Base Editing: The Ever Expanding Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) Tool Kit for Precise Genome Editing in Plants
Abstract
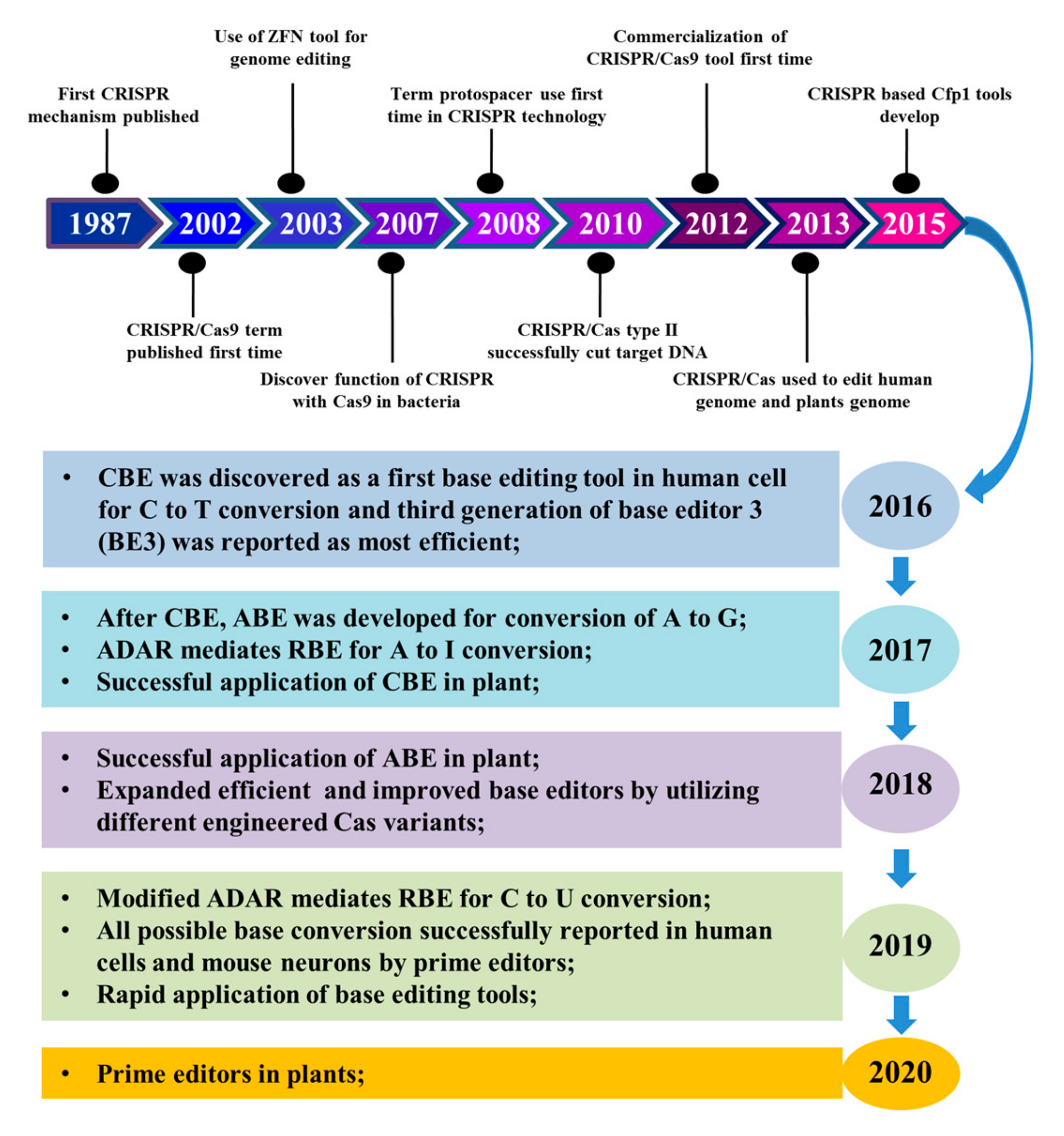
1. Introduction
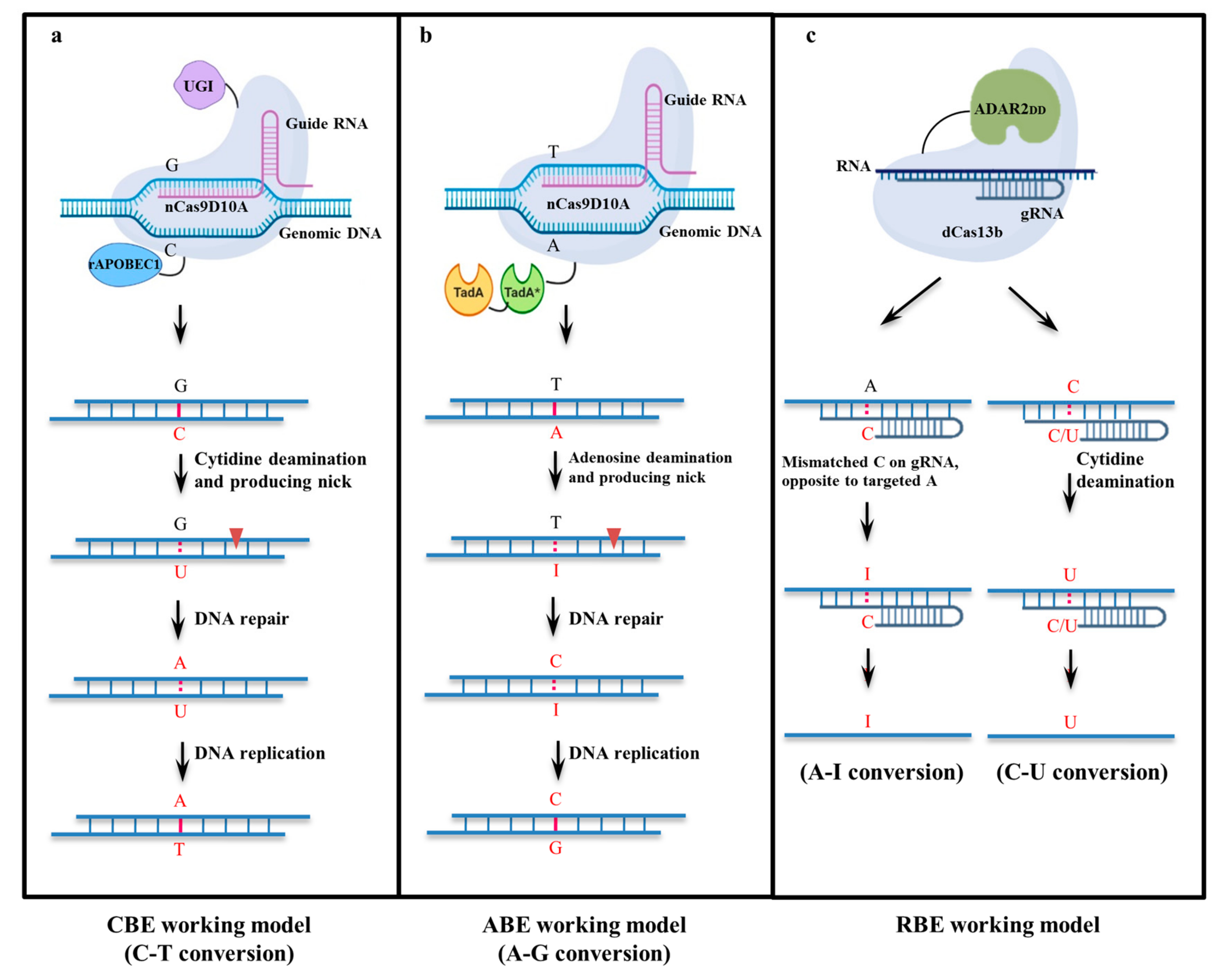
2. Evolution of Base Editors
3. Applications of Base Editing Tools for Plant Improvement
4. Future Perspectives and Limitations
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barman, H.N.; Sheng, Z.; Fiaz, S.; Zhong, M.; Wu, Y.; Cai, Y.; Wang, W.; Jiao, G.; Tang, S.; Wei, X.; et al. Generation of a new thermo-sensitive genic male sterile rice line by targeted mutagenesis of TMS5 gene through CRISPR/Cas9 system. BMC Plant Biol. 2019, 19, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Egelie, K.J.; Graff, G.D.; Strand, S.P.; Johansen, B. The emerging patent landscape of CRISPR-Cas gene editing technology. Nat. Biotechnol. 2016, 34, 1025–1031. [Google Scholar] [CrossRef] [PubMed]
- Shao, G.; Lu, Z.; Xiong, J.; Wang, B.; Jing, Y.; Meng, X.; Liu, G.; Ma, H.; Liang, Y.; Chen, F.; et al. Tiller bud formation regulators MOC1 and MOC3 cooperatively promote tiller bud outgrowth by activating FON1 expression in rice. Mol. Plant 2019, 12, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Mao, B.; Li, Y.; Lv, Q.; Zhang, L.; Chen, C.; He, H.; Wang, W.; Zeng, X.; Shao, Y.; et al. Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Zhou, H.; He, M.; Li, J.; Chen, L.; Huang, Z.; Zheng, S.; Zhu, L.; Ni, E.; Jiang, D.; Zhao, B.; et al. Development of commercial thermo-sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediated TMS5 editing system. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas Genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Mishra, R.; Joshi, R.K.; Zhao, K. Genome editing in rice: Recent advances, challenges, and future implications. Front. Plant Sci. 2018, 9, 1361. [Google Scholar] [CrossRef]
- Zhang, Y.; Massel, K.; Godwin, I.D.; Gao, C. Correction to: Applications and potential of genome editing in crop improvement. Genome Biol. 2019, 20, 13. [Google Scholar] [CrossRef]
- Begemann, M.B.; Gray, B.N.; January, E.; Gordon, G.C.; He, Y.; Liu, H.; Wu, X.; Brutnell, T.P.; Mockler, T.C.; Oufattole, M. Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci. Rep. 2017, 7, 1–6. [Google Scholar] [CrossRef]
- Fiaz, S.; Ahmad, S.; Noor, M.A.; Wang, X.; Younas, A.; Riaz, A.; Riaz, A.; Ali, F. Applications of the CRISPR/Cas9 system for rice grain quality improvement: Perspectives and opportunities. Int. J. Mol. Sci. 2019, 20, 888. [Google Scholar] [CrossRef] [PubMed]
- Smargon, A.A.; Cox, D.B.T.; Pyzocha, N.K.; Zheng, K.; Slaymaker, I.M.; Gootenberg, J.S.; Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S.; et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Massel, K.; Godwin, I.D.; Gao, C. Applications and potential of genome editing in crop improvement. Genome Biol. 2018, 19, 210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-H.; Tee, L.Y.; Wang, X.-G.; Huang, Q.-S.; Yang, S.-H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 2015, 4, e264. [Google Scholar] [CrossRef]
- Mishra, R.; Joshi, R.K.; Zhao, K. Base editing in crops: Current advances, limitations and future implications. Plant Biotechnol. J. 2019, 18, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Ran, Y.; Liang, Z.; Gao, C. Current and future editing reagent delivery systems for plant genome editing. Sci. China Life Sci. 2017, 60, 490–505. [Google Scholar] [CrossRef]
- Huang, T.-K.; Puchta, H. CRISPR/Cas-mediated gene targeting in plants: Finally a turn for the better for homologous recombination. Plant Cell Rep. 2019, 38, 443–453. [Google Scholar] [CrossRef]
- Ali, Z.; Shami, A.; Sedeek, K.; Kamel, R.; Alhabsi, A.; Tehseen, M.; Hassan, N.; Butt, H.; Kababji, A.; Hamdan, S.M.; et al. Fusion of the Cas9 endonuclease and the VirD2 relaxase facilitates homology-directed repair for precise genome engineering in rice. Commun. Biol. 2020, 3, 44. [Google Scholar] [CrossRef]
- Li, J.; Sun, Y.; Du, J.; Zhao, Y.; Xia, L. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant 2017, 10, 526–529. [Google Scholar] [CrossRef]
- Lu, Y.; Zhu, J.K. Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant 2017, 10, 523–525. [Google Scholar] [CrossRef]
- May, A. Base editing on the rise. Nat. Biotechnol. 2017, 35, 428–429. [Google Scholar] [CrossRef] [PubMed]
- Gao, C. The future of CRISPR technologies in agriculture. Nat. Rev. Mol. Cell Biol. 2018, 19, 275–276. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Gao, C.; Qiu, J.L. Progress and prospects in plant genome editing. Nat. Plants 2017, 3, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Davies, K. All about That Base Editing. Genet. Eng. Biotechnol. News 2019, 39, 54–56. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Marx, V. Base editing a CRISPR way. Nat. Methods 2018, 15, 767–770. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of T to G C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J. RNA editing with CRISPR-Cas13. Science 2017, 358, 1–15. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Franklin, B.; Koob, J.; Kellner, M.J.; Joung, J.; Kirchgatterer, P.; Cox, D.B.T.; Zhang, F. A cytosine deaminase for programmable single-base RNA editing. Science 2019, 365, 382–386. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Fonfara, I.; Richter, H.; BratoviÄ, M.; Le Rhun, A.; Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 2016, 532, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; Van Der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided endonuclease of a Class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Hess, G.T.; Tycko, J.; Yao, D.; Bassik, M.C. Methods and applications of CRISPR-mediated base editing in eukaryotic genomes. Mol. Cell 2017, 68, 26–43. [Google Scholar] [CrossRef]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Platt, R.J.; Zhang, F. Therapeutic genome editing: Prospects and challenges. Nat. Med. 2015, 21, 121–131. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, J.; Chai, Z.; Chen, S.; Bai, Y.; Zong, Y.; Chen, K.; Li, J.; Jiang, L.; Gao, C. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat. Plants 2019, 5, 480–485. [Google Scholar] [CrossRef]
- Yang, B.; Yang, L.; Chen, J. Development and application of base editors. CRISPR J. 2019, 2, 91–104. [Google Scholar] [CrossRef]
- Liang, P.; Xie, X.; Zhi, S.; Sun, H.; Zhang, X.; Chen, Y.; Chen, Y.; Xiong, Y.; Ma, W.; Liu, D.; et al. Genome-wide profiling of adenine base editor specificity by EndoV-seq. Nat. Commun. 2019, 10, 67. [Google Scholar] [CrossRef]
- Kim, Y.B.; Komor, A.C.; Levy, J.M.; Packer, M.S.; Zhao, K.T.; Liu, D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017, 35, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zong, Y.; Gao, Q.; Zhu, Z.; Wang, Y.; Qin, P.; Liang, C.; Wang, D.; Qiu, J.L.; Zhang, F.; et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 2019, 364, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Zhang, F.; Karcher, D.; Bock, R. Expanding the genome-targeting scope and the site selectivity of high-precision base editors. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Doman, J.L.; Raguram, A.; Newby, G.A.; Liu, D.R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 2020, 1–9. [Google Scholar] [CrossRef]
- Eid, A.; Alshareef, S.; Mahfouz, M.M. CRISPR base editors: Genome editing without double-stranded breaks. Biochem. J. 2018, 475, 1955–1964. [Google Scholar] [CrossRef]
- Rees, H.A.; Komor, A.C.; Yeh, W.H.; Caetano-Lopes, J.; Warman, M.; Edge, A.S.B.; Liu, D.R. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Molla, K.A.; Yang, Y. CRISPR/Cas-Mediated Base Editing: Technical considerations and practical applications. Trends Biotechnol. 2019, 37, 1121–1142. [Google Scholar] [CrossRef]
- Bharat, S.S.; Li, S.; Li, J.; Yan, L.; Xia, L. Base editing in plants: Current status and challenges. Crop J. 2019. [Google Scholar] [CrossRef]
- Endo, M.; Mikami, M.; Endo, A.; Kaya, H.; Itoh, T.; Nishimasu, H.; Nureki, O.; Toki, S. Genome editing in plants by engineered CRISPR–Cas9 recognizing NG PAM. Nat. Plants 2019, 5, 14–17. [Google Scholar] [CrossRef]
- Hua, K.; Tao, X.; Zhu, J.K. Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol. J. 2019, 17, 499–504. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.W.; Li, Z.; Peterson, R.T.; Yeh, J.R.J.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Qin, R.; Li, J.; Li, H.; Zhang, Y.; Liu, X.; Miao, Y.; Zhang, X.; Wei, P. Developing a highly efficient and wildly adaptive CRISPR- Sa Cas9 toolset for plant genome editing. Plant Biotechnol. J. 2019, 17, 706–708. [Google Scholar] [CrossRef] [PubMed]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xu, W.; Wang, F.; Zhao, S.; Feng, F.; Song, J.; Zhang, C. Increasing Cytosine Base Editing Scope and Efficiency with Engineered Cas9-PmCDA1 Fusions and the Modified sgRNA in Rice. Front. Genet. 2019, 10, 379. [Google Scholar] [CrossRef]
- Xu, W.; Song, W.; Yang, Y.; Wu, Y.; Lv, X.; Yuan, S.; Liu, Y.; Yang, J. Multiplex nucleotide editing by high-fidelity Cas9 variants with improved efficiency in rice. BMC Plant Biol. 2019, 19, 1–10. [Google Scholar] [CrossRef]
- Tan, J.; Zhang, F.; Karcher, D.; Bock, R. Engineering of high-precision base editors for site-specific single nucleotide replacement. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Liu, Y.; Yang, B.; Wang, X.; Wei, J.; Lu, Z.; Zhang, Y.; Wu, J.; Huang, X.; et al. Base editing with a Cpf1-cytidine deaminase fusion. Nat. Biotechnol. 2018, 36, 324–327. [Google Scholar] [CrossRef]
- Koblan, L.W.; Doman, J.L.; Wilson, C.; Levy, J.M.; Tay, T.; Newby, G.A.; Maianti, J.P.; Raguram, A.; Liu, D.R. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018, 36, 843–848. [Google Scholar] [CrossRef]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef]
- Wang, J.; Meng, X.; Hu, X.; Sun, T.; Li, J.; Wang, K.; Yu, H. xCas9 expands the scope of genome editing with reduced efficiency in rice. Plant Biotechnol. J. 2019, 17, 709–711. [Google Scholar] [CrossRef]
- Ge, Z. Engineered xCas9 and SpCas9-NG variants broaden PAM recognition sites to generate mutations in Arabidopsis plants. Plant Biotechnol. J. 2019, 17, 1865–1867. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Wu, S.; Yang, X.; Liu, P.; Xu, Y.; Genetics, M. Expanding the scope of CRISPR/Cas9-mediated genome editing in plants using an xCas9 and Cas9-NG hybrid. J. Integr. Plant Biol. 2019, 62, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Veillet, F.; Perrot, L.; Guyon-debast, A.; Kermarrec, M.; Chauvin, L.; Chauvin, J.; Gallois, J.; Mazier, M.; Nogu, F. Expanding the CRISPR Toolbox in P. patens using SpCas9-NG variant and application for gene and base editing in Solanaceae Crops. Int. J. Mol. Sci. 2020, 21, 1024. [Google Scholar] [CrossRef] [PubMed]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018, 9, 1259–1262. [Google Scholar] [CrossRef]
- Stafforst, T.; Schneider, M.F. An RNA—Deaminase conjugate selectively repairs point mutations. Angew. Chem. Intern. Ed. 2012, 51, 11166–11169. [Google Scholar] [CrossRef]
- Qu, L.; Yi, Z.; Zhu, S.; Wang, C.; Cao, Z.; Zhou, Z.; Yuan, P.; Yu, Y.; Tian, F.; Liu, Z.; et al. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat. Biotechnol. 2019, 37, 1059–1069. [Google Scholar] [CrossRef]
- Zinshteyn, B.; Nishikura, K. Adenosine-to-inosine RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009, 1, 202–209. [Google Scholar] [CrossRef]
- Walkley, C.R.; Li, J.B. Rewriting the transcriptome: Adenosine-to- inosine RNA editing by ADARs. Genome Biol. 2017, 18, 205. [Google Scholar] [CrossRef]
- Zong, Y.; Wang, Y.; Li, C.; Zhang, R.; Chen, K.; Ran, Y.; Qiu, J.L.; Wang, D.; Gao, C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 438–440. [Google Scholar] [CrossRef]
- Tian, S.; Jiang, L.; Cui, X.; Zhang, J.; Guo, S.; Li, M.; Zhang, H.; Ren, Y.; Gong, G.; Zong, M.; et al. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 2018, 37, 1353–1356. [Google Scholar] [CrossRef]
- Zong, Y.; Song, Q.; Li, C.; Jin, S.; Zhang, D.; Wang, Y.; Qiu, J.L.; Gao, C. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 2018, 36, 950. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Yan, F.; Kuang, Y.; Li, N.; Zhang, D.; Lin, H.; Zhou, H. A CRISPR/Cas9 toolkit for efficient targeted base editing to induce genetic variations in rice. Sci. China Life Sci. 2017, 60, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Yan, F.; Kuang, Y.; Li, N.; Zhang, D.; Zhou, X.; Lin, H.; Zhou, H. Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID Mutant. Mol. Plant 2018, 11, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zong, Y.; Wang, Y.; Jin, S.; Zhang, D.; Song, Q.; Zhang, R. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018, 19, 59. [Google Scholar] [CrossRef] [PubMed]
- Negishi, K.; Kaya, H.; Abe, K.; Hara, N.; Saika, H.; Toki, S. An adenine base editor with expanded targeting scope using SpCas9-NGv1 in rice. Plant Biotechnol. J. 2019, 17, 1476–1478. [Google Scholar] [PubMed]
- Yan, F.; Kuang, Y.; Ren, B.; Wang, J.; Zhang, D.; Lin, H.; Yang, B.; Zhou, X.; Zhou, H. Highly Efficient A·T to G·C Base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol. Plant 2018, 11, 631–634. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xiong, X.; Wang, F.; Liang, J.; Li, J.F. Gene disruption through base editing-induced messenger RNA missplicing in plants. New Phytol. 2019, 222, 1139–1148. [Google Scholar] [CrossRef]
- Qin, L.; Li, J.; Wang, Q.; Xu, Z.; Sun, L.; Alariqi, M.; Manghwar, H.; Wang, G.; Li, B.; Ding, X.; et al. High-efficient and precise base editing of C·G to T·A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol. J. 2019, 18, 45–56. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Z.; Ni, H.; Xu, Y.; Chen, Q.; Jiang, L. CRISPR/Cas9-mediated base-editing system efficiently generates gain-of-function mutations in Arabidopsis. Sci. China Life Sci. 2017, 60, 520–523. [Google Scholar] [CrossRef]
- Bastet, A.; Zafirov, D.; Giovinazzo, N.; Guyon-debast, A.; Nogu, F. Mimicking natural polymorphism in eIF4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnol. J. 2019, 17, 1736–1750. [Google Scholar] [CrossRef]
- Kang, B.C.; Yun, J.Y.; Kim, S.T.; Shin, Y.J.; Ryu, J.; Choi, M.; Woo, J.W.; Kim, J.S. Precision genome engineering through adenine base editing in plants. Nat. Plants 2018, 4, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Veillet, F.; Perrot, L.; Chauvin, L.; Kermarrec, M.P.; Guyon-Debast, A.; Chauvin, J.E.; Nogué, F.; Mazier, M. Transgene-free genome editing in tomato and potato plants using Agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci. 2019, 20, 402. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Powles, S.B. Resistance to AHAS inhibitor herbicides: Current understanding. Pest Manag. Sci. 2014, 70, 1340–1350. [Google Scholar] [CrossRef] [PubMed]
- Hua, K.; Tao, X.; Yuan, F.; Wang, D.; Zhu, J.K. Precise A·T to G·C Base editing in the rice genome. Mol. Plant 2018, 11, 627–630. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Willi, M.; Miller, S.M.; Kim, S.; Liu, C.; Liu, D.R.; Hennighausen, L. Targeting fidelity of adenine and cytosine base editors in mouse embryos. Nat. Commun. 2018, 9, 4804. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lim, K.; Kim, S.T.; Yoon, S.H.; Kim, K.; Ryu, S.M.; Kim, J.S. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat. Biotechnol. 2017, 35, 475–480. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Yang, L.; Yang, B.; Chen, J. Previews. One prime for all editing. Cell 2019, 179, 1448–1450. [Google Scholar] [CrossRef]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 1–4. [Google Scholar] [CrossRef]
- Li, C.; Zhang, R.; Meng, X.; Chen, S.; Zong, Y.; Lu, C.; Qiu, J.; Chen, Y.; Li, J.; Gao, C. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 2020, 1–8. [Google Scholar] [CrossRef]
- Wang, L.; Xue, W.; Yan, L.; Li, X.; Wei, J.; Chen, M.; Wu, J.; Yang, B.; Yang, L.; Chen, J. Enhanced base editing by co-expression of free uracil DNA glycosylase inhibitor. Cell Res. 2017, 27, 1289–1292. [Google Scholar] [CrossRef] [PubMed]
- Grünewald, J.; Zhou, R.; Iyer, S.; Lareau, C.A.; Garcia, S.P.; Aryee, M.J.; Joung, J.K. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat. Biotechnol. 2019, 37, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Wei, X.; Sheng, Z.; Hu, P.; Tang, S. CRISPR/Cas9 for development of disease resistance in plants: Recent progress, limitations and future prospects. Brief. Funct. Genom. 2020, 19, 26–39. [Google Scholar] [CrossRef] [PubMed]

| Plant Species | Targeted Gene | Selected PAM | Base Editor | Mutation Efficiency | Editing Window (nt) | Improved Trait or Key Findings | Reference |
|---|---|---|---|---|---|---|---|
| Rice1 | SLR1 NRT1.1B | AGG GGG | APOBEC1-XTEN-Cas9(D10A) | 13.3% 2.7% | 4 to 8 | Reduced plant height; increased nitrogen use efficiency | [20] |
| Rice1 | ACC, ALS, CDC48, DEP1, NRT1.1B OsEV | CCT | ABE7.10 | 3.2–59.1% | 4 to 8 | Development of efficient ABE PABE-7 | [74] |
| Rice1 | ALS, FTIP1e | AGG CCA | Target-AID | 6–89% | −19 to −17 | Develop multiple herbicide resistance | [53] |
| Rice1 | OsAOS1 OsJAR1 OsJAR2 OsCOI2 | CCA TGG | rBE3 rBE9 | 8.3–73.3% | −19 to −13 | Prove editing efficiency of rBE9, which is higher than rBE3 | [73] |
| Rice1 | OsCERK1 OsSERK1 OsSERK2 ipal pi-ta BRI1 | NGA AGTG AGCG | rBE3 | 10.5–38.9% | −19 to −13 | Detect the efficiency of rBE3 | [72] |
| Rice1 | IPA1 (OsSPL14) OsSPL17 OsSPL18 SLR1 | GAG CAG CGA GGA AGCG GGCG | ABE-P1 ABE-P2 ABE-P3 ABE-P4 ABE-P5 | 26% | 3 to 15 | Multiple adenine base editor evaluation | [50] |
| Rice callus1 | sgOs-siteG1 sgOs-site2 sgOs-site3 sgOs-site4 | NGG NGA NGC NGT | ABE7.10 | 29.2–45.8% | 13 to 16 | Develop new ABEs | [75] |
| Rice1 | OsCDC48, OsNRT1.1B OsSPL14 | CGG | pnCas9-PBE | 43.48% | 3 to 9 | Reduce senescence and death | [69] |
| Rice1 | OsNRT1.1B OsCDC48 | NGG CCN | A3A-PBE | 44.1% 82.9% | 1 to 17 | A3A-PBE editor is more efficient than pnCas9-PBE | [71] |
| Rice1 | EPSPS, ALS, DL | NG | Target-AID-NG | 5–95.5% | −9 to −20 | SpCas9-NGv1 application in base editing | [49] |
| Rice1 | MPK6, MPK13, SERK2, WRKY45, Tms9-1 | CCA CCG | ABE7.10 ABE7.8 | 0–62.26% | −17 to −11 | Develop new adenine base editor using fluorescence-tracking | [76] |
| Rice1 | OsACC OsALS OsDEP1 OsNRT1 OsCDC48 OsWx | AGG TGG CCA CCT CGG GGG | Be3 HF1-BE3 ABE(PABE-7) | Off-target mutation is higher in CBE compared to ABE. | [42] | ||
| Rice1 | GL1-1 NAL1 | nCas9-PBE | 58% 68% | 3 to 9 | The mutant with hydrophilic leaf surface and abnormal transcripts of NAL1 | [77] | |
| Wheat2 | TaLOX2 | CGG | pnCas9-PBE | 1.25% | 3 to 9 | Herbicide resistance | [69] |
| Wheat2 | TaALS-P174 | CGG CCT | PBE | 33–75% | 3 to 9 | Increase multiple herbicide resistance | [38] |
| Wheat2 | DEP1, TaEPSPS GW2 | CCT | ABE7.10 | 0.4–1.1% | 4 to 8 | Increase herbicide resistance | [74] |
| Wheat2 | ALS gene | NGG CCN | A3A-PBE | 16.7–22.5% | 1 to 17 | Herbicide resistance and editing efficiency of A3A-PBE | [71] |
| Maize1 | ZmCENH3 | CGG | pnCas9-PBE | 10% | 3 to 9 | Bialaphos-resistant | [69] |
| Cotton | GhCLA GhPEBP | TGG CCA AGG | G. hirsutum-Base Editor 3 (GhBE3) | 26.67–57.78% | −17 to −12 | Point mutation was generated with novel GhBE3 in cotton | [78] |
| Watermelon1 | ALS gene | TGG CGG | BE3 | 23% | 3 to 9 | Herbicides resistance | [70] |
| Arabidopsis1 | ALS gene | TGG | BE3 | 2.7–40% | 4 to 9 | Inheritable herbicides resistance was found | [79] |
| Arabidopsis1 | eIF4E1 | NGG | CBE | 50% | C-to-G base editing generate Clover yellow vein virus resistant plants | [80] | |
| Arabidopsis1 | AtALS AtPDS AtFT AtLFY | TGG AGG GGG CGG | ABE7.10 (pcABE7.10) | 0–85% | 1 to 12 | Plant ABE application | [81] |
| Tomato1 | DELLA ETR1 | AGG CCA | Target-AID | 41–92% | −19 to −17 | Generate marker-free plants | [53] |
| Tomato1 | ALS | TGG | CBE | 71% | −20 to −13 | Obtain of Chlorsulfuron-resistant | [82] |
| Potato3 | StALS StGBSS | NGG CCN | A3A-PBE | 6.5% | 1 to 17 | Widespread use of A3A-PBE in dicotyledons | [71] |
| Potato1 | ALS | TGG | CBE | 100% | −20 to −13 | Herbicide resistant | [82] |
| Rapeseed1 | BnALS BnPDS | TGG AGG GGG CGG | ABE7.10 (pcABE7.10) | 8.8% | 1 to 12 | Plant ABE application | [81] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monsur, M.B.; Shao, G.; Lv, Y.; Ahmad, S.; Wei, X.; Hu, P.; Tang, S. Base Editing: The Ever Expanding Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) Tool Kit for Precise Genome Editing in Plants. Genes 2020, 11, 466. https://doi.org/10.3390/genes11040466
Monsur MB, Shao G, Lv Y, Ahmad S, Wei X, Hu P, Tang S. Base Editing: The Ever Expanding Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) Tool Kit for Precise Genome Editing in Plants. Genes. 2020; 11(4):466. https://doi.org/10.3390/genes11040466
Chicago/Turabian StyleMonsur, Mahmuda Binte, Gaoneng Shao, Yusong Lv, Shakeel Ahmad, Xiangjin Wei, Peisong Hu, and Shaoqing Tang. 2020. "Base Editing: The Ever Expanding Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) Tool Kit for Precise Genome Editing in Plants" Genes 11, no. 4: 466. https://doi.org/10.3390/genes11040466
APA StyleMonsur, M. B., Shao, G., Lv, Y., Ahmad, S., Wei, X., Hu, P., & Tang, S. (2020). Base Editing: The Ever Expanding Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) Tool Kit for Precise Genome Editing in Plants. Genes, 11(4), 466. https://doi.org/10.3390/genes11040466








