A Novel Cosegregating DCTN1 Splice Site Variant in a Family with Bipolar Disorder May Hold the Key to Understanding the Etiology
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Sequencing
2.2. Genomic Alignment
2.3. Analytical Strategy
- Variant calls were restricted to coding and splice regions, and a minimal variant call filter of the probability of not being reference ≥ 90% was applied.
- Variant calls with a minor allele frequency (MAF) ≥ 0.01 in 1000 genomes phase 3 were filtered.
- Using the Venn diagram feature of ArrayStar, only cosegregating variants common to all three affected subjects and not present in any of the controls were retained.
- The variant calls from the sequencing data of ~120,000 exomes within the Genome Aggregation Database v2.02 (gnomAD) [22] were loaded into ArrayStar and all variant calls present in gnomAD were removed by filtering.
2.4. Bioinformatic Predictions
2.5. Sanger Sequencing Validation
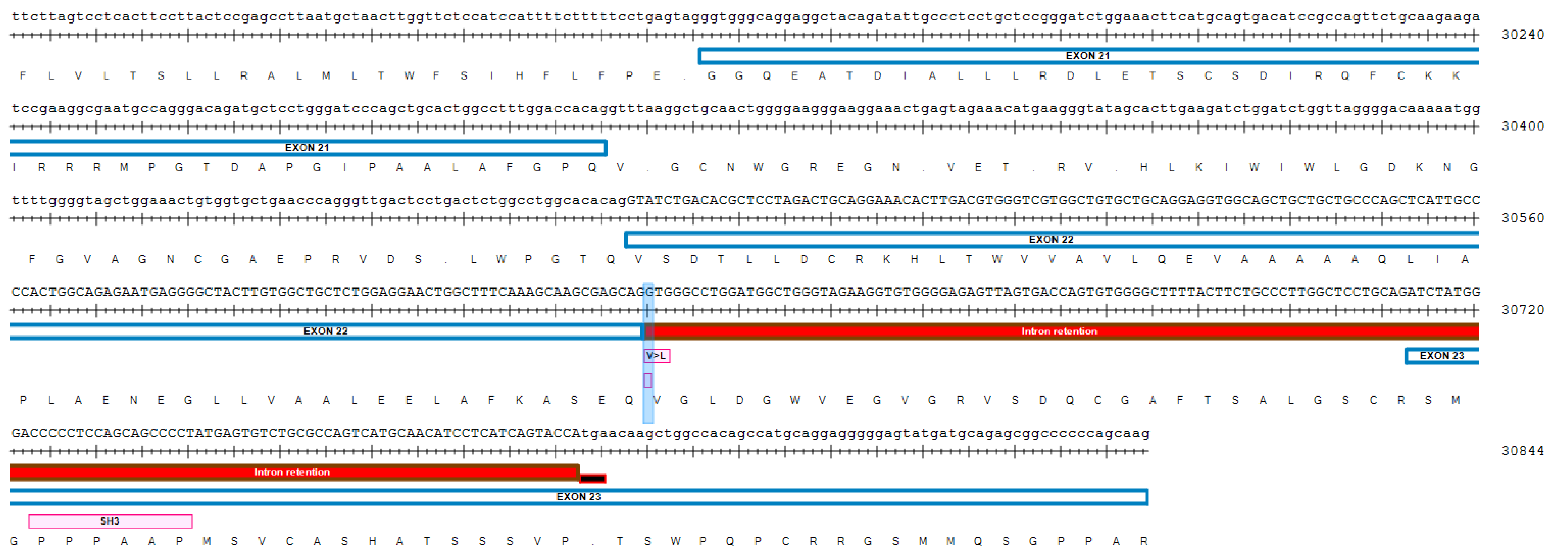
3. Results
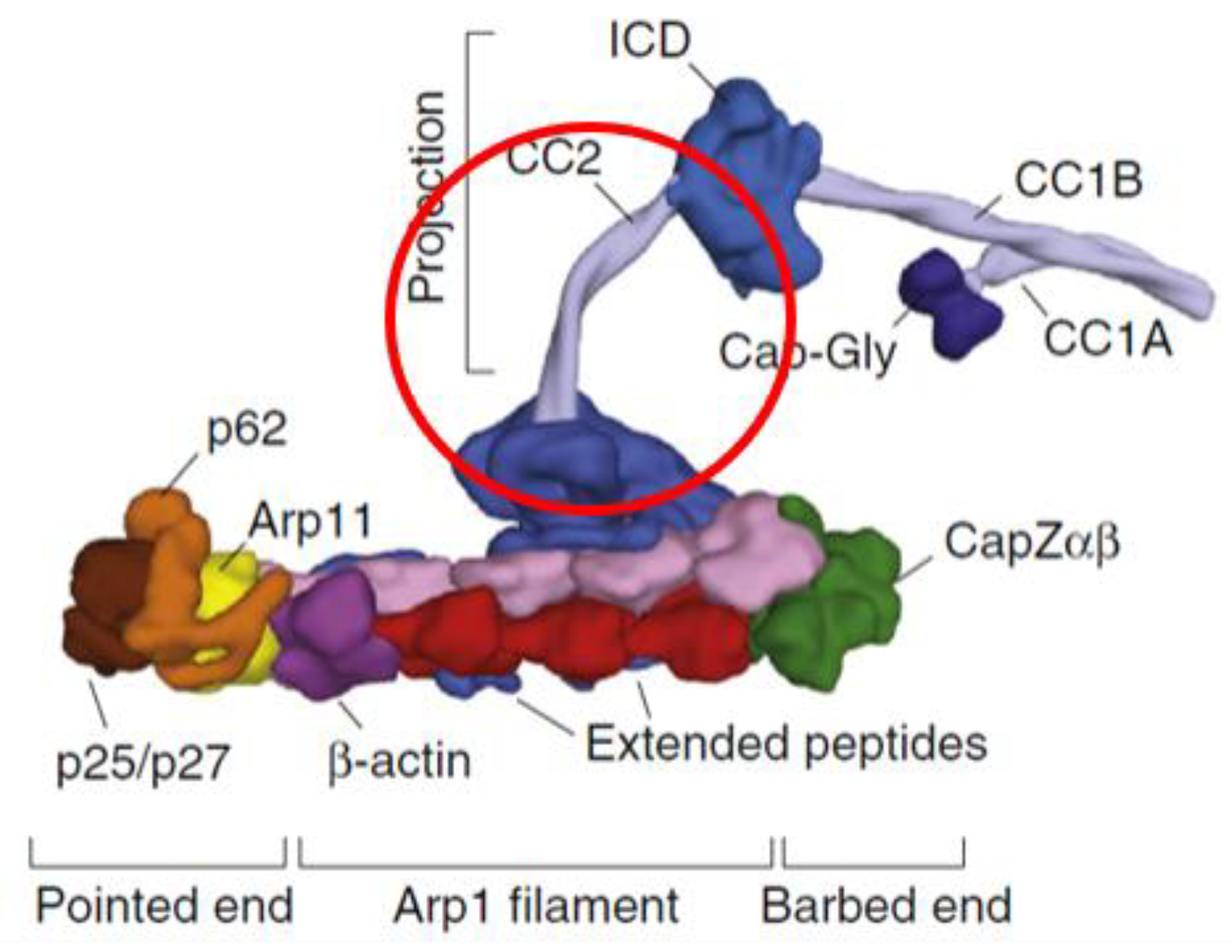
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A

References
- De la Vega, D.; Pina, A.; Peralta, F.J.; Kelly, S.A.; Giner, L. A Review on the General Stability of Mood Disorder Diagnoses Along the Lifetime. Curr. Psychiatry Rep. 2018, 20, 29. [Google Scholar] [CrossRef]
- Van Meter, A.R.; Youngstrom, E.A.; Findling, R.L. Cyclothymic disorder: A critical review. Clin. Psychol. Rev. 2012, 32, 229–243. [Google Scholar] [CrossRef]
- Kleine-Budde, K.; Touil, E.; Moock, J.; Bramesfeld, A.; Kawohl, W.; Rossler, W. Cost of illness for bipolar disorder: A systematic review of the economic burden. Bipolar Disord. 2014, 16, 337–353. [Google Scholar] [CrossRef] [PubMed]
- Boardman, A.P.; Healy, D. Modelling suicide risk in affective disorders. Eur. Psychiatry 2001, 16, 400–405. [Google Scholar] [CrossRef]
- Marangoni, C.; Hernandez, M.; Faedda, G.L. The role of environmental exposures as risk factors for bipolar disorder: A systematic review of longitudinal studies. J. Affect. Disord. 2016, 193, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Uher, R. Gene-environment interactions in severe mental illness. Front. Psychiatry 2014, 5, 48. [Google Scholar] [CrossRef]
- Merikangas, K.R.; Jin, R.; He, J.P.; Kessler, R.C.; Lee, S.; Sampson, N.A.; Viana, M.C.; Andrade, L.H.; Hu, C.; Karam, E.G.; et al. Prevalence and correlates of bipolar spectrum disorder in the world mental health survey initiative. Arch. Gen. Psychiatry 2011, 68, 241–251. [Google Scholar] [CrossRef]
- Patten, S.B.; Williams, J.V.; Lavorato, D.H.; Fiest, K.M.; Bulloch, A.G.; Wang, J. The prevalence of major depression is not changing. Can. J. Psychiatry 2015, 60, 31–34. [Google Scholar] [CrossRef]
- Barnett, J.H.; Smoller, J.W. The genetics of bipolar disorder. Neuroscience 2009, 164, 331–343. [Google Scholar] [CrossRef]
- Smith, D.J.; Harrison, N.; Muir, W.; Blackwood, D.H. The high prevalence of bipolar spectrum disorders in young adults with recurrent depression: Toward an innovative diagnostic framework. J. Affect. Disord. 2005, 84, 167–178. [Google Scholar] [CrossRef]
- Kennedy, N.; Everitt, B.; Boydell, J.; Van Os, J.; Jones, P.B.; Murray, R.M. Incidence and distribution of first-episode mania by age: Results from a 35-year study. Psychol. Med. 2005, 35, 855–863. [Google Scholar] [CrossRef]
- Kroon, J.S.; Wohlfarth, T.D.; Dieleman, J.; Sutterland, A.L.; Storosum, J.G.; Denys, D.; de Haan, L.; Sturkenboom, M.C. Incidence rates and risk factors of bipolar disorder in the general population: A population-based cohort study. Bipolar Disord. 2013, 15, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Tozzi, F.; Prokopenko, I.; Perry, J.D.; Kennedy, J.L.; McCarthy, A.D.; Holsboer, F.; Berrettini, W.; Middleton, L.T.; Chilcoat, H.D.; Muglia, P. Family history of depression is associated with younger age of onset in patients with recurrent depression. Psychol. Med. 2008, 38, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Budde, M.; Forstner, A.J.; Adorjan, K.; Schaupp, S.K.; Nothen, M.M.; Schulze, T.G. Genetics of bipolar disorder. Nervenarzt 2017, 88, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Gordovez, F.J.A.; McMahon, F.J. The genetics of bipolar disorder. Mol. Psychiatry 2020, 25, 544–559. [Google Scholar] [CrossRef] [PubMed]
- Gershon, E.S.; Hamovit, J.; Guroff, J.J.; Dibble, E.; Leckman, J.F.; Sceery, W.; Targum, S.D.; Nurnberger, J.I., Jr.; Goldin, L.R.; Bunney, W.E., Jr. A family study of schizoaffective, bipolar I, bipolar II, unipolar, and normal control probands. Arch. Gen. Psychiatry 1982, 39, 1157–1167. [Google Scholar] [CrossRef]
- Gershon, E.S.; DeLisi, L.E.; Hamovit, J.; Nurnberger, J.I., Jr.; Maxwell, M.E.; Schreiber, J.; Dauphinais, D.; Dingman, C.W., 2nd; Guroff, J.J. A controlled family study of chronic psychoses. Schizophrenia and schizoaffective disorder. Arch. Gen. Psychiatry 1988, 45, 328–336. [Google Scholar] [CrossRef]
- Cirulli, E.T.; Goldstein, D.B. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat. Rev. Genet. 2010, 11, 415–425. [Google Scholar] [CrossRef]
- Kryukov, G.V.; Pennacchio, L.A.; Sunyaev, S.R. Most rare missense alleles are deleterious in humans: Implications for complex disease and association studies. Am. J. Hum. Genet. 2007, 80, 727–739. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O‘Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Davydov, E.V.; Goode, D.L.; Sirota, M.; Cooper, G.M.; Sidow, A.; Batzoglou, S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 2010, 6, e1001025. [Google Scholar] [CrossRef] [PubMed]
- Quang, D.; Chen, Y.; Xie, X. DANN: A deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 2015, 31, 761–763. [Google Scholar] [CrossRef]
- Rogers, M.F.; Shihab, H.A.; Mort, M.; Cooper, D.N.; Gaunt, T.R.; Campbell, C. FATHMM-XF: Accurate prediction of pathogenic point mutations via extended features. Bioinformatics 2018, 34, 511–513. [Google Scholar] [CrossRef] [PubMed]
- Pollard, K.S.; Hubisz, M.J.; Rosenbloom, K.R.; Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010, 20, 110–121. [Google Scholar] [CrossRef]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef]
- Vallee, R.B.; Shpetner, H.S.; Paschal, B.M. The role of dynein in retrograde axonal transport. Trends Neurosci. 1989, 12, 66–70. [Google Scholar] [CrossRef]
- Kardon, J.R.; Vale, R.D. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 2009, 10, 854–865. [Google Scholar] [CrossRef]
- Karki, S.; Holzbaur, E.L. Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr. Opin. Cell Biol. 1999, 11, 45–53. [Google Scholar] [CrossRef]
- Melloni, R.H., Jr.; Tokito, M.K.; Holzbaur, E.L. Expression of the p150Glued component of the dynactin complex in developing and adult rat brain. J. Comp. Neurol. 1995, 357, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Farrer, M.J.; Hulihan, M.M.; Kachergus, J.M.; Dachsel, J.C.; Stoessl, A.J.; Grantier, L.L.; Calne, S.; Calne, D.B.; Lechevalier, B.; Chapon, F.; et al. DCTN1 mutations in Perry syndrome. Nat. Genet. 2009, 41, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Munch, C.; Sedlmeier, R.; Meyer, T.; Homberg, V.; Sperfeld, A.D.; Kurt, A.; Prudlo, J.; Peraus, G.; Hanemann, C.O.; Stumm, G.; et al. Point mutations of the p150 subunit of dynactin (DCTN1) gene in ALS. Neurology 2004, 63, 724–726. [Google Scholar] [CrossRef] [PubMed]
- Stockmann, M.; Meyer-Ohlendorf, M.; Achberger, K.; Putz, S.; Demestre, M.; Yin, H.; Hendrich, C.; Linta, L.; Heinrich, J.; Brunner, C.; et al. The dynactin p150 subunit: Cell biology studies of sequence changes found in ALS/MND and Parkinsonian syndromes. J. Neural. Transm. 2013, 120, 785–798. [Google Scholar] [CrossRef]
- Puls, I.; Oh, S.J.; Sumner, C.J.; Wallace, K.E.; Floeter, M.K.; Mann, E.A.; Kennedy, W.R.; Wendelschafer-Crabb, G.; Vortmeyer, A.; Powers, R.; et al. Distal spinal and bulbar muscular atrophy caused by dynactin mutation. Ann. Neurol. 2005, 57, 687–694. [Google Scholar] [CrossRef]
- Perry, T.L.; Bratty, P.J.; Hansen, S.; Kennedy, J.; Urquhart, N.; Dolman, C.L. Hereditary mental depression and Parkinsonism with taurine deficiency. Arch. Neurol. 1975, 32, 108–113. [Google Scholar] [CrossRef]
- Liu, J.; Juo, S.H.; Dewan, A.; Grunn, A.; Tong, X.; Brito, M.; Park, N.; Loth, J.E.; Kanyas, K.; Lerer, B.; et al. Evidence for a putative bipolar disorder locus on 2p13-16 and other potential loci on 4q31, 7q34, 8q13, 9q31, 10q21-24, 13q32, 14q21 and 17q11-12. Mol. Psychiatry 2003, 8, 333–342. [Google Scholar] [CrossRef]
- Coon, H.; Myles-Worsley, M.; Tiobech, J.; Hoff, M.; Rosenthal, J.; Bennett, P.; Reimherr, F.; Wender, P.; Dale, P.; Polloi, A.; et al. Evidence for a chromosome 2p13-14 schizophrenia susceptibility locus in families from Palau, Micronesia. Mol. Psychiatry 1998, 3, 521–527. [Google Scholar] [CrossRef]
- Goes, F.S.; Zandi, P.P.; Miao, K.; McMahon, F.J.; Steele, J.; Willour, V.L.; Mackinnon, D.F.; Mondimore, F.M.; Schweizer, B.; Nurnberger, J.I., Jr.; et al. Mood-incongruent psychotic features in bipolar disorder: Familial aggregation and suggestive linkage to 2p11-q14 and 13q21-33. Am. J. Psychiatry 2007, 164, 236–247. [Google Scholar] [CrossRef]
- Bulayeva, K.; Lencz, T.; Glatt, S.; Takumi, T.; Gurgenova, F.; Kawakami, H.; Bulayev, O. Mapping genes related to early onset major depressive disorder in Dagestan genetic isolates. Turk. Psikiyatri Derg. 2012, 23, 161–170. [Google Scholar]
- Allen, M.J.; Shan, X.; Caruccio, P.; Froggett, S.J.; Moffat, K.G.; Murphey, R.K. Targeted expression of truncated Glued disrupts giant fiber synapse formation in Drosophila. J. Neurosci. 1999, 19, 9374–9384. [Google Scholar] [CrossRef] [PubMed]
- Waterman-Storer, C.M.; Holzbaur, E.L. The product of the Drosophila gene, Glued, is the functional homologue of the p150Glued component of the vertebrate dynactin complex. J. Biol. Chem. 1996, 271, 1153–1159. [Google Scholar] [CrossRef] [PubMed]
- Garen, A.; Miller, B.R.; Paco-Larson, M.L. Mutations affecting functions of the Drosophila gene glued. Genetics 1984, 107, 645–655. [Google Scholar] [PubMed]
- Harte, P.J.; Kankel, D.R. Genetic analysis of mutations at the Glued locus and interacting loci in Drosophila melanogaster. Genetics 1982, 101, 477–501. [Google Scholar]
- Swaroop, A.; Paco-Larson, M.L.; Garen, A. Molecular genetics of a transposon-induced dominant mutation in the Drosophila locus Glued. Proc. Natl. Acad. Sci. USA 1985, 82, 1751–1755. [Google Scholar] [CrossRef]
- McGrail, M.; Gepner, J.; Silvanovich, A.; Ludmann, S.; Serr, M.; Hays, T.S. Regulation of cytoplasmic dynein function in vivo by the Drosophila Glued complex. J. Cell Biol. 1995, 131, 411–425. [Google Scholar] [CrossRef]
- Fan, S.S.; Ready, D.F. Glued participates in distinct microtubule-based activities in Drosophila eye development. Development 1997, 124, 1497–1507. [Google Scholar]
- Eaton, B.A.; Fetter, R.D.; Davis, G.W. Dynactin is necessary for synapse stabilization. Neuron 2002, 34, 729–741. [Google Scholar] [CrossRef]
- Carter, A.P.; Diamant, A.G.; Urnavicius, L. How dynein and dynactin transport cargos: A structural perspective. Curr. Opin. Struct. Biol. 2016, 37, 62–70. [Google Scholar] [CrossRef]
- Maquat, L.E. Nonsense-mediated mRNA decay in mammals. J. Cell Sci. 2005, 118, 1773–1776. [Google Scholar] [CrossRef]
- Fujiwara, T.; Morimoto, K.; Kakita, A.; Takahashi, H. Dynein and dynactin components modulate neurodegeneration induced by excitotoxicity. J. Neurochem. 2012, 122, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Morgan, S. Psychotic and Bipolar Disorders: Behavioral Disorders in Dementia. FP Essent. 2017, 455, 18–22. [Google Scholar] [PubMed]
- Diniz, B.S.; Teixeira, A.L.; Cao, F.; Gildengers, A.; Soares, J.C.; Butters, M.A.; Reynolds, C.F., 3rd. History of Bipolar Disorder and the Risk of Dementia: A Systematic Review and Meta-Analysis. Am. J. Geriatr. Psychiatry 2017, 25, 357–362. [Google Scholar] [CrossRef]
- Lewerenz, J.; Maher, P. Chronic Glutamate Toxicity in Neurodegenerative Diseases-What is the Evidence? Front. Neurosci. 2015, 9, 469. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Morimoto, K. Cooperative effect of p150Glued and microtubule stabilization to suppress excitotoxicity-induced axon degeneration. Biochem. Biophys. Res. Commun. 2012, 424, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.W. Glutamate neurotoxicity in cortical cell culture is calcium dependent. Neurosci. Lett. 1985, 58, 293–297. [Google Scholar] [CrossRef]
- Nickless, A.; Jackson, E.; Marasa, J.; Nugent, P.; Mercer, R.W.; Piwnica-Worms, D.; You, Z. Intracellular calcium regulates nonsense-mediated mRNA decay. Nat. Med. 2014, 20, 961–966. [Google Scholar] [CrossRef]
- Jun, C.; Choi, Y.; Lim, S.M.; Bae, S.; Hong, Y.S.; Kim, J.E.; Lyoo, I.K. Disturbance of the glutamatergic system in mood disorders. Exp. Neurobiol. 2014, 23, 28–35. [Google Scholar] [CrossRef]
- Rapoport, S.I.; Basselin, M.; Kim, H.W.; Rao, J.S. Bipolar disorder and mechanisms of action of mood stabilizers. Brain Res. Rev. 2009, 61, 185–209. [Google Scholar] [CrossRef]
- Sakabe, N.J.; de Souza, S.J. Sequence features responsible for intron retention in human. BMC Genom. 2007, 8, 59. [Google Scholar] [CrossRef]
- Wang, Z.; Burge, C.B. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. Rna 2008, 14, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Hammesfahr, B.; Kollmar, M. Evolution of the eukaryotic dynactin complex, the activator of cytoplasmic dynein. BMC Evol. Biol. 2012, 12, 95. [Google Scholar] [CrossRef] [PubMed]
- Ikenaka, K.; Kawai, K.; Katsuno, M.; Huang, Z.; Jiang, Y.M.; Iguchi, Y.; Kobayashi, K.; Kimata, T.; Waza, M.; Tanaka, F.; et al. dnc-1/dynactin 1 knockdown disrupts transport of autophagosomes and induces motor neuron degeneration. PLoS ONE 2013, 8, e54511. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.I.; Saiki, S.; Furuya, N.; Imamichi, Y.; Tsuboi, Y.; Hattori, N. p150glued deficiency impairs effective fusion between autophagosomes and lysosomes due to their redistribution to the cell periphery. Neurosci. Lett. 2019, 690, 181–187. [Google Scholar] [CrossRef]
- Sarkar, S.; Rubinsztein, D.C. Inositol and IP3 levels regulate autophagy: Biology and therapeutic speculations. Autophagy 2006, 2, 132–134. [Google Scholar] [CrossRef]
- Tritsaris, K.; Gromada, J.; Jorgensen, T.D.; Nauntofte, B.; Dissing, S. Reduction in the rate of inositol 1,4,5-trisphosphate synthesis in rat parotid acini by lithium. Arch. Oral Biol. 2001, 46, 365–373. [Google Scholar] [CrossRef]
- Li, Y.; McGreal, S.; Zhao, J.; Huang, R.; Zhou, Y.; Zhong, H.; Xia, M.; Ding, W.X. A cell-based quantitative high-throughput image screening identified novel autophagy modulators. Pharmacol. Res. 2016, 110, 35–49. [Google Scholar] [CrossRef]
- Sarkar, S.; Rubinsztein, D.C. Small molecule enhancers of autophagy for neurodegenerative diseases. Mol. Biosyst. 2008, 4, 895–901. [Google Scholar] [CrossRef]
- Park, J.; Chung, S.; An, H.; Kim, J.; Seo, J.; Kim, D.H.; Yoon, S.Y. Haloperidol and clozapine block formation of autophagolysosomes in rat primary neurons. Neuroscience 2012, 209, 64–73. [Google Scholar] [CrossRef]
- Zschocke, J.; Rein, T. Antidepressants encounter autophagy in neural cells. Autophagy 2011, 7, 1247–1248. [Google Scholar] [CrossRef]
- Zschocke, J.; Zimmermann, N.; Berning, B.; Ganal, V.; Holsboer, F.; Rein, T. Antidepressant drugs diversely affect autophagy pathways in astrocytes and neurons--dissociation from cholesterol homeostasis. Neuropsychopharmacology 2011, 36, 1754–1768. [Google Scholar] [CrossRef] [PubMed]
- Kara, N.Z.; Toker, L.; Agam, G.; Anderson, G.W.; Belmaker, R.H.; Einat, H. Trehalose induced antidepressant-like effects and autophagy enhancement in mice. Psychopharmacology 2013, 229, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Kara, N.Z.; Flaisher-Grinberg, S.; Anderson, G.W.; Agam, G.; Einat, H. Mood-stabilizing effects of rapamycin and its analog temsirolimus: Relevance to autophagy. Behav. Pharmacol. 2018, 29, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, B.; Sarkar, S.; Rubinsztein, D.C. Clearance of mutant aggregate-prone proteins by autophagy. Methods Mol. Biol. 2008, 445, 195–211. [Google Scholar] [PubMed]
- Milde, S.; Adalbert, R.; Elaman, M.H.; Coleman, M.P. Axonal transport declines with age in two distinct phases separated by a period of relative stability. Neurobiol. Aging 2015, 36, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, C.; Zhao, Z.; Aungst, S.; Sabirzhanov, B.; Faden, A.I.; Lipinski, M.M. Impaired autophagy flux is associated with neuronal cell death after traumatic brain injury. Autophagy 2014, 10, 2208–2222. [Google Scholar] [CrossRef]
- Zhou, R.; Lu, Y.; Han, Y.; Li, X.; Lou, H.; Zhu, L.; Zhen, X.; Duan, S. Mice heterozygous for cathepsin D deficiency exhibit mania-related behavior and stress-induced depression. Prog. Neuropsychopharmacol. Biol. Psychiatry 2015, 63, 110–118. [Google Scholar] [CrossRef]
- MacLaren, E.J.; Charlesworth, P.; Coba, M.P.; Grant, S.G. Knockdown of mental disorder susceptibility genes disrupts neuronal network physiology in vitro. Mol. Cell. Neurosci. 2011, 47, 93–99. [Google Scholar] [CrossRef]
- Qiu, R.; Zhang, J.; Xiang, X. p25 of the dynactin complex plays a dual role in cargo binding and dynactin regulation. J. Biol. Chem. 2018, 293, 15606–15619. [Google Scholar] [CrossRef]
- Wellcome Trust Case Control, C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007, 447, 661–678. [Google Scholar] [CrossRef]
- Rao, A.R.; Yourshaw, M.; Christensen, B.; Nelson, S.F.; Kerner, B. Rare deleterious mutations are associated with disease in bipolar disorder families. Mol. Psychiatry 2017, 22, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Settembre, C.; Fraldi, A.; Jahreiss, L.; Spampanato, C.; Venturi, C.; Medina, D.; de Pablo, R.; Tacchetti, C.; Rubinsztein, D.C.; Ballabio, A. A block of autophagy in lysosomal storage disorders. Hum. Mol. Genet. 2008, 17, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Staretz-Chacham, O.; Choi, J.H.; Wakabayashi, K.; Lopez, G.; Sidransky, E. Psychiatric and behavioral manifestations of lysosomal storage disorders. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010, 153, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- Wender, M.; Pruchnik-Wolinska, D.; Paprzycki, W.; Czartoryska, B. Familial metachromatic leukodystrophy as a cause of psychotic manifestations in young adults. Psychiatr. Pol. 1998, 32, 113–119. [Google Scholar] [PubMed]
- Maubert, A.; Hanon, C.; Metton, J.P. Adult onset Niemann-Pick type C disease and psychosis: Literature review. Encephale 2013, 39, 315–319. [Google Scholar] [CrossRef] [PubMed]
- MacQueen, G.M.; Rosebush, P.I.; Mazurek, M.F. Neuropsychiatric aspects of the adult variant of Tay-Sachs disease. J. Neuropsychiatry Clin. Neurosci. 1998, 10, 10–19. [Google Scholar] [CrossRef]
- Saxena, A.; Scaini, G.; Bavaresco, D.V.; Leite, C.; Valvassoria, S.S.; Carvalho, A.F.; Quevedo, J. Role of Protein Kinase C in Bipolar Disorder: A Review of the Current Literature. Mol. Neuropsychiatry 2017, 3, 108–124. [Google Scholar] [CrossRef]
- Zarate, C.A.; Manji, H.K. Protein kinase C inhibitors: Rationale for use and potential in the treatment of bipolar disorder. CNS Drugs 2009, 23, 569–582. [Google Scholar] [CrossRef]
- Abrial, E.; Betourne, A.; Etievant, A.; Lucas, G.; Scarna, H.; Lambas-Senas, L.; Haddjeri, N. Protein kinase C inhibition rescues manic-like behaviors and hippocampal cell proliferation deficits in the sleep deprivation model of mania. Int. J. Neuropsychopharmacol. 2014, 18, 31. [Google Scholar] [CrossRef]
- Yildiz, A.; Guleryuz, S.; Ankerst, D.P.; Ongur, D.; Renshaw, P.F. Protein kinase C inhibition in the treatment of mania: A double-blind, placebo-controlled trial of tamoxifen. Arch. Gen. Psychiatry 2008, 65, 255–263. [Google Scholar] [CrossRef]
- Jiang, H.; Cheng, D.; Liu, W.; Peng, J.; Feng, J. Protein kinase C inhibits autophagy and phosphorylates LC3. Biochem. Biophys. Res. Commun. 2010, 395, 471–476. [Google Scholar] [CrossRef]
- Jin, A.; Neufeld, T.P.; Choe, J. Kibra and aPKC regulate starvation-induced autophagy in Drosophila. Biochem. Biophys. Res. Commun. 2015, 468, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Liu, M. Atypical protein kinase C in cell motility. Cell. Mol. Life Sci. 2013, 70, 3057–3066. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Johns, L.A.; Allen, M.J. A modifier screen in the Drosophila eye reveals that aPKC interacts with Glued during central synapse formation. BMC Genet. 2009, 10, 77. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, A.A.; Sternberg, M.J.; Makarov, A.A. Polyproline-II helix in proteins: Structure and function. J. Mol. Biol. 2013, 425, 2100–2132. [Google Scholar] [CrossRef]
- Baures, P.W.; Pradhan, A.; Ojala, W.H.; Gleason, W.B.; Mishra, R.K.; Johnson, R.L. Synthesis and dopamine receptor modulating activity of unsubstituted and substituted triproline analogues of L-prolyl-L-leucyl-glycinamide (PLG). Bioorg. Med. Chem. Lett. 1999, 9, 2349–2352. [Google Scholar] [CrossRef]
- Raghavan, B.; Skoblenick, K.J.; Bhagwanth, S.; Argintaru, N.; Mishra, R.K.; Johnson, R.L. Allosteric modulation of the dopamine D2 receptor by Pro-Leu-Gly-NH2 peptidomimetics constrained in either a polyproline II helix or a type II β-turn conformation. J. Med. Chem. 2009, 52, 2043–2051. [Google Scholar] [CrossRef]
- Wang, D.; Ji, X.; Liu, J.; Li, Z.; Zhang, X. Dopamine Receptor Subtypes Differentially Regulate Autophagy. Int. J. Mol. Sci. 2018, 19, 1540. [Google Scholar] [CrossRef]
- Kurian, S.M.; Le-Niculescu, H.; Patel, S.D.; Bertram, D.; Davis, J.; Dike, C.; Yehyawi, N.; Lysaker, P.; Dustin, J.; Caligiuri, M.; et al. Identification of blood biomarkers for psychosis using convergent functional genomics. Mol. Psychiatry 2011, 16, 37–58. [Google Scholar] [CrossRef]
- Ashok, A.H.; Marques, T.R.; Jauhar, S.; Nour, M.M.; Goodwin, G.M.; Young, A.H.; Howes, O.D. The dopamine hypothesis of bipolar affective disorder: The state of the art and implications for treatment. Mol. Psychiatry 2017, 22, 666–679. [Google Scholar] [CrossRef]
- Seeman, P.; Schwarz, J.; Chen, J.F.; Szechtman, H.; Perreault, M.; McKnight, G.S.; Roder, J.C.; Quirion, R.; Boksa, P.; Srivastava, L.K.; et al. Psychosis pathways converge via D2high dopamine receptors. Synapse 2006, 60, 319–346. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Kralovics, R.; Rudzki, Z.; Grabowska, B.; Buser, A.S.; Olcaydu, D.; Gisslinger, H.; Tiedt, R.; Frank, P.; Okon, K.; et al. A de novo splice donor mutation in the thrombopoietin gene causes hereditary thrombocythemia in a Polish family. Haematologica 2008, 93, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Attanasio, C.; de Moerloose, P.; Antonarakis, S.E.; Morris, M.A.; Neerman-Arbez, M. Activation of multiple cryptic donor splice sites by the common congenital afibrinogenemia mutation, FGA IVS4 + 1 G-->T. Blood 2001, 97, 1879–1881. [Google Scholar] [CrossRef] [PubMed]
- Villate, O.; Ibarluzea, N.; Fraile-Bethencourt, E.; Valenzuela, A.; Velasco, E.A.; Grozeva, D.; Raymond, F.L.; Botella, M.P.; Tejada, M.I. Functional Analyses of a Novel Splice Variant in the CHD7 Gene, Found by Next Generation Sequencing, Confirm Its Pathogenicity in a Spanish Patient and Diagnose Him with CHARGE Syndrome. Front. Genet. 2018, 9, 7. [Google Scholar] [CrossRef] [PubMed]
- Seoighe, C.; Gehring, C. Heritability in the efficiency of nonsense-mediated mRNA decay in humans. PLoS ONE 2010, 5, e11657. [Google Scholar] [CrossRef] [PubMed]
- Zetoune, A.B.; Fontaniere, S.; Magnin, D.; Anczukow, O.; Buisson, M.; Zhang, C.X.; Mazoyer, S. Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC Genet. 2008, 9, 83. [Google Scholar] [CrossRef]
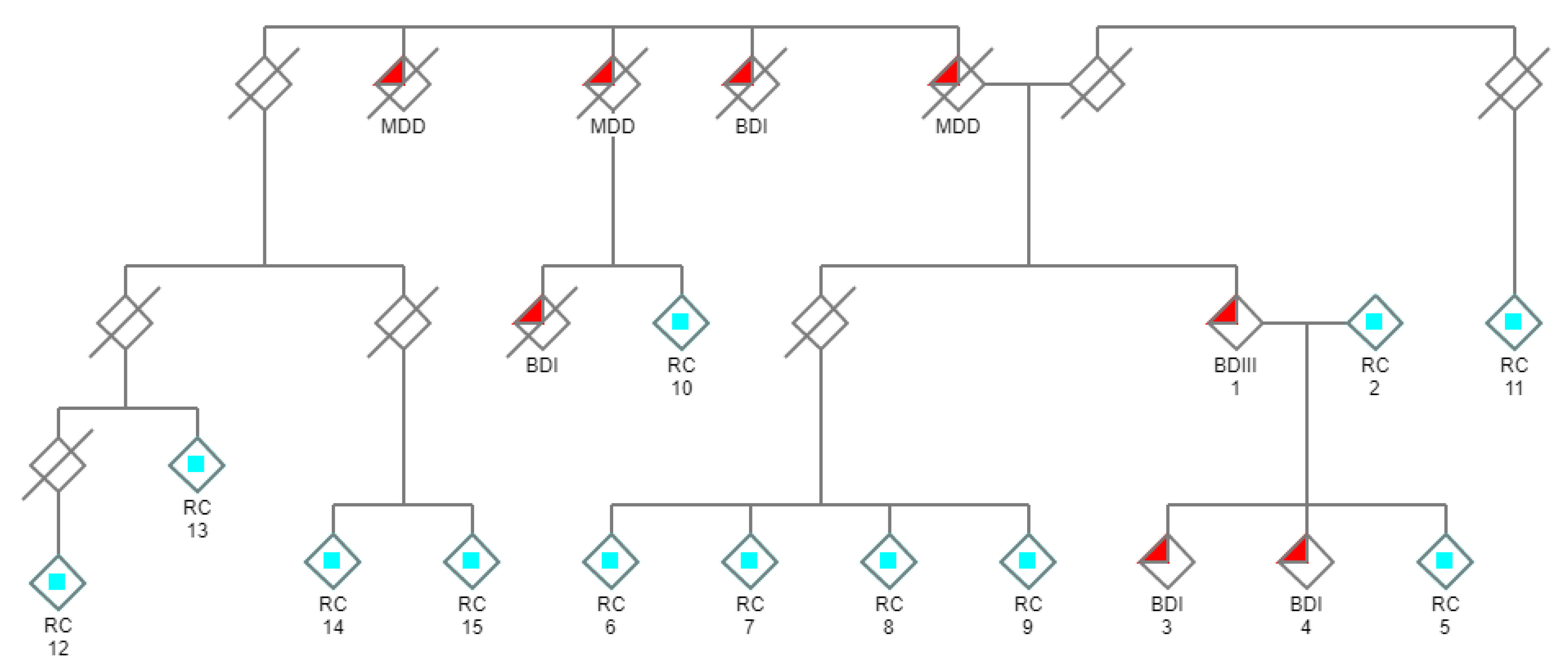
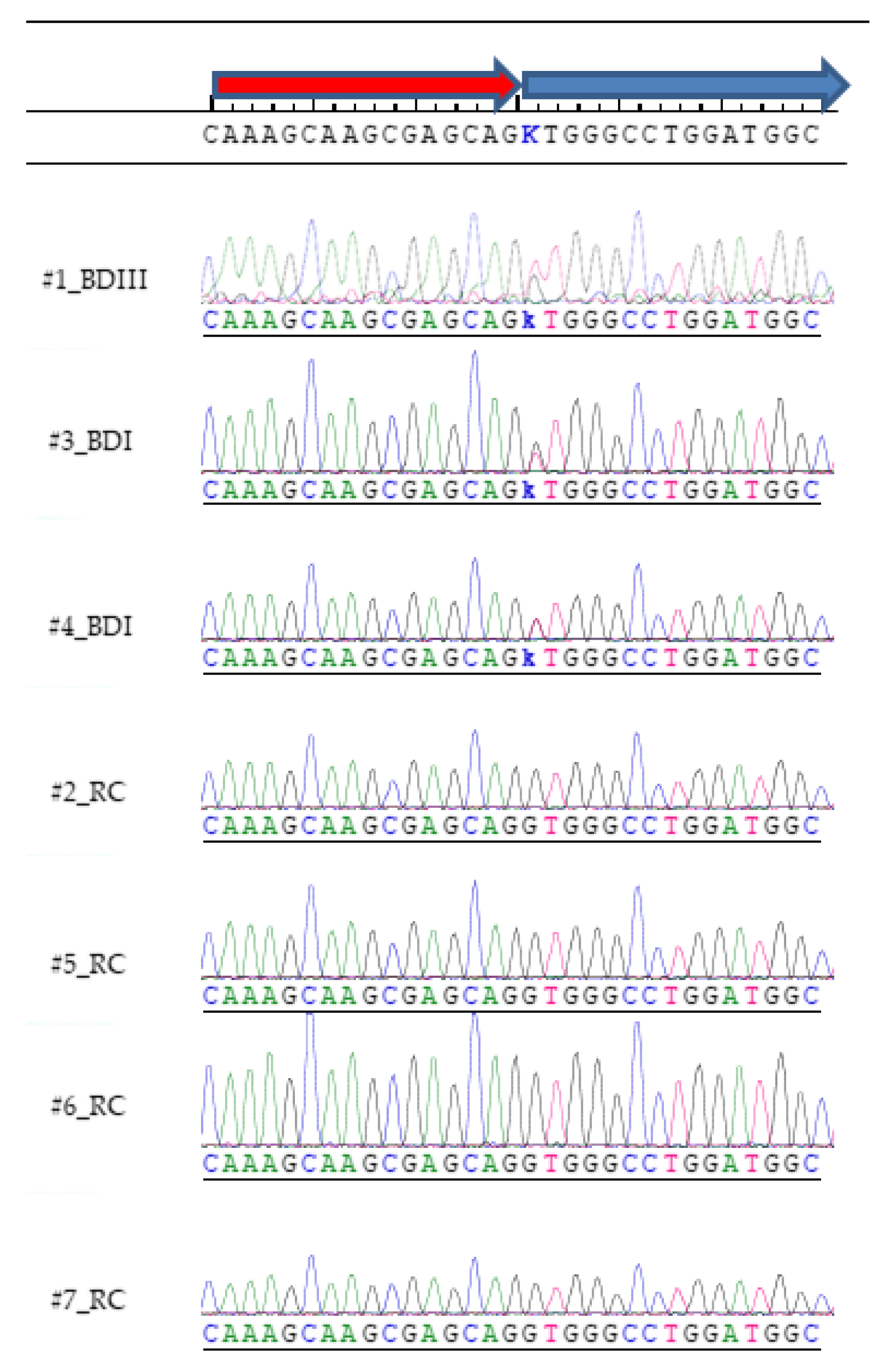
| Subject # | Diagnosis | DCTN1 Count C | DCTN1 Count A |
|---|---|---|---|
| 1 | BDIII | 31 | 51 |
| 3 | BDI | 51 | 58 |
| 4 | BDI | 76 | 64 |
| GERP++RS | PhastCons100way_Vertebrate | PhyloP100way_Vertebrate | |
|---|---|---|---|
| Score | 5.08 (range 1–6.18) | 1 (range 0–1) | 5.75 (range—20–10) |
| Conservation | Highly conserved | Highly conserved | Highly conserved |
| CADD | DANN | FATHMM-XF5 | MutationTaster | |
|---|---|---|---|---|
| Score | 25.6 [> 20 = 1% most pathogenic] | 0.9946 [range 0–1] | 0.996 [range 0–1] | 1.0 [range 0–1] |
| Prediction | Deleterious | Deleterious | Deleterious | Deleterious |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hallen, A.; Cooper, A.J.L. A Novel Cosegregating DCTN1 Splice Site Variant in a Family with Bipolar Disorder May Hold the Key to Understanding the Etiology. Genes 2020, 11, 446. https://doi.org/10.3390/genes11040446
Hallen A, Cooper AJL. A Novel Cosegregating DCTN1 Splice Site Variant in a Family with Bipolar Disorder May Hold the Key to Understanding the Etiology. Genes. 2020; 11(4):446. https://doi.org/10.3390/genes11040446
Chicago/Turabian StyleHallen, André, and Arthur J. L. Cooper. 2020. "A Novel Cosegregating DCTN1 Splice Site Variant in a Family with Bipolar Disorder May Hold the Key to Understanding the Etiology" Genes 11, no. 4: 446. https://doi.org/10.3390/genes11040446
APA StyleHallen, A., & Cooper, A. J. L. (2020). A Novel Cosegregating DCTN1 Splice Site Variant in a Family with Bipolar Disorder May Hold the Key to Understanding the Etiology. Genes, 11(4), 446. https://doi.org/10.3390/genes11040446





