A Reference Genome of Bursaphelenchus mucronatus Provides New Resources for Revealing Its Displacement by Pinewood Nematode
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Cultivation, and Growth Synchronization
2.2. Library Construction and Sequencing
2.3. Genome Size Estimation
2.4. Genome Assembly
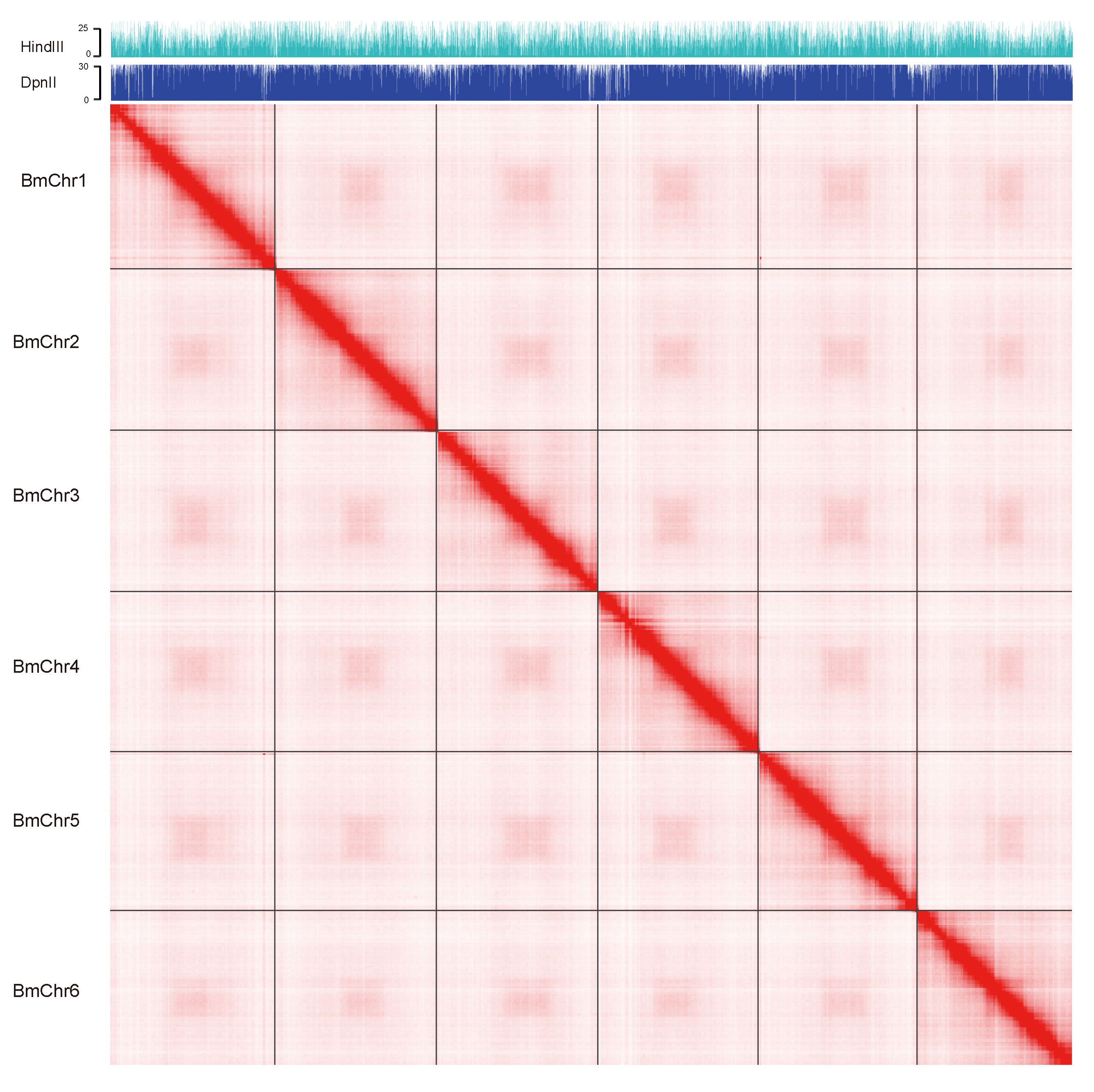
2.5. Pseudo-Chromosomal Level Scaffolding with Hi-C Data
2.6. Genome Annotation
2.6.1. Protein-Coding Gene Annotation
2.6.2. Repeat and Non-Coding RNA Identification
2.7. Evolution and Comparative Analysis
2.7.1. Reconstruction of Gene Families
2.7.2. Reconstruction of Phylogenetic Tree
2.7.3. Collinear and Similarity Analysis of B. mucronatus and B. xylophilus
2.8. Identification and Comparison of GPCR, Cellulase, and Pectin Genes of B. xylophilus and B. mucronatus
3. Results and Discussion
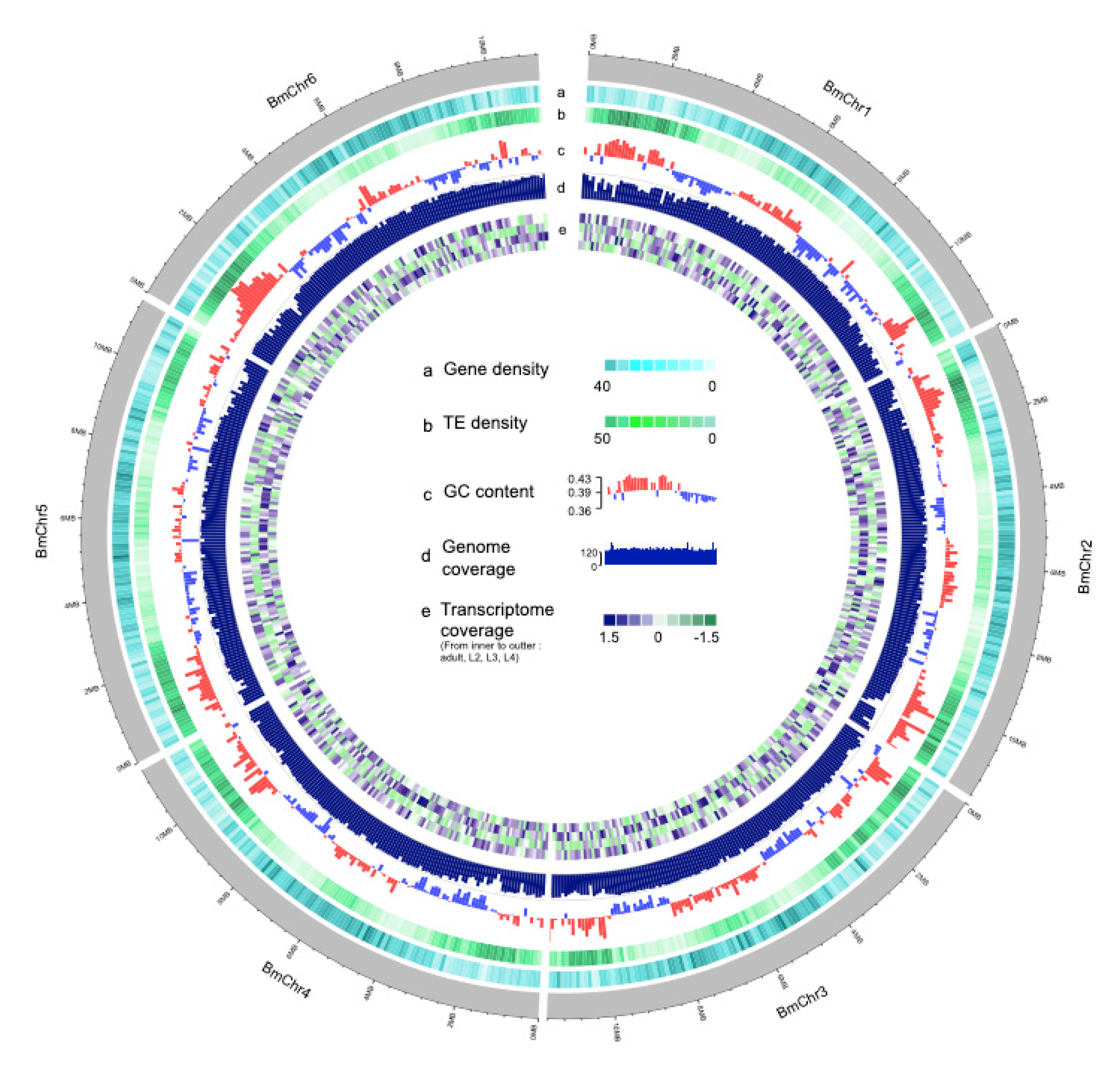
3.1. Genome Assembly and Quality Assessment
3.2. Repeat and Non-Coding RNA Identification
3.3. Gene Annotation and Functional Analysis
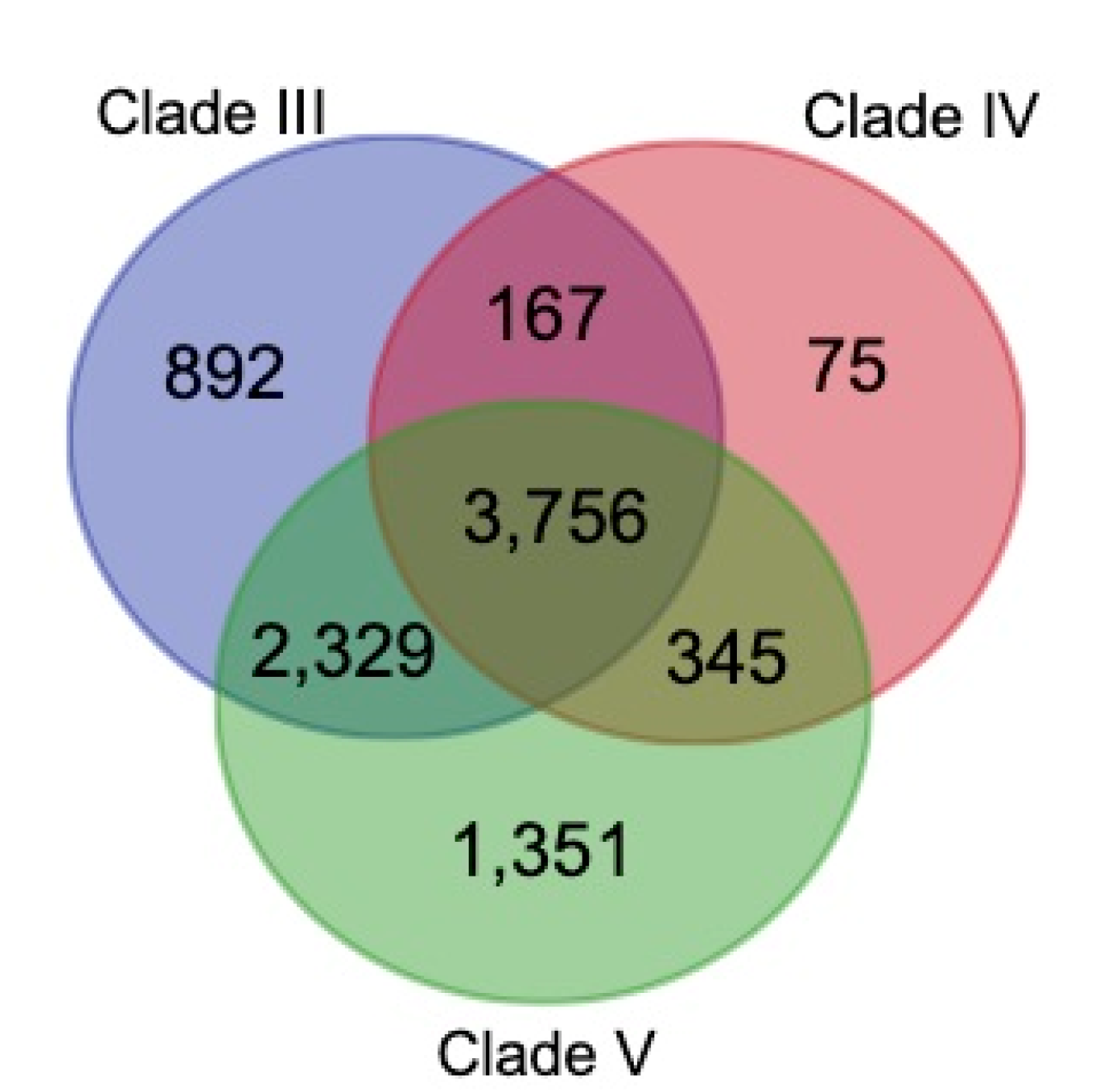
3.4. Construction of Gene families
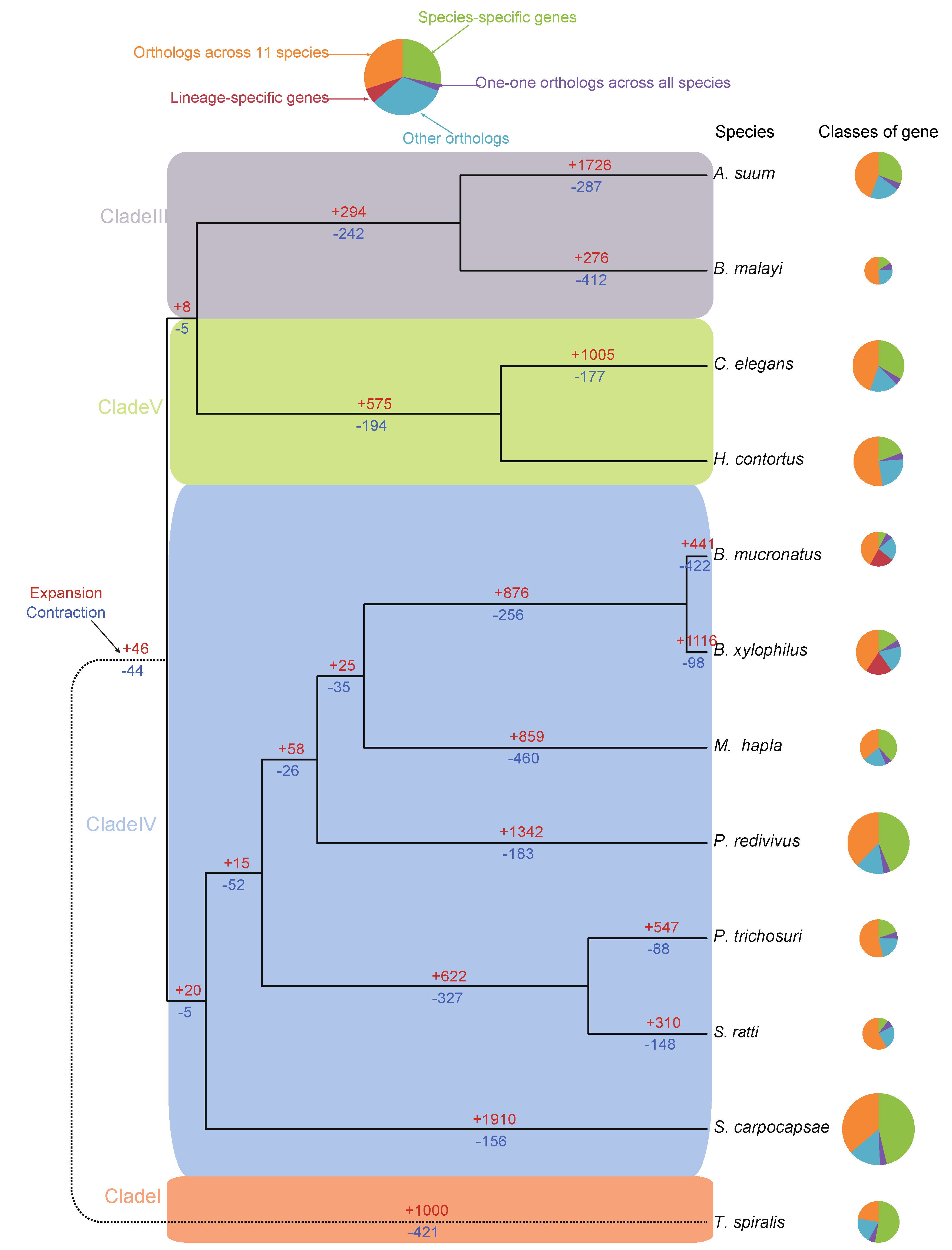
3.5. Genome Phylogeny of B. mucronatus
3.6. Dynamics of the Gene Families
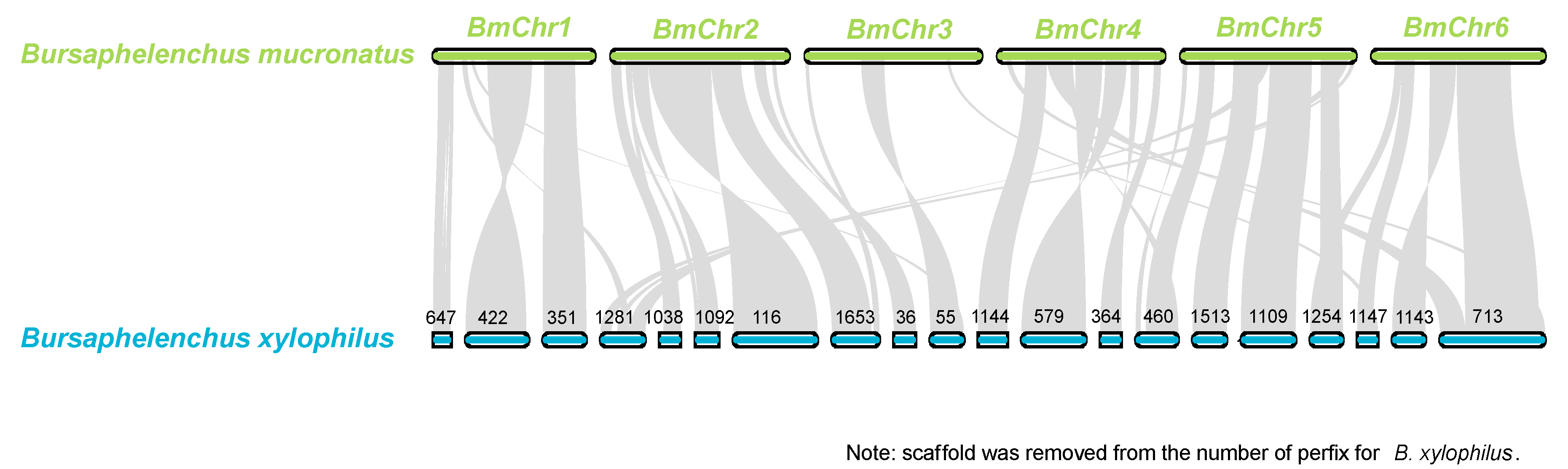
3.7. Genome Comparison of B. mucronatus and B. xylophilus
3.8. GPCR Contractions May Be the Reason That B. mucronatus Was Displaced by B. xylophilus
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhao, L.; Mota, M.; Vieira, P.; Butcher, R.A.; Sun, J. Interspecific communication between pinewood nematode, its insect vector, and associated microbes. Trends Parasitol. 2014, 30, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, X.; Wei, Y.; Zhou, J.; Zhang, W.; Qin, P.; Chinta, S.; Kong, X.; Liu, Y.; Yu, H.; et al. Ascarosides coordinate the dispersal of a plant-parasitic nematode with the metamorphosis of its vector beetle. Nat. Commun. 2016, 7, 12341. [Google Scholar] [CrossRef] [PubMed]
- Guiran, G.D.; Bruguier, N. Hybridization and Phylogeny of the Pine Wood Nematode (Bursaphelenchus Spp.). Nematologica 1989, 35, 321–330. [Google Scholar] [CrossRef]
- Yan, X.; Cheng, X.Y.; Wang, Y.S.; Luo, J.; Mao, Z.C.; Ferris, V.R.; Xie, B.Y. Comparative transcriptomics of two pathogenic pinewood nematodes yields insights into parasitic adaptation to life on pine hosts. Gene 2012, 505, 81–90. [Google Scholar] [CrossRef]
- Pereira, F.; Moreira, C.; Fonseca, L.; van Asch, B.; Mota, M.; Abrantes, I.; Amorim, A. New insights into the phylogeny and worldwide dispersion of two closely related nematode species, Bursaphelenchus xylophilus and Bursaphelenchus mucronatus. PLoS ONE 2013, 8, e56288. [Google Scholar] [CrossRef]
- Mamiya, Y.; Enda, N. Bursaphelenchus Mucronatus N. Sp. (Nematoda: Aphelenchoididae) from pine wood and its biology and pathogenicity to pine trees. Nematologica 1979, 25, 353–361. [Google Scholar] [CrossRef]
- Niu, H.T.; Zhao, L.L.; Sun, J.H. Phenotypic plasticity of reproductive traits in response to food availability in invasive and native species of nematode. Biol. Invasions 2013, 15, 1407–1415. [Google Scholar] [CrossRef]
- Nickle, W.R.; Golden, A.M.; Mamiya, Y.; Wergin, W.P. On the Taxonomy and Morphology of the Pine Wood Nematode, Bursaphelenchus xylophilus (Steiner &Buhrer 1934) Nickle 1970. J. Nematol. 1981, 13, 385–392. [Google Scholar]
- Mamiya, Y.; Kiyohara, T.; Mamiya, Y.; Kiyohara, T. Description of Bursaphelenchus Lignicolus N. Sp. (Nematoda: Aphelenchoididae) From Pine Wood and Histopathology of Nematode-Infested Trees. Nematologica 1972, 18, 120–124. [Google Scholar] [CrossRef]
- Kishi, Y. Pine Wood Nematode and the Japanese Pine Sawyer; Thomas Company: Tokyo, Japan, 1995. [Google Scholar]
- Cheng, X.Y.; Xie, P.Z.; Cheng, F.X.; Xu, R.M.; Xie, B.Y. Competitive displacement of the native species Bursaphelenchus mucronatus by an alien species Bursaphelenchus xylophilus (Nematoda: Aphelenchida: Aphelenchoididae): A case of successful invasion. Biol. Invasions. 2009, 11, 205–213. [Google Scholar] [CrossRef]
- Jikumaru, S.; Togashi, K. Inhibitory Effect of Bursaphelenchus mucronatus (Nematoda: Aphelenchoididae) on B. xylophilus Boarding Adult Monochamus alternatus (Coleoptera: Cerambycidae). J. Nematol. 2004, 36, 95–99. [Google Scholar] [PubMed]
- Vincent, B.; Altemayer, V.; Rouxmorabito, G.; Naves, P.; Sousa, E.; Lieutier, F. Competitive interaction between Bursaphelenchus xylophilus and the closely related species Bursaphelenchus mucronatus. Nematology 2008, 10, 219–230. [Google Scholar]
- Wan, F.; Yin, C.; Tang, R.; Chen, M.; Wu, Q.; Huang, C.; Qian, W.; Rota-Stabelli, O.; Yang, N.; Wang, S.; et al. A chromosome-level genome assembly of Cydia pomonella provides insights into chemical ecology and insecticide resistance. Nat. Commun. 2019, 10, 4237. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Zhang, S.; Li, X.; Cao, Y.; Liu, X.; Wang, Q.; Liu, Q.; Liu, H.; Hu, X.; Zhou, X.J.; et al. Fall webworm genomes yield insights into rapid adaptation of invasive species. Nat. Ecol. Evol. 2019, 3, 105–115. [Google Scholar] [CrossRef]
- Kingan, S.B.; Urban, J.; Lambert, C.C.; Baybayan, P.; Childers, A.K.; Coates, B.; Scheffler, B.; Hackett, K.; Korlach, J.; Geib, S.M. A high-quality genome assembly from a single, field-collected spotted lanternfly (Lycorma delicatula) using the PacBio Sequel II system. Gigascience 2019, 8, 10. [Google Scholar] [CrossRef]
- Hunt, V.L.; Tsai, I.J.; Coghlan, A.; Reid, A.J.; Holroyd, N.; Foth, B.J.; Tracey, A.; Cotton, J.A.; Stanley, E.J.; Beasley, H.; et al. The genomic basis of parasitism in the Strongyloides clade of nematodes. Nat. Genet. 2016, 48, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, T.; Cotton, J.A.; Dalzell, J.J.; Hasegawa, K.; Kanzaki, N.; McVeigh, P.; Takanashi, T.; Tsai, I.J.; Assefa, S.A.; Cock, P.J.; et al. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011, 7, e1002219. [Google Scholar] [CrossRef] [PubMed]
- Marcais, G.; Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Vurture, G.W.; Sedlazeck, F.J.; Nattestad, M.; Underwood, C.J.; Fang, H.; Gurtowski, J.; Schatz, M.C. GenomeScope: Fast reference-free genome profiling from short reads. Bioinformatics 2017, 33, 2202–2204. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Durand, N.C.; Robinson, J.T.; Shamim, M.S.; Machol, I.; Mesirov, J.P.; Lander, E.S.; Aiden, E.L. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 2016, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Dudchenko, O.; Batra, S.S.; Omer, A.D.; Nyquist, S.K.; Hoeger, M.; Durand, N.C.; Shamim, M.S.; Machol, I.; Lander, E.S.; Aiden, A.P.; et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 2017, 356, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Niknafs, Y.S.; Pandian, B.; Iyer, H.K.; Chinnaiyan, A.M.; Iyer, M.K. TACO produces robust multisample transcriptome assemblies from RNA-seq. Nat. Methods 2017, 14, 68–70. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Haas, B.J.; Delcher, A.L.; Mount, S.M.; Wortman, J.R.; Smith, R.K., Jr.; Hannick, L.I.; Maiti, R.; Ronning, C.M.; Rusch, D.B.; Town, C.D.; et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003, 31, 5654–5666. [Google Scholar] [CrossRef]
- Stanke, M.; Diekhans, M.; Baertsch, R.; Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 2008, 24, 637–644. [Google Scholar] [CrossRef]
- Borodovsky, M.; Mills, R.; Besemer, J.; Lomsadze, A. Prokaryotic gene prediction using GeneMark and GeneMark.hmm. Curr. Protoc. Bioinform. 2003, 1, 4.5.1–4.5.16. [Google Scholar] [CrossRef]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L.; et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Coordinators, N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef]
- The UniProt, C. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Finn, R.D.; Attwood, T.K.; Babbitt, P.C.; Bateman, A.; Bork, P.; Bridge, A.J.; Chang, H.Y.; Dosztanyi, Z.; El-Gebali, S.; Fraser, M.; et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017, 45, D190–D199. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, R.M.; Seppey, M.; Simao, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef]
- Kalvari, I.; Argasinska, J.; Quinones-Olvera, N.; Nawrocki, E.P.; Rivas, E.; Eddy, S.R.; Bateman, A.; Finn, R.D.; Petrov, A.I. Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018, 46, D335–D342. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef]
- Han, M.V.; Thomas, G.W.; Lugo-Martinez, J.; Hahn, M.W. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 2013, 30, 1987–1997. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Kielbasa, S.M.; Wan, R.; Sato, K.; Horton, P.; Frith, M.C. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011, 21, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.; Tang, A.; O’Loughlin, C.T.; Balistreri, A.; Chang, E.; Coto Villa, D.; Li, J.; Varshney, A.; Jimenez, V.; Pyle, J.; et al. C. elegans avoids toxin-producing Streptomyces using a seven transmembrane domain chemosensory receptor. Elife 2017, 6, e23770. [Google Scholar] [CrossRef]
- Tan, F.J.; Fire, A.Z.; Hill, R.B. Regulation of apoptosis by C. elegans CED-9 in the absence of the C-terminal transmembrane domain. Cell Death Differ. 2007, 14, 1925–1935. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rodelsperger, C.; Prabh, N.; Sommer, R.J. New Gene Origin and Deep Taxon Phylogenomics: Opportunities and Challenges. Trends Genet. 2019, 35, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Prabh, N.; Roeseler, W.; Witte, H.; Eberhardt, G.; Sommer, R.J.; Rodelsperger, C. Deep taxon sampling reveals the evolutionary dynamics of novel gene families in Pristionchus nematodes. Genome Res. 2018, 28, 1664–1674. [Google Scholar] [CrossRef]
- Vidal, B.; Aghayeva, U.; Sun, H.S.; Wang, C.; Glenwinkel, L.; Bayer, E.A.; Hobert, O. An atlas of Caenorhabditis elegans chemoreceptor expression. PLoS Biol. 2018, 16, e2004218. [Google Scholar] [CrossRef]
- Son, J.A.; Jung, C.S.; Han, H.R. Migration and Attacking Ability of Bursaphelenchus mucronatus in Pinus thunbergii Stem Cuttings. Plant Pathol. J. 2016, 32, 340–346. [Google Scholar] [CrossRef]
- Frooninckx, L.; Rompay, L.V.; Temmerman, L.; Sinay, E.V.; Beets, I.; Janssen, T.; Husson, S.J.; Schoofs, L. Neuropeptide GPCRs in C. elegans. Front. Endocrinol. (Lausanne) 2012, 3, 167. [Google Scholar] [CrossRef]
- Meng, J.; Wickham, J.D.; Ren, W.; Zhao, L.; Sun, J. Species displacement facilitated by ascarosides between two sympatric sibling species: A native and invasive nematode. J. Pest Sci. 2020, 93, 1059–1071. [Google Scholar] [CrossRef]
- Smythe, A.B.; Holovachov, O.; Kocot, K.M. Improved phylogenomic sampling of free-living nematodes enhances resolution of higher-level nematode phylogeny. BMC Evol. Biol. 2019, 19, 121. [Google Scholar] [CrossRef] [PubMed]
- van den Elsen, S.; Holovachov, O.; Karssen, G.; van Megen, H.; Helder, J.; Bongers, T.; Bakker, J.; Holterman, M.; Mooyman, P. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology 2009, 11, 927–950. [Google Scholar] [CrossRef]
- Desjardins, C.A.; Cerqueira, G.C.; Goldberg, J.M.; Dunning Hotopp, J.C.; Haas, B.J.; Zucker, J.; Ribeiro, J.M.; Saif, S.; Levin, J.Z.; Fan, L.; et al. Genomics of Loa loa, a Wolbachia-free filarial parasite of humans. Nat. Genet. 2013, 45, 495–500. [Google Scholar] [CrossRef]
- International Helminth Genomes Consortium. Comparative genomics of the major parasitic worms. Nat. Genet. 2019, 51, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, T.A.; Riga, E.; Webster, J.M. Temperature-mediated Behavioral Relationships in Bursaphelenchus xylophilus, B. mucronatus, and Their Hybrids. J. Nematol. 1992, 24, 40–44. [Google Scholar] [PubMed]
- Ji, N.; Middelkoop, T.C.; Mentink, R.A.; Betist, M.C.; Tonegawa, S.; Mooijman, D.; Korswagen, H.C.; van Oudenaarden, A. Feedback control of gene expression variability in the Caenorhabditis elegans Wnt pathway. Cell 2013, 155, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Raw, J.L.; Miranda, N.A.; Perissinotto, R. Chemical cues released by an alien invasive aquatic gastropod drive its invasion success. PLoS ONE 2013, 8, e64071. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, S.; Gao, S.; Wang, S.; Meng, J.; Wickham, J.; Luo, S.; Tan, X.; Yu, H.; Xiang, Y.; Hu, S.; et al. A Reference Genome of Bursaphelenchus mucronatus Provides New Resources for Revealing Its Displacement by Pinewood Nematode. Genes 2020, 11, 570. https://doi.org/10.3390/genes11050570
Wu S, Gao S, Wang S, Meng J, Wickham J, Luo S, Tan X, Yu H, Xiang Y, Hu S, et al. A Reference Genome of Bursaphelenchus mucronatus Provides New Resources for Revealing Its Displacement by Pinewood Nematode. Genes. 2020; 11(5):570. https://doi.org/10.3390/genes11050570
Chicago/Turabian StyleWu, Shuangyang, Shenghan Gao, Sen Wang, Jie Meng, Jacob Wickham, Sainan Luo, Xinyu Tan, Haiying Yu, Yujia Xiang, Songnian Hu, and et al. 2020. "A Reference Genome of Bursaphelenchus mucronatus Provides New Resources for Revealing Its Displacement by Pinewood Nematode" Genes 11, no. 5: 570. https://doi.org/10.3390/genes11050570
APA StyleWu, S., Gao, S., Wang, S., Meng, J., Wickham, J., Luo, S., Tan, X., Yu, H., Xiang, Y., Hu, S., Zhao, L., & Sun, J. (2020). A Reference Genome of Bursaphelenchus mucronatus Provides New Resources for Revealing Its Displacement by Pinewood Nematode. Genes, 11(5), 570. https://doi.org/10.3390/genes11050570





