Genomic Identification, Evolution, and Expression Analysis of Collagen Genes Family in Water Buffalo during Lactation
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of the Buffalo Collagen Genes
2.2. Sequence Analysis
2.3. Comparative Transcriptomic Analyses for the Milk Samples of Buffalo and Cattle
2.4. Real-time Quantitative PCR
3. Results
3.1. Genomic Identification of Buffalo Collagen Genes
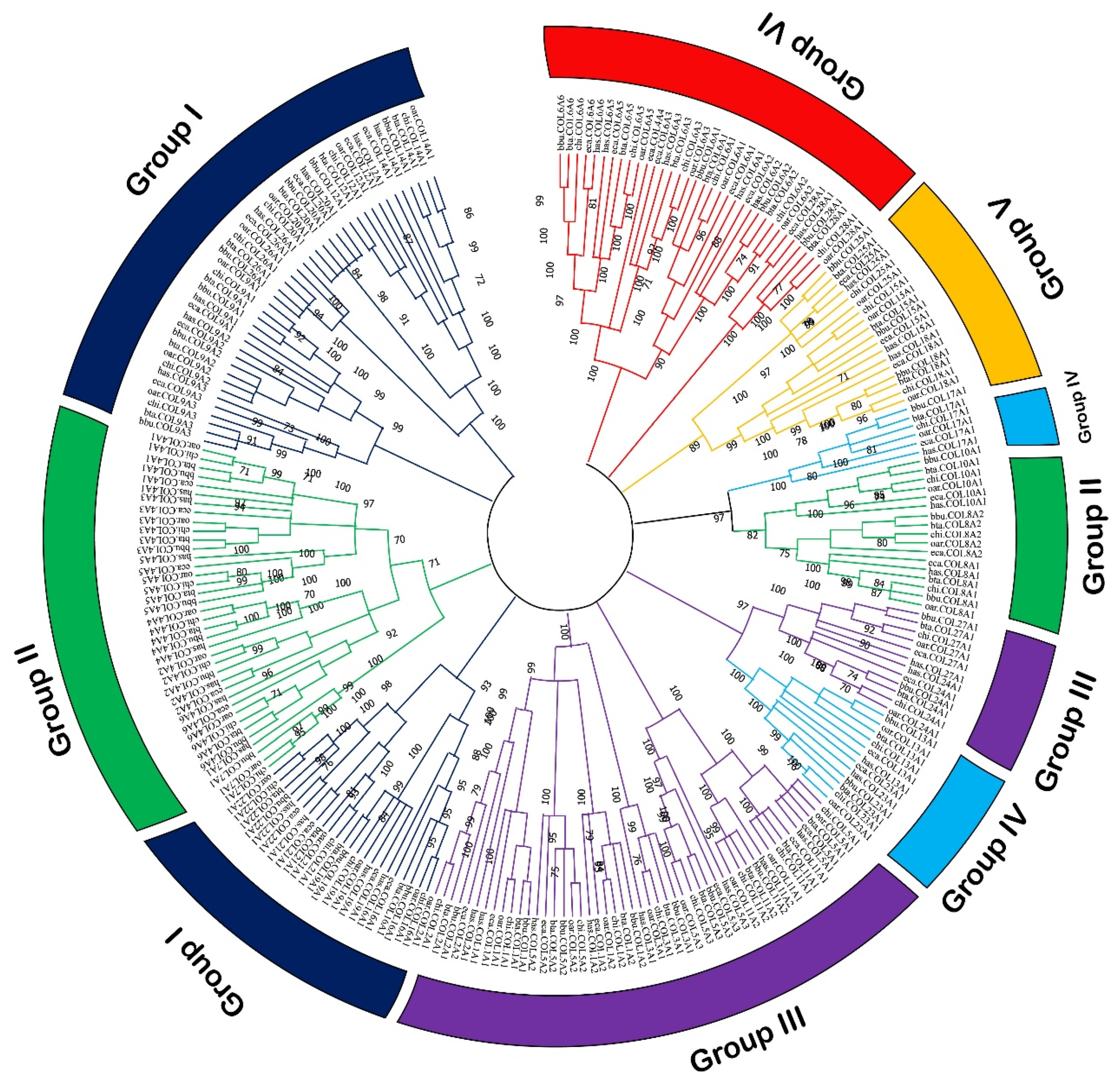
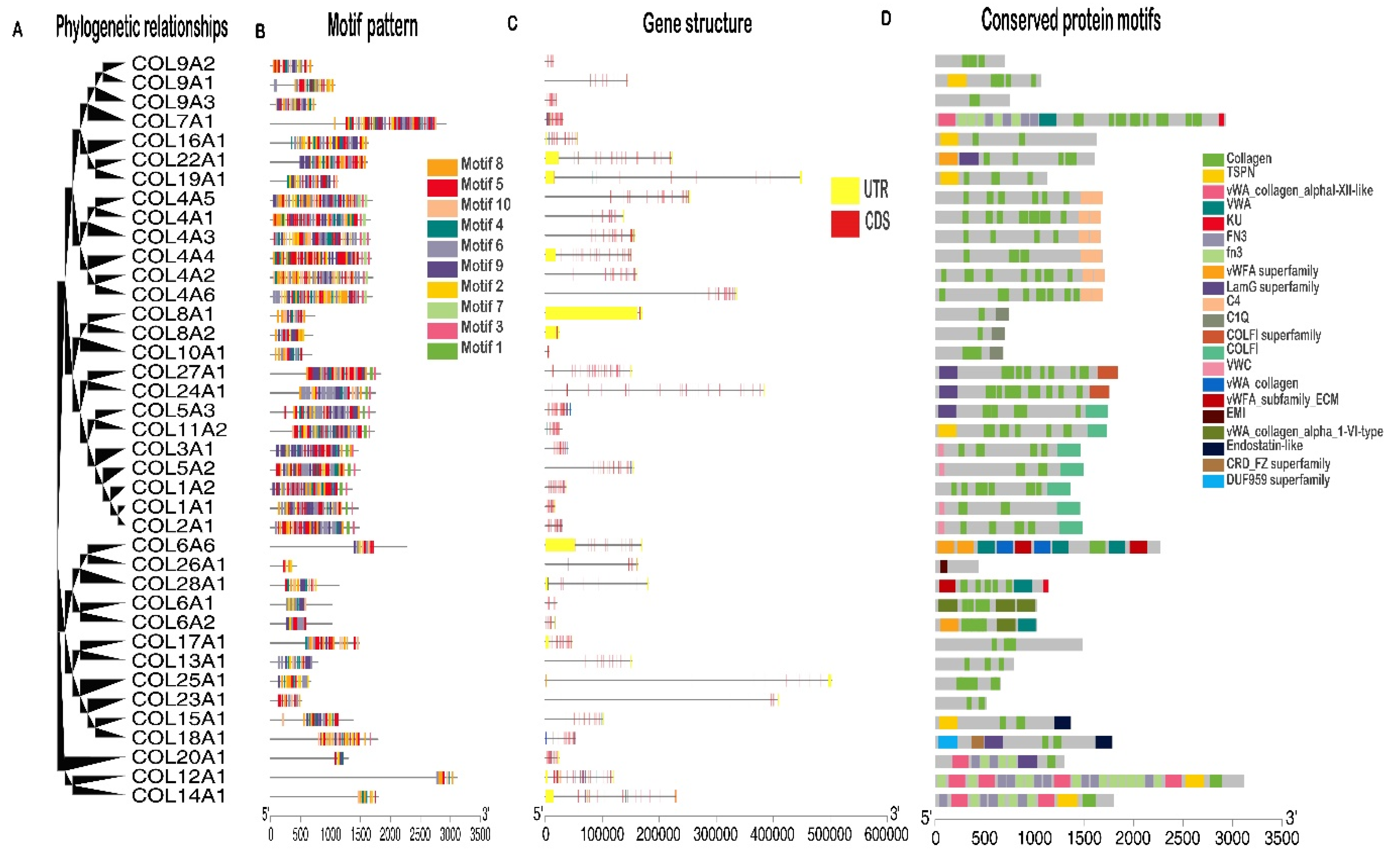
3.2. Sequence Analysis of Buffalo Collagen Genes
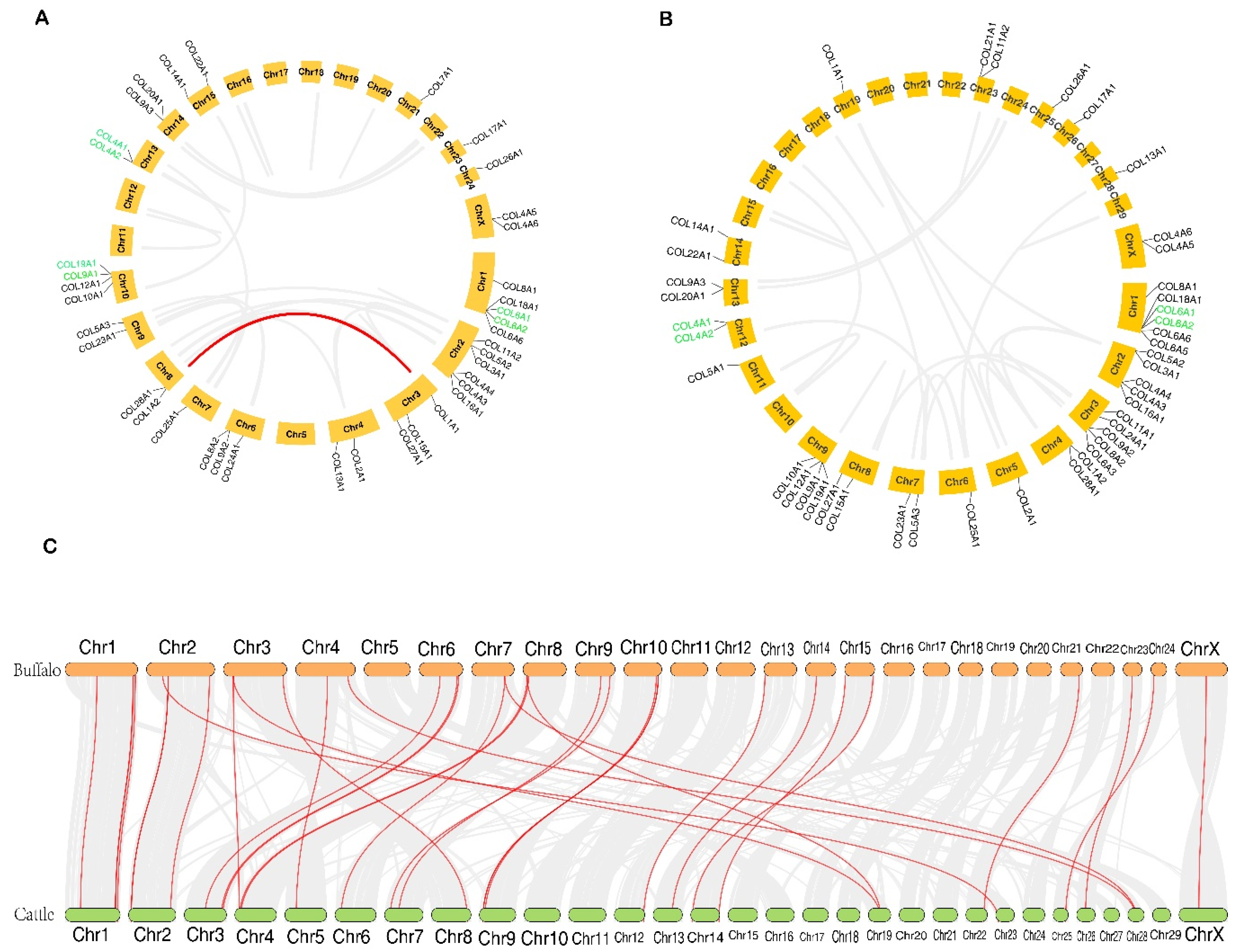
3.3. Chromosomal Distribution and Collinearity Analysis of Collagen Genes
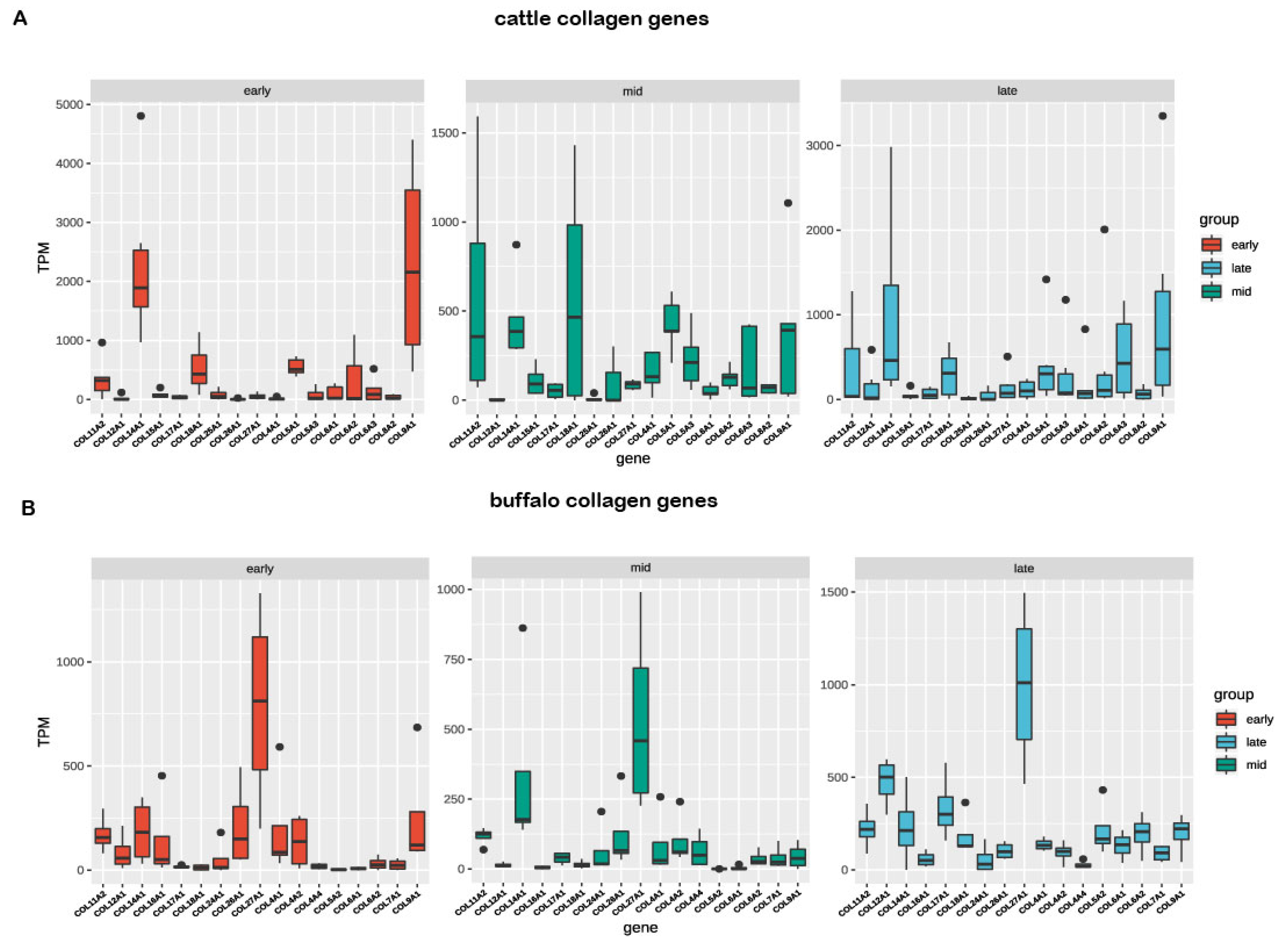
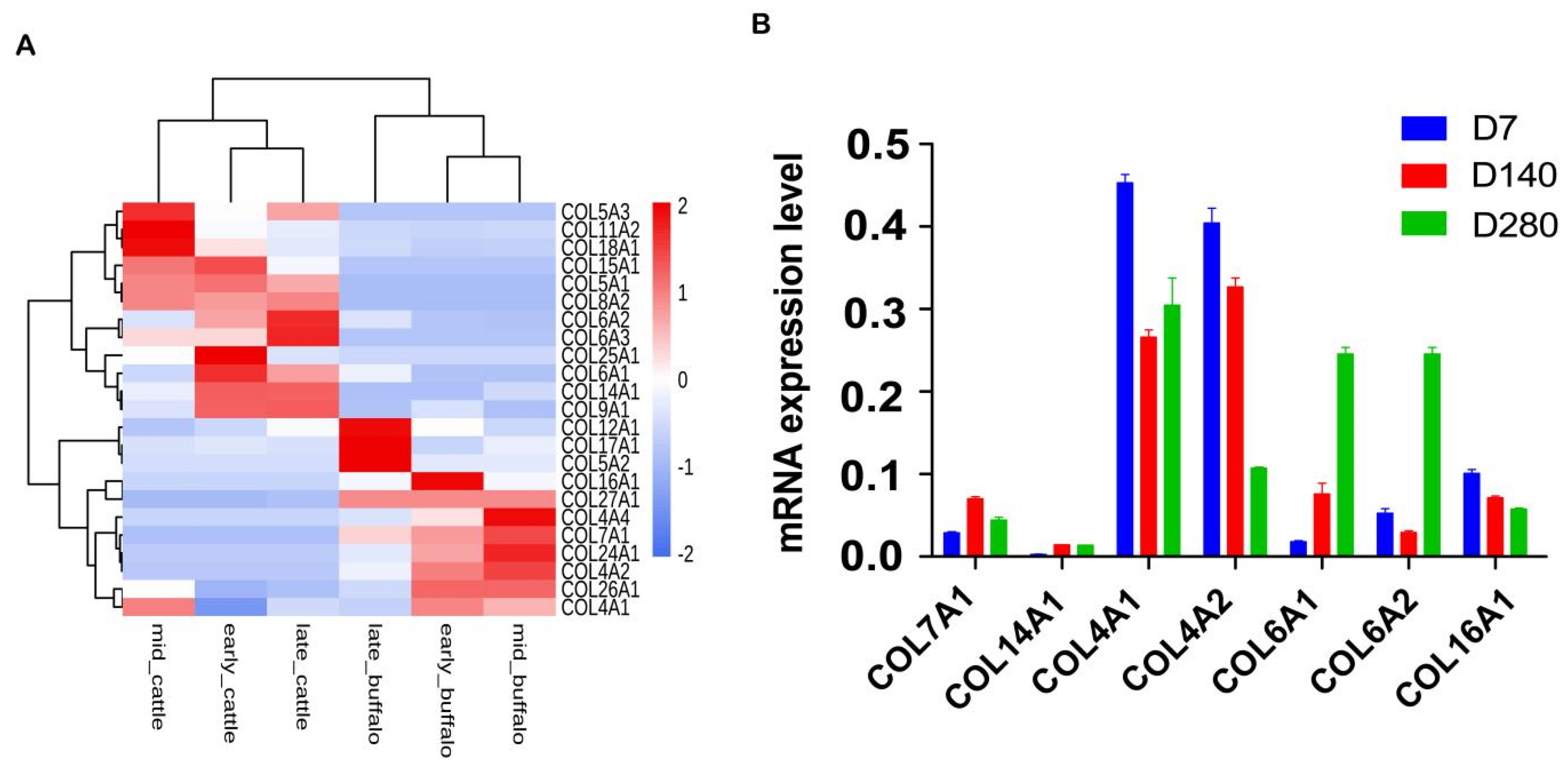
3.4. Comparative Transcriptomic Analyses of Orthologous Collagens between Buffalo and Cattle
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gelse, K. Collagens—structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 2003, 55, 1531–1546. [Google Scholar] [CrossRef] [PubMed]
- Novaro, V.; Roskelley, C.D.; Bissell, M.J. Collagen-IV and laminin-1 regulate estrogen receptor alpha expression and function in mouse mammary epithelial cells. J. Cell Sci. 2003, 116, 2975–2986. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yano, T.; Aso, H.; Sakamoto, K.; Kobayashi, Y.; Hagino, A.; Katoh, K.; Obara, Y. Laminin and collagen IV enhanced casein synthesis in bovine mammary epithelial cells. J. Anim. Feed. Sci. 2004, 13, 579–582. [Google Scholar] [CrossRef]
- Chen, Q.; He, G.; Zhang, W.; Xu, T.; Qi, H.; Li, J.; Zhang, Y.; Gao, M.-Q. Stromal fibroblasts derived from mammary gland of bovine with mastitis display inflammation-specific changes. Sci. Rep. 2016, 6, 27462. [Google Scholar] [CrossRef]
- Crisà, A.; Ferre’, F.; Chillemi, G.; Moioli, B. RNA-Sequencing for profiling goat milk transcriptome in colostrum and mature milk. BMC Veter- Res. 2016, 12, 264. [Google Scholar] [CrossRef]
- Anderson, B.M.; MacLennan, M.B.; Hillyer, L.M.; Ma, D.W. Lifelong exposure to n-3 PUFA affects pubertal mammary gland development. Appl. Physiol. Nutr. Metab. 2014, 39, 699–706. [Google Scholar] [CrossRef]
- Inman, J.L.; Robertson, C.; Mott, J.D.; Bissell, M.J. Mammary gland development: Cell fate specification, stem cells and the microenvironment. Development 2015, 142, 1028–1042. [Google Scholar] [CrossRef]
- Mintoo, A.A.; Zhang, H.; Chen, C.; Moniruzzaman, M.; Deng, T.; Anam, M.; Huque, Q.M.E.; Guang, X.; Wang, P.; Zhong, Z.; et al. Draft genome of the river water buffalo. Ecol. Evol. 2019, 9, 3378–3388. [Google Scholar] [CrossRef]
- Hazra, T.; Sharma, V.; Sharma, R.; De, S.; Arora, S.; Lal, D. Detection of cow milk paneer in mixed/buffalo milk paneer through conventional species specific polymerase chain reaction. Indian J. Anim. Res. 2016, 522–528. [Google Scholar] [CrossRef]
- Iamartino, D.; Nicolazzi, E.L.; Van Tassell, C.P.; Reecy, J.M.; Fritz-Waters, E.R.; Koltes, J.E.; Biffani, S.; Sonstegard, T.S.; Schroeder, S.G.; Ajmone-Marsan, P.; et al. Design and validation of a 90K SNP genotyping assay for the water buffalo (Bubalus bubalis). PLoS ONE 2017, 12, e0185220. [Google Scholar] [CrossRef]
- De Camargo, G.M.F.; Aspilcueta-Borquis, R.R.; Fortes, M.R.; Porto-Neto, L.R.; Cardoso, D.; Santos, D.J.D.A.; Lehnert, S.; Reverter, A.; Moore, S.; Tonhati, H. Prospecting major genes in dairy buffaloes. BMC Genom. 2015, 16, 872. [Google Scholar] [CrossRef] [PubMed]
- Strazzullo, M.; Gasparrini, B.; Neglia, G.; Balestrieri, M.L.; Francioso, R.; Rossetti, C.; Nassa, G.; De Filippo, M.R.; Weisz, A.; Di Francesco, S.; et al. Global transcriptome profiles of Italian Mediterranean buffalo embryos with normal and retarded growth. PLoS ONE 2014, 9, e90027. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Fu, Q.; Pu, L.; Zhang, P.; Huang, D.; Hou, Z.; Xu, Z.; Chen, N.; Huang, F.; Deng, T.; et al. Integrated analysis of quantitative proteome and transcriptional profiles reveals the dynamic function of maternally expressed proteins after parthenogenetic activation of buffalo oocyte. Mol. Cell. Proteom. 2018, 17, 1875–1891. [Google Scholar] [CrossRef] [PubMed]
- Low, W.Y.; Tearle, R.; Bickhart, D.; Rosen, B.D.; Kingan, S.B.; Swale, T.; Thibaud-Nissen, F.; Murphy, T.D.; Young, R.; Lefevre, L.; et al. Chromosome-level assembly of the water buffalo genome surpasses human and goat genomes in sequence contiguity. Nat. Commun. 2019, 10, 260. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/genome (accessed on 6 April 2020).
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. BioRxiv 2018. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; SÃ nchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sã, N.-G.A. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- FastQC. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 6 April 2020).
- Kim, D.; Langmead, B.; Salzberg, S. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Boil. 2014, 15, 31. [Google Scholar] [CrossRef]
- Yu, L.; Wang, G.-D.; Ruan, J.; Chen, Y.-B.; Yang, C.-P.; Cao, X.; Wu, H.; Liu, Y.-H.; Du, Z.-L.; Wang, X.-P.; et al. Genomic analysis of snub-nosed monkeys (Rhinopithecus) identifies genes and processes related to high-altitude adaptation. Nat. Genet. 2016, 48, 947–952. [Google Scholar] [CrossRef]
- Schmitz, S.; Pfaffl, M.; Meyer, H.H.D.; Bruckmaier, R.M. Short-term changes of mRNA expression of various inflammatory factors and milk proteins in mammary tissue during LPS-induced mastitis. Domest. Anim. Endocrinol. 2004, 26, 111–126. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D.J.M. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Gordon, M.K.; Hahn, R.A. Collagens. Cell Tissue Res. 2010, 339, 247. [Google Scholar] [CrossRef]
- Hu, G.; Li, L.; Xu, W. Extracellular matrix in mammary gland development and breast cancer progression. Front. Lab. Med. 2017, 1, 36–39. [Google Scholar] [CrossRef]
- Ricard-Blum, S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Exposito, J.-Y.; Valcourt, U.; Cluzel, C.; Lethias, C. The fibrillar collagen family. Int. J. Mol. Sci. 2010, 11, 407–426. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Carrique, L.; Goff, S.V.-L.; Mariano, N.; Georges, R.-N.; Delolme, F.; Koivunen, P.; Myllyharju, J.; Moali, C.; Aghajari, N.; et al. Structural basis of homo- and heterotrimerization of collagen I. Nat. Commun. 2017, 8, 14671. [Google Scholar] [CrossRef]
- Hughes, A.L. The evolution of functionally novel proteins after gene duplication. Proc. Biol. Sci. 1994, 256, 119–124. [Google Scholar]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Boil. 2009, 60, 433–453. [Google Scholar] [CrossRef]
- Liu, G.; Ventura, M.; Cellamare, A.; Chen, L.; Cheng, Z.; Zhu, B.; Li, C.-J.; Song, J.; Eichler, E.E. Analysis of recent segmental duplications in the bovine genome. BMC Genom. 2009, 10, 571. [Google Scholar] [CrossRef]
- Feng, X.; Jiang, J.; Padhi, A.; Ning, C.; Fu, J.; Wang, A.; Mrode, R.; Liu, J.-F. Characterization of genome-wide segmental duplications reveals a common genomic feature of association with immunity among domestic animals. BMC Genom. 2017, 18, 293. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, D.; Wang, R.; Kong, N.; Zhang, C.; Yang, C.; Wu, W.; Ma, H.; Chen, Q. Genome-wide analysis of the potato Hsp20 gene family: Identification, genomic organization and expression profiles in response to heat stress. BMC Genom. 2018, 19, 61. [Google Scholar] [CrossRef]
- Ghajar, C.M.; Bissell, M.J. Extracellular matrix control of mammary gland morphogenesis and tumorigenesis: Insights from imaging. Histochem. Cell Boil. 2008, 130, 1105–1118. [Google Scholar] [CrossRef]
- Zhu, J.; Xiong, G.; Trinkle, C.; Xu, R. Integrated extracellular matrix signaling in mammary gland development and breast cancer progression. Histol. Histopathol. 2014, 29, 1083–1092. [Google Scholar] [PubMed]
- Liu, J.; Wang, Z.; Li, J.; Li, H.; Yang, L. Genome-wide identification of diacylglycerol acyltransferases (DGAT) family genes influencing milk production in buffalo. BMC Genet. 2020, 21, 26. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Zou, Y.-X.; Liu, J.; Liu, H.; White, R.R. Transcriptomic profiles of the bovine mammary gland during lactation and the dry period. Funct. Integr. Genom. 2017, 18, 125–140. [Google Scholar] [CrossRef] [PubMed]
| Motif | Protein Sequence | Length | Pfam Domain |
|---|---|---|---|
| MEME-1 | GLPGLKGEKGEAGLPGFKGEKGVKGEKGE | 29 | - |
| MEME-2 | KGEDGLPGLPGEKGEKGEKGDPGPPGPPG | 29 | - |
| MEME-3 | GEKGERGLPGLPGKKGAKGEPGIPGAKGEKGPPGPPGPPGE | 41 | Collagen |
| MEME-4 | GPPGPPGPPGPPGPPGLPGPPGPPGLPGPP | 30 | - |
| MEME-5 | PGPPGPKGPRGEKGDPGSTGPPGEPGLPGLQGPPGEKGDKG | 41 | Collagen |
| MEME-6 | GPKGERGPKGQKGEKGQPGEP | 21 | - |
| MEME-7 | TGPPGPIGLPGLPGPKGEKGE | 21 | - |
| MEME-8 | GEPGJPGEKGEPGLPGPPGLPGEKGPKGK | 29 | - |
| MEME-9 | GEQGERGPKGEKGEA | 15 | - |
| MEME-10 | RGEPGLPGPPGPPGP | 15 | - |
| Gene Pairs | CHR | Ka | Ks | Ka/Ks Ratio |
|---|---|---|---|---|
| COL1A1/COL1A2 | 3/8 | 0.2872 | 1.2576 | 0.2283 |
| COL4A2/ COL4A1 | 13 | 0.5336 | 1.9459 | 0.2742 |
| COL6A1/ COL6A2 | 1 | 0.7068 | 1.0659 | 0.6631 |
| COL9A1/ COL19A1 | 10 | 0.6218 | 2.6177 | 0.2375 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, X.; Duan, A.; Liang, S.; Ma, X.; Deng, T. Genomic Identification, Evolution, and Expression Analysis of Collagen Genes Family in Water Buffalo during Lactation. Genes 2020, 11, 515. https://doi.org/10.3390/genes11050515
Lu X, Duan A, Liang S, Ma X, Deng T. Genomic Identification, Evolution, and Expression Analysis of Collagen Genes Family in Water Buffalo during Lactation. Genes. 2020; 11(5):515. https://doi.org/10.3390/genes11050515
Chicago/Turabian StyleLu, Xingrong, Anqin Duan, Shasha Liang, Xiaoya Ma, and Tingxian Deng. 2020. "Genomic Identification, Evolution, and Expression Analysis of Collagen Genes Family in Water Buffalo during Lactation" Genes 11, no. 5: 515. https://doi.org/10.3390/genes11050515
APA StyleLu, X., Duan, A., Liang, S., Ma, X., & Deng, T. (2020). Genomic Identification, Evolution, and Expression Analysis of Collagen Genes Family in Water Buffalo during Lactation. Genes, 11(5), 515. https://doi.org/10.3390/genes11050515





