureSelector: A web based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Takezaki, N. Estimation of genetic distances and phylogenetic trees from DNA analysis. In Proceedings of the 5th World Congress on Genetics Applied to Livestock Production; The Pennsylvania State University: State College, PA, USA, 1983; p. 16802. [Google Scholar]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Rédei, K.; Keserű, Z.; Csiha, I.; Rásó, J.; Bakti, B.; Takács, M. Improvement of black locust (Robinia pseudoacacia L.) growing under marginal site conditions in Hungary: Case studies. Acta Agrar. Debr. 2018, 30, 129–133. [Google Scholar] [CrossRef]
- Van Leeuwen, M.; Hilker, T.; Coops, N.C.; Frazer, G.; Wulder, M.A.; Newnham, G.J.; Culvenor, D.S. Assessment of standing wood and fiber quality using ground and airborne laser scanning: A review. For. Ecol. Manag. 2011, 261, 1467–1478. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, R.; Jin, G.; Feng, Z.; Zhou, Z. Assessing the genetic diversity and genealogical reconstruction of cypress (Cupressus funebris Endl.) breeding parents using SSR markers. Forests 2016, 7, 160. [Google Scholar] [CrossRef] [Green Version]
- Hao, L.; Zhang, G.; Lu, D.; Hu, J.; Jia, H. Analysis of the genetic diversity and population structure of Salix psammophila based on phenotypic traits and simple sequence repeat markers. PeerJ 2019, 7, e6419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West-Eberhard, M.J. Developmental plasticity and the origin of species differences. PNAS 2005, 102 (Suppl. 1), 6543–6549. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Ma, W.; Lu, N.; Wang, Z.; Wang, N.; Zhai, W.; Kong, L.; Qu, G.; Wang, Q.; Wang, J. Genetic variation of growth traits and genotype-by-environment interactions in clones of Catalpa bungei and Catalpa fargesii f. duclouxii. Forests 2019, 10, 57. [Google Scholar] [CrossRef] [Green Version]
- Matsushita, M.; Takata, K.; Hitsuma, G.; Yagihashi, T.; Noguchi, M.; Shibata, M.; Masaki, T. A novel growth model evaluating age–size effect on long-term trends in tree growth. Funct. Ecol. 2015, 29, 1250–1259. [Google Scholar] [CrossRef]
- Yang, H.; Weng, Q.; Li, F.; Zhou, C.; Li, M.; Chen, S.; Ji, H.; Gan, S. Genotypic variation and genotype-by-environment interactions in growth and wood properties in a cloned Eucalyptus urophylla × E. tereticornis family in Southern China. For. Sci. 2018, 64, 225–232. [Google Scholar] [CrossRef]
- Kenis, K.; Keulemans, J. Study of tree architecture of apple (Malus × domestica Borkh.) by QTL analysis of growth traits. Mol. Breed. 2007, 19, 193–208. [Google Scholar] [CrossRef]
- Kaviriri, D.K.; Liu, X.; Fan, Z.; Wang, J.; Wang, Q.; Wang, L.; Wang, L.; Khasa, D.; Zhao, X. Genetic Variation in Growth and Cone Traits of Pinus Koraiensis Half-Sib Families in Northeast China. Phyton 2020, 89, 57–69. [Google Scholar] [CrossRef]
- Cheng, X.; Xie, H.; Zhang, L.; Wang, M.; Li, C.; Yu, M.; He, Z. A comparative study of growth and leaf trait variation in twenty Cornus wilsoniana W. families in southeastern China. iForest 2017, 10, 759–765. [Google Scholar] [CrossRef] [Green Version]
- Hamrick, J.L.; Milton, J.; Linhart, Y.B. Levels of genetic variation in trees: Influence of life history characteristics. In General Technical Report PSW-GTR-48; Pacific Southwest Forest and Range Exp. Stn, Forest Service, US Department of Agriculture: Berkeley, CA, USA, 1981; Volume 48, pp. 35–41. [Google Scholar]
- Nicotra, A.B.; Chazdon, R.L.; Schlichting, C.D. Patterns of genotypic variation and phenotypic plasticity of light response in two tropical Piper (Piperaceae) species. Am. J. Bot. 1997, 84, 1542–1552. [Google Scholar] [CrossRef] [PubMed]
- Ganopoulos, I.; Moysiadis, T.; Xanthopoulou, A.; Ganopoulou, M.; Avramidou, E.; Aravanopoulos, F.A.; Tani, E.; Madesis, P.; Tsaftaris, A.; Kazantzis, K. Diversity of morpho-physiological traits in worldwide sweet cherry cultivars of GeneBank collection using multivariate analysis. Sci. Hortic. 2015, 197, 381–391. [Google Scholar] [CrossRef]
- Volk, G.M.; Henk, A.D.; Richards, C.M. Genetic diversity among US garlic clones as detected using AFLP methods. J. Am. Soc. Hortic. Sci. 2004, 129, 559–569. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Xie, Y.; Bi, Y.; Liu, J.; Amombo, E.; Hu, T.; Fu, J. Comparative study of diversity based on heat tolerant-related morpho-physiological traits and molecular markers in tall fescue accessions. Sci. Rep. 2015, 5, 18213. [Google Scholar] [CrossRef] [PubMed]
- Cholastova, T.; Knotova, D. Using morphological and microsatellite (SSR) markers to assess the genetic diversity in Alfalfa (Medicago sativa L.). Int. J. Biol. Biomol. Agric. Food Biotechnol. Eng. 2012, 69, 856–862. [Google Scholar]
- Lande, R.; Thompson, R. Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 1990, 124, 743–756. [Google Scholar]
- Stinchcombe, J.R.; Rutter, M.T.; Burdick, D.S.; Tiffin, P.; Rausher, M.D.; Mauricio, R. Testing for environmentally induced bias in phenotypic estimates of natural selection: Theory and practice. Am. Nat. 2002, 160, 511–523. [Google Scholar] [CrossRef]
- Kim, Z.; Lee, S.; Hwang, J. Genetic diversity and structure of natural populations of Pinus thunbergii in Korea. Silvae Genet. 1997, 46, 120–123. [Google Scholar]
- Yu, J.-H.; Chen, C.-M.; Tang, Z.-H.; Yuan, S.-S.; Wang, C.-J.; Zu, Y.-G. Isolation and characterization of 13 novel polymorphic microsatellite markers for Pinus koraiensis (Pinaceae). Am. J. Bot. 2012, 99, e421–e424. [Google Scholar] [CrossRef]
- Yun, J.; Juan, Z.; Ying, W.; Wei-Bing, F.; Gui-Fang, Z.; Zhong-Hu, L. Effects of geological and environmental events on the diversity and genetic divergence of four closely related Pines: Pinus koraiensis, P. armandii, P. griffithii, and P. pumila. Front. Plant. Sci. 2018, 9, 1264. [Google Scholar]
- Feng, F.J.; Sui, X.; Chen, M.M.; Zhao, D.; Han, H.J.; Li, M.H. Mode of pollen spread in clonal seed orchard of Pinus koraiensis. J. Biophys. Chem. 2010, 1, 33–39. [Google Scholar] [CrossRef] [Green Version]
- Potenko, V. Genetic diversity and mating system of Korean pine in Russia. In Breeding and Genetic Resources of Five-Needle Pines: Growth, Adaptability and Pest Resistance; USDA Forest Service Proceedings RMRS-P-32; US Department of Agriculture, Forest Service, Rocky Mountain Research Station: Fort Collins, CO, USA, 2004; p. 11. [Google Scholar]
- Potenko, V.; Velikov, A. Genetic diversity and differentiation of natural populations of Pinus koraiensis (Sieb. et Zucc.) in Russia. Silvae Genet. 1998, 47, 202–207. [Google Scholar]
- Feng, F.J.; Han, S.J.; Wang, H.M. Genetic diversity and genetic differentiation of natural Pinus koraiensis population. J. Res. 2006, 17, 21–24. [Google Scholar] [CrossRef]
- Arnaud-Haond, S.; Teixeira, S.; Massa, S.I.; Billot, C.; Saenger, P.; Coupland, G.; Duarte, C.M.; Serrao, E. Genetic structure at range edge: Low diversity and high inbreeding in Southeast Asian mangrove (Avicennia marina) populations. Mol. Ecol. 2006, 15, 3515–3525. [Google Scholar] [CrossRef] [PubMed]
- Ellstrand, N.C. Current knowledge of gene flow in plants: Implications for transgene flow. Philosophical Transactions of the Royal Society of London. Ser. B Biol. Sci. 2003, 358, 1163–1170. [Google Scholar] [CrossRef] [Green Version]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Leimu, R.; Mutikainen, P.; Koricheva, J.; Fischer, M. How general are positive relationships between plant population size, fitness and genetic variation? J. Ecol. 2006, 94, 942–952. [Google Scholar] [CrossRef]
- Suwarni, E.; Omondi, S.; Dancik, B.; Khasa, D. Estimation of pollen contamination and mating system in Pinus merkusii seedling seed orchard using allozyme markers. J. Trop. Sci. 2018, 30, 95–105. [Google Scholar]
- Lowe, A.; Boshier, D.; Ward, M.; Bacles, C.; Navarro, C. Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicted theory for neotropical trees. Heredity 2005, 95, 255–273. [Google Scholar] [CrossRef] [Green Version]
- Kremer, A.; Ronce, O.; Robledo-Arnuncio, J.J.; Guillaume, F.; Bohrer, G.; Nathan, R.; Bridle, J.R.; Gomulkiewicz, R.; Klein, E.K.; Ritland, K. Long-distance gene flow and adaptation of forest trees to rapid climate change. Ecol. Lett. 2012, 15, 378–392. [Google Scholar] [CrossRef] [Green Version]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar] [PubMed]
- Legendre, P.; Fortin, M.J. Comparison of the Mantel test and alternative approaches for detecting complex multivariate relationships in the spatial analysis of genetic data. Mol. Ecol. Resour. 2010, 10, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Franco, J.; Crossa, J.; Ribaut, J.; Bertran, J.; Warburton, M.; Khairallah, M. A method for combining molecular markers and phenotypic attributes for classifying plant genotypes. Theor. Appl. Genet. 2001, 103, 944–952. [Google Scholar] [CrossRef]
- Corrado, G.; La Mura, M.; Ambrosino, O.; Pugliano, G.; Varricchio, P.; Rao, R. Relationships of Campanian olive cultivars: Comparative analysis of molecular and phenotypic data. Genome 2009, 52, 692–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roldán-Ruiz, I.; Van Euwijk, F.; Gilliland, T.; Dubreuil, P.; Dillmann, C.; Lallemand, J.; De Loose, M.; Baril, C. A comparative study of molecular and morphological methods of describing relationships between perennial ryegrass (Lolium perenne L.) varieties. Theor. Appl. Genet. 2001, 103, 1138–1150. [Google Scholar] [CrossRef]
- Avval, S.E. Assessing polymorphism information content (PIC) using SSR molecular markers on local species of Citrullus colocynthis. Case study: Iran, Sistan-Balouchestan province. J. Mol. Biol. Res. 2017, 7, 42–49. [Google Scholar] [CrossRef] [Green Version]

| Traits | Units | SV | SS | df | MS | F | PCV | H2 | QST |
|---|---|---|---|---|---|---|---|---|---|
| Tree height | (m) | Clones | 620.899 | 109 | 5.696 | 2.965 *** | 8.36 | 0.663 | 0.0236 |
| Blocks | 89.767 | 9 | 9.974 | 5.191 *** | |||||
| Basal diameter | (cm) | Clones | 7287.222 | 109 | 66.855 | 3.981 *** | 9.05 | 0.749 | 0.0761 |
| Blocks | 384.485 | 9 | 42.721 | 2.544 ** | |||||
| Diameter at breast height | (cm) | Clones | 6810.669 | 109 | 62.483 | 4.260 *** | 9.3 | 0.765 | 0.0036 |
| Blocks | 118.065 | 9 | 13.118 | 0.894 *** | |||||
| Diameter at 3-m height | (cm) | Clones | 6228.944 | 109 | 57.146 | 3.685 *** | 9.55 | 0.729 | 0.0258 |
| Blocks | 162.950 | 9 | 18.106 | 1.167 * | |||||
| Stem volume | (m3) | Clones | 6.985 | 109 | 0.064 | 4.490 *** | 23.54 | 0.777 | 0.0003 |
| Blocks | 0.254 | 9 | 0.028 | 1.976 ** | |||||
| Burk thickness | (mm) | Clones | 542.443 | 109 | 4.977 | 1.976 *** | 12.2 | 0.494 | 0.0122 |
| Blocks | 159.142 | 9 | 17.682 | 7.020 *** | |||||
| Stem straightness degree | Clones | 5.469 | 109 | 109 | 2.022 *** | 4.44 | 0.505 | 0.0001 | |
| Blocks | 8.701 | 9 | 0.967 | 38.963 *** | |||||
| Branch angle | (°) | Clones | 114,639.207 | 109 | 1051.736 | 2.063 *** | 9.18 | 0.515 | 0.6407 |
| Blocks | 3209.167 | 9 | 356.574 | 0.699N | |||||
| Crown breath | (m) | Clones | 77.354 | 109 | 0.710 | 2.168 *** | 8.03 | 0.539 | 0.0003 |
| Blocks | 46.356 | 9 | 5.151 | 15.738 *** | |||||
| Crown height | (m) | Clones | 601.50 | 109 | 5.52 | 2.72 *** | 10.69 | 0.633 | 0.0076 |
| Blocks | 52.15 | 9 | 5.79 | 2.86 *** | |||||
| Branch number per node | Clones | 94.641 | 109 | 109 | 1.455 ** | 13.01 | 0.313 | 0.0034 | |
| Blocks | 12.895 | 9 | 1.433 | 2.400 * | |||||
| Wood density | (g/cm3) | Clones | 1.025 | 109 | 0.009 | 1.991 *** | 13.20 | 0.498 | 2.00 × 10−5 |
| Blocks | 0.016 | 9 | 0.008 | 1.644N | |||||
| Fiber length | (µm) | Clones | 281,044,500.967 | 109 | 2,578,389.917 | 4.664 *** | 11.13 | 0.786 | 0.0992 |
| Blocks | 2,491,085,275.474 | 9 | 85,899,492.258 | 155.372 *** | |||||
| Fiber width | (µm) | Clones | 12,701.725 | 109 | 116.530 | 2.324 *** | 5.71 | 0.570 | 0.1901 |
| Blocks | 57,119.958 | 9 | 1969.654 | 39.277 *** | |||||
| Cone number | Clones | 925.100 | 109 | 109 | 0.686N | 17.98 | 0.457 | 0.0183 | |
| Blocks | 60.502 | 9 | 6.722 | 0.539N | |||||
| Cone length | (mm) | Clones | 27,1671.646 | 109 | 2492.400 | 13.650 *** | 8.91 | 0.927 | 0.0869 |
| Blocks | 6518.537 | 9 | 210.275 | 1.152 * | |||||
| Cone width | (mm) | Clones | 16,5848.289 | 109 | 1521.544 | 43.426 *** | 13.25 | 0.977 | 0.4178 |
| Blocks | 1231.719 | 9 | 39.733 | 1.134 * | |||||
| Cone weight | (g) | Clones | 2,370,353.219 | 109 | 21,746.360 | 10.751 *** | 16.04 | 0.907 | 0.9809 |
| Blocks | 53,424.308 | 9 | 1723.365 | 0.852N | |||||
| Layer number | Clones | 4149.593 | 109 | 109 | 10.607 *** | 11.16 | 0.906 | 0.2264 | |
| Blocks | 208.822 | 9 | 6.736 | 1.877 *** | |||||
| Seed number per cone | Clones | 352,729.157 | 109 | 109 | 5.892 *** | 9.73 | 0.830 | 0.4171 | |
| Blocks | 46,422.283 | 9 | 1497.493 | 2.727 *** | |||||
| Seed length | (mm) | Clones | 1510.20 | 109 | 13.86 | 10.87 *** | 5.86 | 0.908 | 0.0388 |
| Blocks | 191.65 | 9 | 13.69 | 10.74 *** | |||||
| Seed width | (mm) | Clones | 739.32 | 109 | 6.78 | 5.84 *** | 6.28 | 0.829 | 0.0059 |
| Blocks | 96.10 | 9 | 6.86 | 5.91 *** | |||||
| Seed weight | (g) | Clones | 20.32 | 109 | 0.19 | 16.37 *** | 15.66 | 0.939 | 0.0002 |
| Blocks | 2.40 | 9 | 0.17 | 15.07 *** | |||||
| Nut length | (mm) | Clones | 1242.77 | 109 | 11.40 | 12.22 *** | 6.51 | 0.918 | 0.0188 |
| Blocks | 126.41 | 9 | 9.03 | 9.68 *** | |||||
| Nut width | (mm) | Clones | 279.18 | 109 | 2.56 | 5.62 *** | 5.84 | 0.822 | 0.0004 |
| Blocks | 31.92 | 9 | 2.28 | 5.01 *** | |||||
| Nut weight | (g) | Clones | 3.55 | 109 | 0.03 | 6.72 *** | 17.17 | 0.851 | 3.00 × 10−5 |
| Blocks | 0.44 | 9 | 0.03 | 6.50 *** | |||||
| Coat thickness | (mm) | Clones | 6.65 | 109 | 0.06 | 5.04 *** | 6.97 | 0.801 | 2.00 × 10−6 |
| Blocks | 0.51 | 9 | 0.04 | 3.03 *** | |||||
| 1000 seed weight | (g) | Clones | 3,082,247.35 | 109 | 28,277.50 | 11.47 *** | 13.37 | 0.913 | 0.9958 |
| Blocks | 6090.76 | 9 | 1015.13 | 0.41N |
| Locus | Na | Ne | I | Ho | He | uHe | F | Fst |
|---|---|---|---|---|---|---|---|---|
| PCP45071 | 7.00 ± 0.00 | 4.10 ± 0.88 | 1.59 ± 0.11 | 0.92 ± 0.02 | 0.75 ± 0.05 | 0.76 ± 0.05 | −0.24 ± 0.07 | 0.042 |
| Pt79951 | 3.00 ± 1.00 | 2.02 ± 0.09 | 0.75 ± 0.08 | 0.58 ± 0.02 | 0.51 ± 0.02 | 0.51 ± 0.02 | −0.16 ± 0.09 | 0.003 |
| 10F/RR | 8.00 ± 0.00 | 4.35 ± 0.42 | 1.66 ± 0.08 | 0.79 ± 0.11 | 0.77 ± 0.02 | 0.78 ± 0.03 | −0.03 ± 0.12 | 0.053 |
| P11 | 3.00 ± 1.00 | 1.98 ± 0.12 | 0.77 ± 0.11 | 0.55 ± 0.03 | 0.49 ± 0.03 | 0.50 ± 0.02 | −0.11 ± 0.07 | 0.002 |
| P25 | 2.00 ± 0.00 | 1.98 ± 0.09 | 0.69 ± 0.01 | 0.55 ± 0.03 | 0.49 ± 0.01 | 0.50 ± 0.01 | −0.11 ± 0.01 | 0.002 |
| P44 | 3.50 ± 0.50 | 2.26 ± 0.24 | 0.96 ± 0.02 | 0.66 ± 0.14 | 0.55 ± 0.05 | 0.56 ± 0.05 | −0.19 ± 0.15 | 0.034 |
| P49 | 4.00 ± 0.00 | 3.23 ± 0.28 | 1.26 ± 0.06 | 0.71 ± 0.04 | 0.69 ± 0.03 | 0.69 ± 0.03 | −0.04 ± 0.01 | 0.034 |
| P60 | 9.00 ± 1.00 | 4.37 ± 0.45 | 1.75 ± 0.06 | 0.92 ± 0.03 | 0.77 ± 0.02 | 0.78 ± 0.02 | −0.20 ± 0.08 | 0.048 |
| P60 | 9.00 ± 2.00 | 4.22 ± 0.15 | 1.69 ± 0.01 | 0.94 ± 0.04 | 0.76 ± 0.01 | 0.78 ± 0.02 | −0.24 ± 0.07 | 0.001 |
| P62 | 3.00 ± 0.00 | 2.20 ± 0.03 | 0.90 ± 0.01 | 0.60 ± 0.10 | 0.55 ± 0.01 | 0.55 ± 0.01 | −0.10 ± 0.20 | 0.020 |
| P63 | 5.00 ± 0.00 | 3.25 ± 0.55 | 1.32 ± 0.12 | 0.81 ± 0.09 | 0.68 ± 0.05 | 0.69 ± 0.06 | −0.19 ± 0.04 | 0.029 |
| P79 | 5.00 ± 1.00 | 2.97 ± 0.64 | 1.24 ± 0.25 | 0.81 ± 0.06 | 0.65 ± 0.07 | 0.66 ± 0.07 | −0.27 ± 0.05 | 0.003 |
| P74 | 2.00 ± 0.00 | 1.53 ± 0.06 | 0.53 ± 0.03 | 0.41 ± 0.01 | 0.35 ± 0.03 | 0.35 ± 0.02 | −0.20 ± 0.05 | 0.024 |
| P82 | 4.00 ± 1.00 | 3.19 ± 0.78 | 1.29 ± 0.24 | 0.69 ± 0.04 | 0.67 ± 0.08 | 0.68 ± 0.08 | −0.04 ± 0.07 | 0.077 |
| P90 | 4.00 ± 0.00 | 2.47 ± 0.20 | 1.11 ± 0.05 | 0.56 ± 0.11 | 0.59 ± 0.03 | 0.60 ± 0.03 | 0.06 ± 0.13 | 0.003 |
| P92 | 3.00 ± 0.00 | 2.55 ± 0.27 | 0.99 ± 0.07 | 0.43 ± 0.23 | 0.60 ± 0.04 | 0.61 ± 0.05 | 0.26 ± 0.43 | 0.044 |
| Mean | 4.67 ± 0.43 | 2.916 ± 0.18 | 1.15 ± 0.07 | 0.69 ± 0.04 | 0.62 ± 0.02 | 0.63 ± 0.02 | −0.11 ± 0.03 | 0.03 ± 0.01 |
| Source | df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Among collection sites | 1 | 19.680 | 19.680 | 0.224 | 4% |
| Within collection sites | 218 | 1094.361 | 5.020 | 5.020 | 96% |
| Total | 219 | 1114.041 | − | 5.244 | 100% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaviriri, D.K.; Zhang, Q.; Zhang, X.; Jiang, L.; Zhang, J.; Wang, J.; Khasa, D.P.; You, X.; Zhao, X. Phenotypic Variability and Genetic Diversity in a Pinus koraiensis Clonal Trial in Northeastern China. Genes 2020, 11, 673. https://doi.org/10.3390/genes11060673
Kaviriri DK, Zhang Q, Zhang X, Jiang L, Zhang J, Wang J, Khasa DP, You X, Zhao X. Phenotypic Variability and Genetic Diversity in a Pinus koraiensis Clonal Trial in Northeastern China. Genes. 2020; 11(6):673. https://doi.org/10.3390/genes11060673
Chicago/Turabian StyleKaviriri, David Kombi, Qinhui Zhang, Xinxin Zhang, Luping Jiang, Jinpeng Zhang, Jingyuan Wang, Damase P. Khasa, Xiangling You, and Xiyang Zhao. 2020. "Phenotypic Variability and Genetic Diversity in a Pinus koraiensis Clonal Trial in Northeastern China" Genes 11, no. 6: 673. https://doi.org/10.3390/genes11060673
APA StyleKaviriri, D. K., Zhang, Q., Zhang, X., Jiang, L., Zhang, J., Wang, J., Khasa, D. P., You, X., & Zhao, X. (2020). Phenotypic Variability and Genetic Diversity in a Pinus koraiensis Clonal Trial in Northeastern China. Genes, 11(6), 673. https://doi.org/10.3390/genes11060673





