Characterization of the IS200/IS605 Insertion Sequence Family in Halanaerobium Hydrogeniformans
Abstract
1. Introduction
2. Materials and Methods
2.1. Insertion Sequence Identification
2.2. Alignments
2.3. Phylogenetic Analysis
2.4. Secondary Structure Identification
3. Results
3.1. Insertion Sequence Identification
3.2. TnpA
3.3. IS605 TnpB
3.4. IS605 TnpB Inserts
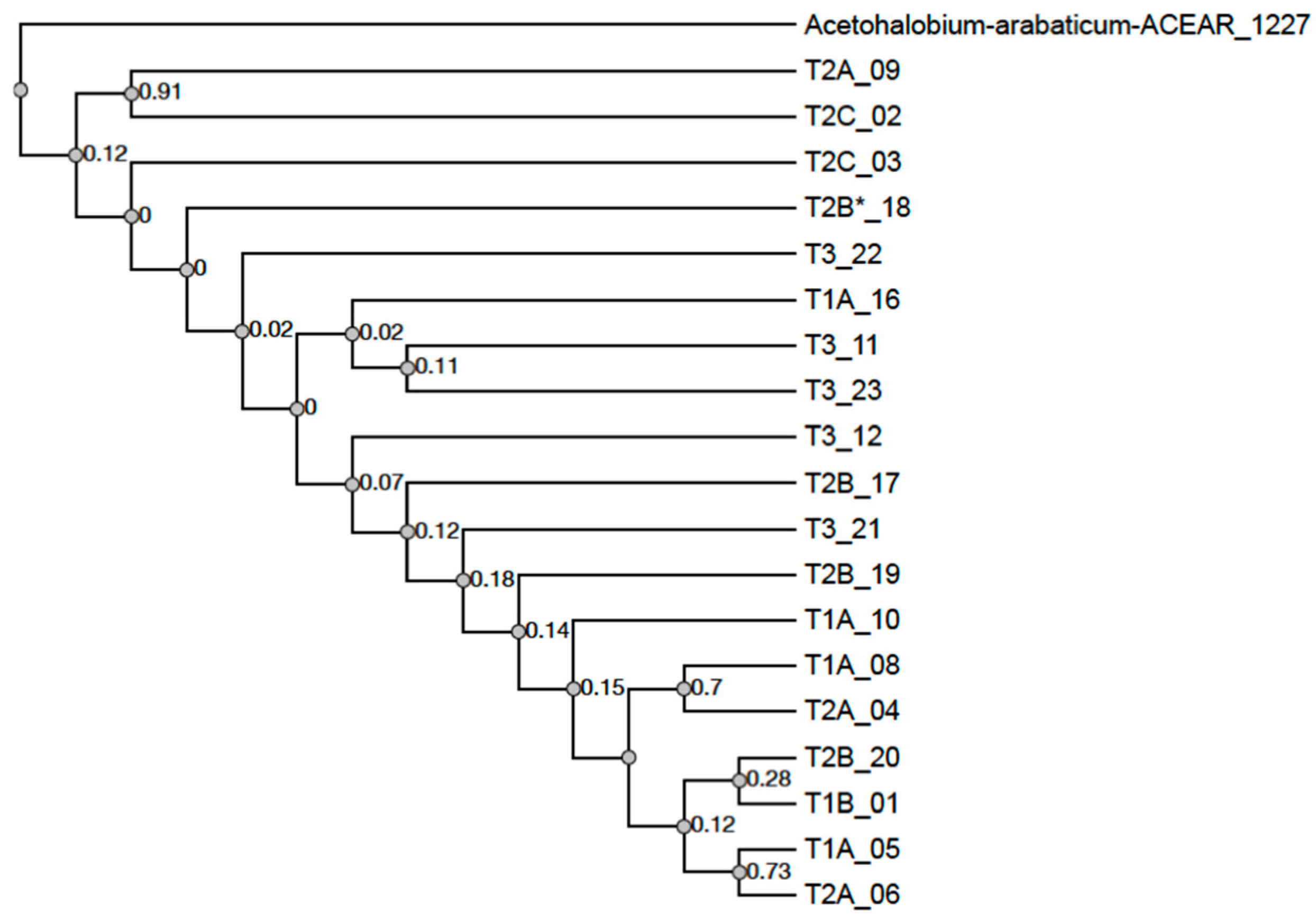
3.5. IS605 TnpB Phylogeny
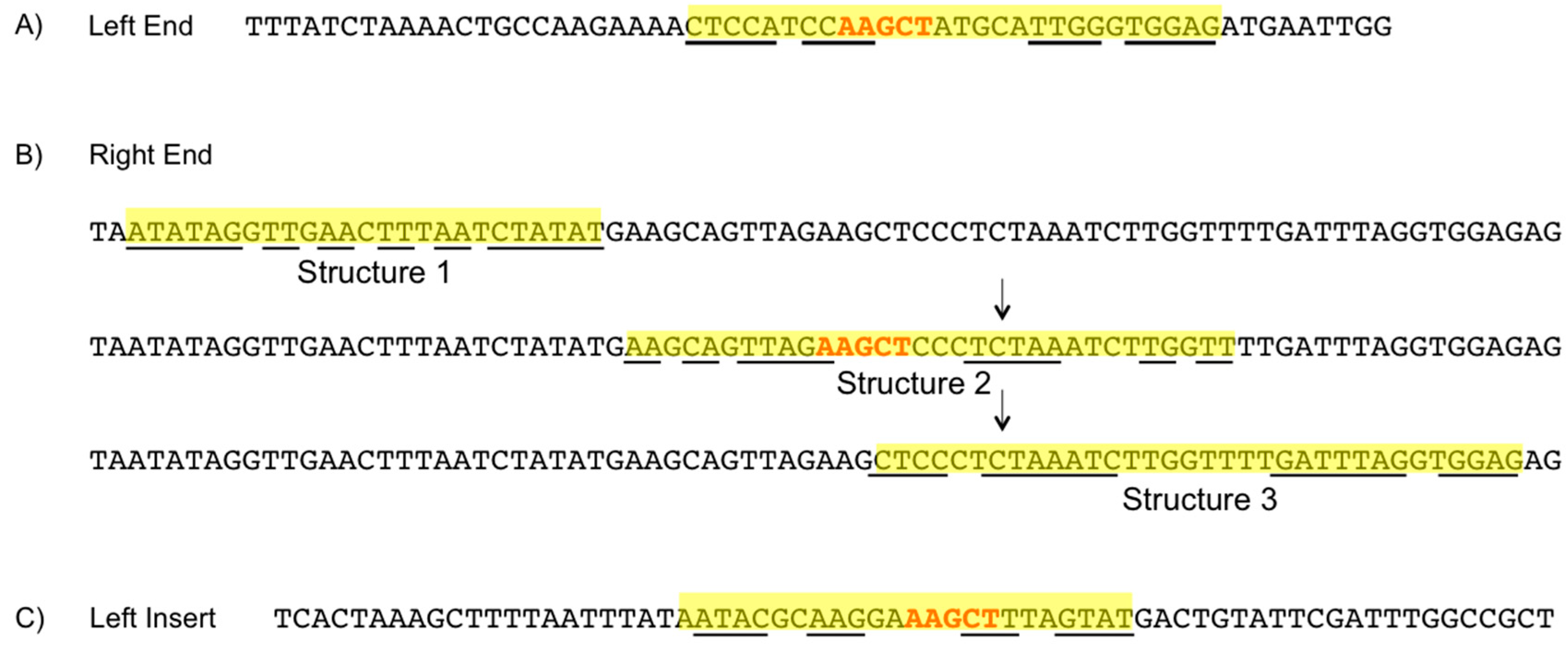
3.6. Element Ends
3.7. Hairpin Structures
3.8. PATE-Like Structure
4. Discussion
4.1. Insertion Sequence Identification
4.2. IS605 TnpB Phylogeny
4.3. Type 5 TnpA
4.4. IS605 TnpB Inserts
4.5. Element Ends
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mormile, M.R. Going from microbial ecology to genome data and back: Studies on a haloalkaliphilic bacterium isolated from Soap Lake, Washington State. Front. Microbiol. 2014, 5, 628. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Filée, J.; Chandler, M. Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 2006, 9, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Ronning, D.R.; Guynet, C.; Ton-Hoang, B.; Perez, Z.N.; Ghirlando, R.; Chandler, M.; Dyda, F. Active Site Sharing and Subterminal Hairpin Recognition in a New Class of DNA Transposases. Mol. Cell 2005, 20, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Kersulyte, D.; Akopyants, N.S.; Clifton, S.W.; Roe, B.A.; Berg, D.E. Novel sequence organization and insertion specificity of IS605 and IS606: Chimaeric transposable elements of Helicobacter pylori. Gene 1998, 223, 175–186. [Google Scholar] [CrossRef]
- Barabas, O.; Ronning, D.R.; Guynet, C.; Hickman, A.B.; Ton-Hoang, B.; Chandler, M.; Dyda, F. Mechanism of IS200/IS605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection. Cell 2008, 132, 208–220. [Google Scholar] [CrossRef]
- Kersulyte, D.; Velapatino, B.; Dailide, G.; Mukhopadhyay, A.K.; Ito, Y.; Cahuayme, L.; Parkinson, A.J.; Gilman, R.H.; Berg, D.E. Transposable Element ISHp608 of Helicobacter pylori: Nonrandom Geographic Distribution, Functional Organization, and Insertion Specificity. J. Bacteriol. 2002, 184, 992–1002. [Google Scholar] [CrossRef]
- Ton-Hoang, B.; Guynet, C.; Ronning, D.R.; Cointin-Marty, B.; Dyda, F.; Chandler, M. Transposition of ISHp608, member of an unusual family of bacterial insertion sequences. EMBO J. 2005, 24, 3325–3338. [Google Scholar] [CrossRef]
- Pasternak, C.; Dulermo, R.; Ton-Hoang, B.; Debuchy, R.; Siguier, P.; Coste, G.; Chandler, M.; Sommer, S. ISDra2transposition inDeinococcus radioduransis downregulated by TnpB. Mol. Microbiol. 2013, 88, 443–455. [Google Scholar] [CrossRef]
- Cerveau, N.; Leclercq, S.; Bouchon, D.; Cordaux, R. Evolutionary Dynamics and Genomic Impact of Prokaryote Transposable Elements. In Evolutionary Biology–Concepts, Biodiversity, Macroevolution and Genome Evolution; Pontarotti, P., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 291–312. [Google Scholar]
- Wagner, A. Periodic Extinctions of Transposable Elements in Bacterial Lineages: Evidence from Intragenomic Variation in Multiple Genomes. Mol. Biol. Evol. 2006, 23, 723–733. [Google Scholar] [CrossRef]
- Wagner, A.; Lewis, C.; Bichsel, M. A survey of bacterial insertion sequences using IScan. Nucleic Acids Res. 2007, 35, 5284–5293. [Google Scholar] [CrossRef]
- Cerveau, N.; Leclercq, S.; Leroy, E.; Bouchon, D.; Cordaux, R. Short- and Long-term Evolutionary Dynamics of Bacterial Insertion Sequences: Insights from Wolbachia Endosymbionts. Genome Biol. Evol. 2011, 3, 1175–1186. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [PubMed]
- Varani, A.M.; Siguier, P.; Gourbeyre, E.; Charneau, V.; Chandler, M. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 2011, 12, R30. [Google Scholar] [CrossRef]
- Li, W.; Cowley, A.; Uludag, M.; Gur, T.; McWilliam, H.; Squizzato, S.; Park, Y.M.; Buso, N.; Lopez, R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015, 43, W580–W584. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Cordaux, R. Gene Conversion Maintains Nonfunctional Transposable Elements in an Obligate Mutualistic Endosymbiont. Mol. Biol. Evol. 2009, 26, 1679–1682. [Google Scholar] [CrossRef]
- Guynet, C.; Hickman, A.B.; Barabas, O.; Dyda, F.; Chandler, M.; Ton-Hoang, B. In Vitro Reconstitution of a Single-Stranded Transposition Mechanism of IS608. Mol. Cell 2008, 29, 302–312. [Google Scholar] [CrossRef]
- Ton-Hoang, B.; Pasternak, C.; Siguier, P.; Guynet, C.; Hickman, A.B.; Dyda, F.; Sommer, S.; Chandler, M. Single-Stranded DNA Transposition Is Coupled to Host Replication. Cell 2010, 142, 398–408. [Google Scholar] [CrossRef] [PubMed]


| Locus | TnpA Type | TnpB Type | LE | RE | TnpA Locus ID | TnpB Locus ID | Leading/Lagging |
|---|---|---|---|---|---|---|---|
| 1 | Type 2 | 1B | consensus | type 2 | HALSA_RS01255 | HALSA_RS01260 | Lead |
| 2 | Type 5 | 2C | consensus | type 1 | N/A | HALSA_RS01330 | Lead |
| 3 | Type 5 | 2C | consensus | type 1 | N/A | HALSA_RS01515 | Lag |
| 4 | Type 5 | 2A | consensus | type 2 | N/A | HALSA_RS01645 | Lead |
| 5 | Type 5 | 1A | consensus | type 1 | HALSA_RS02280 | HALSA_RS02275 | Lead |
| 6 | Type 5 | 2A | consensus | type 2 | HALSA_RS02590 | HALSA_RS02585 | Lag |
| 7 | IS200 | N/A | unknown | unknown | HALSA_RS03110 | N/A | Lag |
| 8 | Type 5 | 1A | consensus | type 1 | N/A | HALSA_RS03165 | Lag |
| 9 | Type 1 | 2A | consensus | type 1 | HALSA_RS03745 | HALSA_RS03750 | Lag |
| 10 | Type 5 | 1A | consensus | type 1 | N/A | HALSA_RS04080 | Lag |
| 11 | Type 5 | 3 | consensus | type 1 | N/A | HALSA_RS12615 | Lag |
| 12 | Type 5 | 3 | consensus | type 1 | N/A | HALSA_RS12630 | Lag |
| 13 | Type 3 | 2A’ | consensus | type 1 | HALSA_RS12635 | HALSA_RS05500 | Lag |
| 14 | Type 5 | MISC | consensus | type 2 | N/A | N/A | Lead |
| 15 | Type 4 | 3’ | consensus | MISC | HALSA_RS12645 | HALSA_RS12715 | Lead |
| 16 | Type 5 | 1A | consensus | type 1 | N/A | HALSA_RS06215 | Lag |
| 17 | Type 5 | 2B | consensus | type 1 | N/A | HALSA_RS07530 | Lag |
| 18 | Type 5 | 2B’ | consensus | type 1 | N/A | HALSA_RS08275 | Lead |
| 19 | Type 5 | 2B | consensus | type 1 | N/A | HALSA_RS08865 | Lag |
| 20 | Type 2 | 2B | consensus | type 1 | HALSA_RS11165 | HALSA_RS11170 | Lead |
| 21 | Type 5 | 3 | consensus | type 2 | N/A | HALSA_RS12685 | Lead |
| 22 | Type 5 | 3 | consensus | type 1 | N/A | HALSA_RS12690 | Lead |
| 23 | Type 5 | 3 | consensus | type 1 | N/A | HALSA_RS12700 | Lag |
| PATE | NA | NA | Hairpin | type 2 | N/A | N/A | Lag |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadler, M.; R. Mormile, M.; L. Frank, R. Characterization of the IS200/IS605 Insertion Sequence Family in Halanaerobium Hydrogeniformans. Genes 2020, 11, 484. https://doi.org/10.3390/genes11050484
Sadler M, R. Mormile M, L. Frank R. Characterization of the IS200/IS605 Insertion Sequence Family in Halanaerobium Hydrogeniformans. Genes. 2020; 11(5):484. https://doi.org/10.3390/genes11050484
Chicago/Turabian StyleSadler, Michael, Melanie R. Mormile, and Ronald L. Frank. 2020. "Characterization of the IS200/IS605 Insertion Sequence Family in Halanaerobium Hydrogeniformans" Genes 11, no. 5: 484. https://doi.org/10.3390/genes11050484
APA StyleSadler, M., R. Mormile, M., & L. Frank, R. (2020). Characterization of the IS200/IS605 Insertion Sequence Family in Halanaerobium Hydrogeniformans. Genes, 11(5), 484. https://doi.org/10.3390/genes11050484





