Meta-Analysis of Transcriptomic Data of Dorsolateral Prefrontal Cortex and of Peripheral Blood Mononuclear Cells Identifies Altered Pathways in Schizophrenia
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Selection and Analysis
2.2. Evalution of the Involvement of Different Brain Cell Types in SCZ
2.3. Statistical Analysis
3. Results
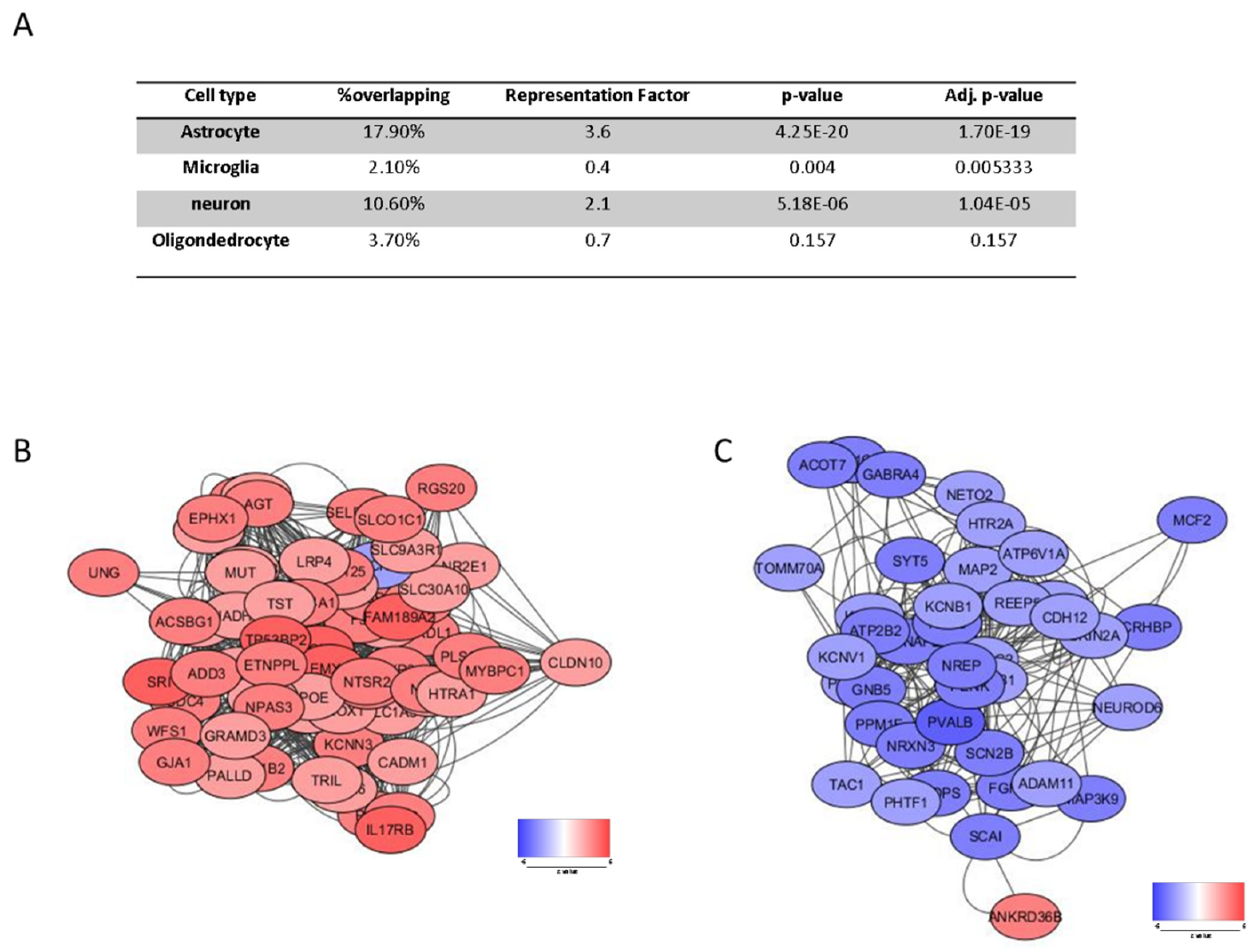
3.1. Identification of a SCZ Gene Profile in BA46
3.2. Cell Type Specific Enrichment
3.3. Perturbed Gene Expression Pattern in SCZ PBMCs and Functional Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mueser, K.T.; McGurk, S.R. Schizophrenia. Lancet 2004, 363, 2063–2072. [Google Scholar] [CrossRef]
- McGrath, J.; Saha, S.; Chant, D.; Welham, J. Schizophrenia: A concise overview of incidence, prevalence, and mortality. Epidemiol. Rev. 2008, 30, 67–76. [Google Scholar] [CrossRef]
- Guillozet-Bongaarts, A.L.; Hyde, T.M.; Dalley, R.A.; Hawrylycz, M.J.; Henry, A.; Hof, P.R.; Hohmann, J.; Jones, A.R.; Kuan, C.L.; Royall, J.; et al. Altered gene expression in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol. Psychiatry 2014, 19, 478–485. [Google Scholar] [CrossRef]
- Duan, J.; Sanders, A.R.; Gejman, P.V. Genome-wide approaches to schizophrenia. Brain Res. Bull. 2010, 83, 93–102. [Google Scholar] [CrossRef][Green Version]
- Kleinman, J.E.; Law, A.J.; Lipska, B.K.; Hyde, T.M.; Ellis, J.K.; Harrison, P.J.; Weinberger, D.R. Genetic neuropathology of schizophrenia: New approaches to an old question and new uses for postmortem human brains. Biol. Psychiatry 2011, 69, 140–145. [Google Scholar] [CrossRef]
- Selemon, L.D.; Rajkowska, G.; Goldman-Rakic, P.S. Abnormally high neuronal density in the schizophrenic cortex. A morphometric analysis of prefrontal area 9 and occipital area 17. Arch. Gen. Psychiatry 1995, 52, 805–818. [Google Scholar] [CrossRef]
- Nathaniel-James, D.A.; Frith, C.D. The role of the dorsolateral prefrontal cortex: Evidence from the effects of contextual constraint in a sentence completion task. Neuroimage 2002, 16, 1094–1102. [Google Scholar] [CrossRef]
- Snow, P.J. The structural and functional organization of cognition. Front. Hum. Neurosci. 2016, 10, 501. [Google Scholar] [CrossRef]
- Goldman-Rakic, P.S.; Selemon, L.D. Functional and anatomical aspects of prefrontal pathology in schizophrenia. Schizophr. Bull. 1997, 23, 437–458. [Google Scholar] [CrossRef]
- Tamminga, C.A.; Selemon, L.D. Increased cortical neuronal density in schizophrenia. Am. J. Psychiatry 2004, 161, 1564. [Google Scholar]
- Selemon, L.D.; Mrzljak, J.; Kleinman, J.E.; Herman, M.M.; Goldman-Rakic, P.S. Regional specificity in the neuropathologic substrates of schizophrenia: A morphometric analysis of Broca’s area 44 and area 9. Arch. Gen. Psychiatry 2003, 60, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Selemon, L.D.; Rajkowska, G.; Goldman-Rakic, P.S. Elevated neuronal density in prefrontal area 46 in brains from schizophrenic patients: Application of a three-dimensional, stereologic counting method. J. Comp. Neurol. 1998, 392, 402–412. [Google Scholar] [CrossRef]
- Jones, L.B. Recent cytoarchitechtonic changes in the prefrontal cortex of schizophrenics. Front. Biosci. 2001, 6, E148–E153. [Google Scholar] [CrossRef]
- Forbes, N.F.; Carrick, L.A.; McIntosh, A.M.; Lawrie, S.M. Working memory in schizophrenia: A meta-analysis. Psychol. Med. 2009, 39, 889–905. [Google Scholar] [CrossRef]
- Heinrichs, R.W.; Zakzanis, K.K. Neurocognitive deficit in schizophrenia: A quantitative review of the evidence. Neuropsychology 1998, 12, 426–445. [Google Scholar] [CrossRef]
- Kaminski, J.; Gleich, T.; Fukuda, Y.; Katthagen, T.; Gallinat, J.; Heinz, A.; Schlagenhauf, F. Association of cortical glutamate and working memory activation in patients with schizophrenia: A multimodal proton magnetic resonance spectroscopy and functional magnetic resonance imaging study. Biol. Psychiatry 2020, 87, 225–233. [Google Scholar] [CrossRef]
- Lee, J.; Folley, B.S.; Gore, J.; Park, S. Origins of spatial working memory deficits in schizophrenia: An event-related FMRI and near-infrared spectroscopy study. PLoS ONE 2008, 3, e1760. [Google Scholar] [CrossRef]
- Mexal, S.; Berger, R.; Logel, J.; Ross, R.G.; Freedman, R.; Leonard, S. Differential regulation of alpha7 nicotinic receptor gene (CHRNA7) expression in schizophrenic smokers. J. Mol. Neurosci. 2010, 40, 185–195. [Google Scholar] [CrossRef]
- Lanz, T.A.; Reinhart, V.; Sheehan, M.J.; Rizzo, S.J.S.; Bove, S.E.; James, L.C.; Volfson, D.; Lewis, D.A.; Kleiman, R.J. Postmortem transcriptional profiling reveals widespread increase in inflammation in schizophrenia: A comparison of prefrontal cortex, striatum, and hippocampus among matched tetrads of controls with subjects diagnosed with schizophrenia, bipolar or major depressive disorder. Transl. Psychiatry 2019, 9, 151. [Google Scholar]
- Iwamoto, K.; Bundo, M.; Kato, T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum. Mol. Genet. 2005, 14, 241–253. [Google Scholar] [CrossRef]
- Mattioli, M.; Agnelli, L.; Fabris, S.; Baldini, L.; Morabito, F.; Bicciato, S.; Verdelli, D.; Intini, D.; Nobili, L.; Cro, L.; et al. Gene expression profiling of plasma cell dyscrasias reveals molecular patterns associated with distinct IGH translocations in multiple myeloma. Oncogene 2005, 24, 2461–2473. [Google Scholar] [CrossRef] [PubMed]
- Toro-Domínguez, D.; Martorell-Marugán, J.; López-Domínguez, R.; García-Moreno, A.; González-Rumayor, V.; Alarcón-Riquelme, M.E.; Carmona-Sáez, P. ImaGEO: Integrative gene expression meta-analysis from GEO database. Bioinformatics 2019, 35, 880–882. [Google Scholar] [CrossRef] [PubMed]
- Fatemi, S.H.; Reutiman, T.J.; Folsom, T.D.; Bell, C.; Nos, L.; Fried, P.; Pearce, D.A.; Singh, S.; Siderovski, D.P.; Willard, F.S.; et al. Chronic olanzapine treatment causes differential expression of genes in frontal cortex of rats as revealed by DNA microarray technique. Neuropsychopharmacology 2006, 31, 1888–1899. [Google Scholar] [CrossRef]
- Bousman, C.A.; Chana, G.; Glatt, S.J.; Chandler, S.D.; Lucero, G.R.; Tatro, E.; May, T.; Lohr, J.B.; Kremen, W.S.; Tsuang, M.T.; et al. Preliminary evidence of ubiquitin proteasome system dysregulation in schizophrenia and bipolar disorder: Convergent pathway analysis findings from two independent samples. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2010, 153, 494–502. [Google Scholar] [CrossRef]
- Van Beveren, N.J.M.; Buitendijk, G.H.S.; Swagemakers, S.; Krab, L.C.; Röder, C.; de Haan, L.; van der Spek, P.; Elgersma, Y. Marked reduction of AKT1 expression and deregulation of AKT1-associated pathways in peripheral blood mononuclear cells of schizophrenia patients. PLoS ONE 2012, 7, e32618. [Google Scholar] [CrossRef]
- McKenzie, A.T.; Wang, M.; Hauberg, M.E.; Fullard, J.F.; Kozlenkov, A.; Keenan, A.; Hurd, Y.L.; Dracheva, S.; Casaccia, P.; Roussos, P.; et al. Brain cell type specific gene expression and co-expression network architectures. Sci. Rep. 2018, 8, 8868. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Lindenberg, A.; Miletich, R.S.; Kohn, P.D.; Esposito, G.; Carson, R.E.; Quarantelli, M.; Weinberger, D.R.; Berman, K.F. Reduced prefrontal activity predicts exaggerated striatal dopaminergic function in schizophrenia. Nat. Neurosci. 2002, 5, 267–271. [Google Scholar] [CrossRef]
- Sivarajah, A.; Collino, M.; Yasin, M.; Benetti, E.; Gallicchio, M.; Mazzon, E.; Cuzzocrea, S.; Fantozzi, R.; Thiemermann, C. Anti-apoptotic and anti-inflammatory effects of hydrogen sulfide in a rat model of regional myocardial I/R. Shock 2009, 31, 267–274. [Google Scholar] [CrossRef]
- Henseler, I.; Falkai, P.; Gruber, O. Disturbed functional connectivity within brain networks subserving domain-specific subcomponents of working memory in schizophrenia: Relation to performance and clinical symptoms. J. Psychiatr. Res. 2010, 44, 364–372. [Google Scholar] [CrossRef]
- Altar, C.A.; Jurata, L.W.; Charles, V.; Lemire, A.; Liu, P.; Bukhman, Y.; Young, T.A.; Bullard, J.; Yokoe, H.; Webster, M.J.; et al. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol. Psychiatry 2005, 58, 85–96. [Google Scholar] [CrossRef]
- Maycox, P.R.; Kelly, F.; Taylor, A.; Bates, S.; Reid, J.; Logendra, R.; Barnes, M.R.; Larminie, C.; Jones, N.; Lennon, M.; et al. Analysis of gene expression in two large schizophrenia cohorts identifies multiple changes associated with nerve terminal function. Mol. Psychiatry 2009, 14, 1083–1094. [Google Scholar] [CrossRef]
- Barnes, M.R.; Huxley-Jones, J.; Maycox, P.R.; Lennon, M.; Thornber, A.; Kelly, F.; Bates, S.; Taylor, A.; Reid, J.; Jones, N.; et al. Transcription and pathway analysis of the superior temporal cortex and anterior prefrontal cortex in schizophrenia. J. Neurosci. Res. 2011, 89, 1218–1227. [Google Scholar] [CrossRef]
- Middleton, F.A.; Peng, L.; Lewis, D.A.; Levitt, P.; Mirnics, K. Altered expression of 14-3-3 genes in the prefrontal cortex of subjects with schizophrenia. Neuropsychopharmacology 2005, 30, 974–983. [Google Scholar] [CrossRef][Green Version]
- Candido, S.; Lupo, G.; Pennisi, M.; Basile, M.; Anfuso, C.; Petralia, M.; Gattuso, G.; Vivarelli, S.; Spandidos, D.; Libra, M.; et al. The analysis of miRNA expression profiling datasets reveals inverse microRNA patterns in glioblastoma and Alzheimer’s disease. Oncol. Rep. 2019, 42, 911–922. [Google Scholar] [CrossRef]
- Lombardo, S.D.; Mazzon, E.; Basile, M.S.; Cavalli, E.; Bramanti, P.; Nania, R.; Fagone, P.; Nicoletti, F.; Petralia, M.C. Upregulation of IL-1 receptor antagonist in a mouse model of migraine. Brain Sci. 2019, 9, 172. [Google Scholar] [CrossRef]
- Lombardo, S.D.; Presti, M.; Mangano, K.; Petralia, M.C.; Basile, M.S.; Libra, M.; Candido, S.; Fagone, P.; Mazzon, E.; Nicoletti, F.; et al. Prediction of PD-L1 expression in neuroblastoma via computational modeling. Brain Sci. 2019, 9, 221. [Google Scholar] [CrossRef]
- Petralia, M.C.; Mazzon, E.; Fagone, P.; Falzone, L.; Bramanti, P.; Nicoletti, F.; Basile, M.S. Retrospective follow-up analysis of the transcriptomic patterns of cytokines, cytokine receptors and chemokines at preconception and during pregnancy, in women with post-partum depression. Exp. Ther. Med. 2019, 18, 2055–2062. [Google Scholar] [CrossRef]
- Lombardo, S.D.; Mazzon, E.; Mangano, K.; Basile, M.S.; Cavalli, E.; Mammana, S.; Fagone, P.; Nicoletti, F.; Petralia, M.C. Transcriptomic analysis reveals involvement of the macrophage migration inhibitory factor gene network in duchenne muscular dystrophy. Genes (Basel) 2019, 10, 939. [Google Scholar] [CrossRef]
- Lombardo, S.D.; Mazzon, E.; Basile, M.S.; Campo, G.; Corsico, F.; Presti, M.; Bramanti, P.; Mangano, K.; Petralia, M.C.; Nicoletti, F.; et al. Modulation of tetraspanin 32 (TSPAN32) expression in T cell-mediated immune responses and in multiple sclerosis. Int. J. Mol. Sci. 2019, 20, 4323. [Google Scholar] [CrossRef]
- Petralia, M.C.; Mazzon, E.; Basile, M.S.; Cutuli, M.; Di Marco, R.; Scandurra, F.; Saraceno, A.; Fagone, P.; Nicoletti, F.; Mangano, K. Effects of treatment with the hypomethylating agent 5-aza-2’-deoxycytidine in murine type II collagen-induced arthritis. Pharmaceuticals 2019, 12, 174. [Google Scholar] [CrossRef]
- Fagone, P.; Mazzon, E.; Mammana, S.; Di Marco, R.; Spinasanta, F.; Basile, M.; Petralia, M.; Bramanti, P.; Nicoletti, F.; Mangano, K. Identification of CD4+ T cell biomarkers for predicting the response of patients with relapsing-remitting multiple sclerosis to natalizumab treatment. Mol. Med. Rep. 2019. [Google Scholar] [CrossRef]
- Nicoletti, F.; Mazzon, E.; Fagone, P.; Mangano, K.; Mammana, S.; Cavalli, E.; Basile, M.S.; Bramanti, P.; Scalabrino, G.; Lange, A.; et al. Prevention of clinical and histological signs of MOG-induced experimental allergic encephalomyelitis by prolonged treatment with recombinant human EGF. J. Neuroimmunol. 2019, 332, 224–232. [Google Scholar] [CrossRef]
- Cavalli, E.; Mazzon, E.; Basile, M.S.; Mangano, K.; Di Marco, R.; Bramanti, P.; Nicoletti, F.; Fagone, P.; Petralia, M.C. Upregulated expression of macrophage migration inhibitory factor, its analogue D-dopachrome tautomerase, and the CD44 receptor in peripheral CD4 T cells from clinically isolated syndrome patients with rapid conversion to clinical defined multiple sclerosis. Medicina (Buenos Aires) 2019, 55, 667. [Google Scholar] [CrossRef]
- Cavalli, E.; Mazzon, E.; Basile, M.S.; Mammana, S.; Pennisi, M.; Fagone, P.; Kalfin, R.; Martinovic, V.; Ivanovic, J.; Andabaka, M.; et al. In silico and in vivo analysis of IL37 in multiple sclerosis reveals its probable homeostatic role on the clinical activity, disability, and treatment with fingolimod. Molecules 2019, 25, 20. [Google Scholar] [CrossRef]
- Günther, S.; Fagone, P.; Jalce, G.; Atanasov, A.G.; Guignabert, C.; Nicoletti, F. Role of MIF and D-DT in immune-inflammatory, autoimmune, and chronic respiratory diseases: From pathogenic factors to therapeutic targets. Drug Discov. Today 2019, 24, 428–439. [Google Scholar] [CrossRef]
- Basile, M.S.; Mazzon, E.; Mangano, K.; Pennisi, M.; Petralia, M.C.; Lombardo, S.D.; Nicoletti, F.; Fagone, P.; Cavalli, E. Impaired expression of tetraspanin 32 (TSPAN32) in memory T cells of patients with multiple sclerosis. Brain Sci. 2020, 10, 52. [Google Scholar] [CrossRef]
- Fagone, P.; Mangano, K.; Mammana, S.; Cavalli, E.; Di Marco, R.; Barcellona, M.L.; Salvatorelli, L.; Magro, G.; Nicoletti, F. Carbon monoxide-releasing molecule-A1 (CORM-A1) improves clinical signs of experimental autoimmune uveoretinitis (EAU) in rats. Clin. Immunol. 2015, 157, 198–204. [Google Scholar] [CrossRef]
- Cavalli, E.; Mazzon, E.; Mammana, S.; Basile, M.S.; Lombardo, S.D.; Mangano, K.; Bramanti, P.; Nicoletti, F.; Fagone, P.; Petralia, M.C. Overexpression of macrophage migration inhibitory factor and its homologue D-dopachrome tautomerase as negative prognostic factor in neuroblastoma. Brain Sci. 2019, 9, 284. [Google Scholar] [CrossRef] [PubMed]
- Petralia, M.C.; Mazzon, E.; Fagone, P.; Russo, A.; Longo, A.; Avitabile, T.; Nicoletti, F.; Reibaldi, M.; Basile, M.S. Characterization of the pathophysiological role of CD47 in uveal melanoma. Molecules 2019, 24, 2450. [Google Scholar] [CrossRef] [PubMed]
- Mangano, K.; Cavalli, E.; Mammana, S.; Basile, M.S.; Caltabiano, R.; Pesce, A.; Puleo, S.; Atanasov, A.G.; Magro, G.; Nicoletti, F.; et al. Involvement of the Nrf2/HO-1/CO axis and therapeutic intervention with the CO-releasing molecule CORM-A1, in a murine model of autoimmune hepatitis. J. Cell. Physiol. 2018, 233, 4156–4165. [Google Scholar] [CrossRef]
- Mammana, S.; Bramanti, P.; Mazzon, E.; Cavalli, E.; Basile, M.S.; Fagone, P.; Petralia, M.C.; McCubrey, J.A.; Nicoletti, F.; Mangano, K. Preclinical evaluation of the PI3K/Akt/mTOR pathway in animal models of multiple sclerosis. Oncotarget 2018, 9, 8263–8277. [Google Scholar] [CrossRef]
- Mammana, S.; Fagone, P.; Cavalli, E.; Basile, M.S.; Petralia, M.C.; Nicoletti, F.; Bramanti, P.; Mazzon, E. The role of macrophages in neuroinflammatory and neurodegenerative pathways of alzheimer’s disease, amyotrophic lateral sclerosis, and multiple sclerosis: Pathogenetic cellular effectors and potential therapeutic targets. Int. J. Mol. Sci. 2018, 19, 831. [Google Scholar] [CrossRef]
- Petralia, M.C.; Mazzon, E.; Fagone, P.; Basile, M.S.; Lenzo, V.; Quattropani, M.C.; Bendtzen, K.; Nicoletti, F. Pathogenic contribution of the Macrophage migration inhibitory factor family to major depressive disorder and emerging tailored therapeutic approaches. J. Affect. Disord. 2020, 263, 15–24. [Google Scholar] [CrossRef]
- Fagone, P.; Mangano, K.; Mammana, S.; Pesce, A.; Pesce, A.; Caltabiano, R.; Giorlandino, A.; Rosanna, C.; Portale, T.R.; Cavalli, E.; et al. Identification of novel targets for the diagnosis and treatment of liver fibrosis. Int. J. Mol. Med. 2015, 36. [Google Scholar] [CrossRef]
- Basile, M.; Mazzon, E.; Krajnovic, T.; Draca, D.; Cavalli, E.; Al-Abed, Y.; Bramanti, P.; Nicoletti, F.; Mijatovic, S.; Maksimovic-Ivanic, D. Anticancer and differentiation properties of the nitric oxide derivative of lopinavir in human glioblastoma cells. Molecules 2018, 23, 2463. [Google Scholar] [CrossRef]
- Petralia, M.C.; Battaglia, G.; Bruno, V.; Pennisi, M.; Mangano, K.; Lombardo, S.D.; Fagone, P.; Cavalli, E.; Saraceno, A.; Nicoletti, F.; et al. The role of macrophage migration inhibitory factor in alzheimer’s disease: Conventionally pathogenetic or unconventionally protective? Molecules 2020, 25, 291. [Google Scholar] [CrossRef]
- Mammana, S.; Cavalli, E.; Gugliandolo, A.; Silvestro, S.; Pollastro, F.; Bramanti, P.; Mazzon, E. Could the combination of two non-psychotropic cannabinoids counteract neuroinflammation? Effectiveness of cannabidiol associated with cannabigerol. Medicina (Kaunas) 2019, 55, 747. [Google Scholar] [CrossRef]
- Schepici, G.; Cavalli, E.; Bramanti, P.; Mazzon, E. Autism spectrum disorder and miRNA: An overview of experimental models. Brain Sci. 2019, 9, 265. [Google Scholar] [CrossRef]
- Gusev, A.; Mancuso, N.; Won, H.; Kousi, M.; Finucane, H.K.; Reshef, Y.; Song, L.; Safi, A.; McCarroll, S.; Neale, B.M.; et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 2018, 50, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Huckins, L.M.; Dobbyn, A.; Ruderfer, D.M.; Hoffman, G.; Wang, W.; Pardiñas, A.F.; Rajagopal, V.M.; Als, T.D.; Nguyen, H.T.; Girdhar, K.; et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat. Genet. 2019, 51, 659–674. [Google Scholar] [CrossRef] [PubMed]
- Gandal, M.J.; Zhang, P.; Hadjimichael, E.; Walker, R.L.; Chen, C.; Liu, S.; Won, H.; Van Bakel, H.; Varghese, M.; Wang, Y.; et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 2018, 362. [Google Scholar] [CrossRef] [PubMed]
- Fromer, M.; Roussos, P.; Sieberts, S.K.; Johnson, J.S.; Kavanagh, D.H.; Perumal, T.M.; Ruderfer, D.M.; Oh, E.C.; Topol, A.; Shah, H.R.; et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016, 19, 1442–1453. [Google Scholar] [CrossRef] [PubMed]
- Hall, L.S.; Medway, C.W.; Pain, O.; Pardiñas, A.F.; Rees, E.G.; Escott-Price, V.; Pocklington, A.; Bray, N.J.; Holmans, P.A.; Walters, J.T.R.; et al. A transcriptome-wide association study implicates specific pre- and post-synaptic abnormalities in schizophrenia. Hum. Mol. Genet. 2020, 29, 159–167. [Google Scholar] [CrossRef]
- Kyosseva, S.V. Differential expression of mitogen-activated protein kinases and immediate early genes fos and jun in thalamus in schizophrenia. Prog. Neuropsychopharmacol. Biol. Psychiatry 2004, 28, 997–1006. [Google Scholar] [CrossRef]
- Kyosseva, S.V.; Elbein, A.D.; Griffin, W.S.; Mrak, R.E.; Lyon, M.; Karson, C.N. Mitogen-activated protein kinases in schizophrenia. Biol. Psychiatry 1999, 46, 689–696. [Google Scholar] [CrossRef]
- David Sweatt, J. The neuronal MAP kinase cascade: A biochemical signal integration system subserving synaptic plasticity and memory. J. Neurochem. 2001, 76, 1–10. [Google Scholar] [CrossRef]
- Gelinas, J.N.; Banko, J.L.; Peters, M.M.; Klann, E.; Weeber, E.J.; Nguyen, P.V. Activation of exchange protein activated by cyclic-AMP enhances long-lasting synaptic potentiation in the hippocampus. Learn. Mem. 2008, 15, 403–411. [Google Scholar] [CrossRef]
- Reichenberg, A. The assessment of neuropsychological functioning in schizophrenia. Dialogues Clin. Neurosci. 2010, 12, 383–392. [Google Scholar] [PubMed]
- Sands, W.A.; Palmer, T.M. Regulating gene transcription in response to cyclic AMP elevation. Cell Signal. 2008, 20, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Winchester, C.L.; Ohzeki, H.; Vouyiouklis, D.A.; Thompson, R.; Penninger, J.M.; Yamagami, K.; Norrie, J.D.; Hunter, R.; Pratt, J.A.; Morris, B.J. Converging evidence that sequence variations in the novel candidate gene MAP2K7 (MKK7) are functionally associated with schizophrenia. Hum. Mol. Genet. 2012, 21, 4910–4921. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.J.; Lee, B.D.; Kim, C. Pilot study for family-based association analysis of schizophrenia in a Korean population: Analysis for candidate genes positionally on chromosome 18q21. Asia Pac. Psychiatry 2015, 7, 268–275. [Google Scholar] [CrossRef]
- Funk, A.J.; McCullumsmith, R.E.; Haroutunian, V.; Meador-Woodruff, J.H. Abnormal activity of the MAPK- and cAMP-associated signaling pathways in frontal cortical areas in postmortem brain in schizophrenia. Neuropsychopharmacology 2012, 37, 896–905. [Google Scholar] [CrossRef]
- Hirayama-Kurogi, M.; Takizawa, Y.; Kunii, Y.; Matsumoto, J.; Wada, A.; Hino, M.; Akatsu, H.; Hashizume, Y.; Yamamoto, S.; Kondo, T.; et al. Downregulation of GNA13-ERK network in prefrontal cortex of schizophrenia brain identified by combined focused and targeted quantitative proteomics. J. Proteom. 2017, 158, 31–42. [Google Scholar] [CrossRef]
- Coulthard, L.R.; Taylor, J.C.; Eyre, S.; Robinson, J.I.; Wilson, A.G.; Isaacs, J.D.; Hyrich, K.; Emery, P.; Barton, A.; Barrett, J.H.; et al. Genetic variants within the MAP kinase signaling network and anti-TNF treatment response in Rheumatoid arthritis patients. Ann. Rheum. Dis. 2011, 70, 98–103. [Google Scholar] [CrossRef]
- Kanbe, K.; Chen, Q.; Nakamura, A.; Hobo, K. Inhibition of MAP kinase in synovium by treatment with tocilizumab in rheumatoid arthritis. Clin. Rheumatol. 2011, 30, 1407–1413. [Google Scholar] [CrossRef]
- Volin, M.V.; Huynh, N.; Klosowska, K.; Chong, K.K.; Woods, J.M. Fractalkine is a novel chemoattractant for rheumatoid arthritis fibroblast-like synoviocyte signaling through MAP kinases and Akt. Arthritis Rheum. 2007, 56, 2512–2522. [Google Scholar] [CrossRef]
- Kunisch, E.; Gandesiri, M.; Fuhrmann, R.; Roth, A.; Winter, R.; Kinne, R.W. Predominant activation of MAP kinases and pro-destructive/pro-inflammatory features by TNF α in early-passage synovial fibroblasts via TNF receptor-1: Failure of p38 inhibition to suppress matrix metalloproteinase-1 in rheumatoid arthritis. Ann. Rheum. Dis. 2007, 66, 1043–1051. [Google Scholar] [CrossRef]
- Chen, S.-F.; Wang, L.-Y.; Chiang, J.-H.; Hsu, C.-Y.; Shen, Y.-C. Assessing whether the association between rheumatoid arthritis and schizophrenia is bidirectional: A nationwide population-based cohort study. Sci. Rep. 2019, 9, 4493. [Google Scholar] [CrossRef] [PubMed]
- Malavia, T.A.; Chaparala, S.; Wood, J.; Chowdari, K.; Prasad, K.M.; McClain, L.; Jegga, A.G.; Ganapathiraju, M.K.; Nimgaonkar, V.L. Generating testable hypotheses for schizophrenia and rheumatoid arthritis pathogenesis by integrating epidemiological, genomic, and protein interaction data. NPJ Schizophr. 2017, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Byrne, E.M.; Hultman, C.M.; Kähler, A.; Vinkhuyzen, A.A.E.; Ripke, S.; Andreassen, O.A.; Frisell, T.; Gusev, A.; Hu, X.; et al. New data and an old puzzle: The negative association between schizophrenia and rheumatoid arthritis. Int. J. Epidemiol. 2015, 44, 1706–1721. [Google Scholar] [CrossRef] [PubMed]
- Grinshpoon, A.; Barchana, M.; Ponizovsky, A.; Lipshitz, I.; Nahon, D.; Tal, O.; Weizman, A.; Levav, I. Cancer in schizophrenia: Is the risk higher or lower? Schizophr. Res. 2005, 73, 333–341. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, N.; Li, H.; Liu, S.; Chen, X.; Yu, S.; Wu, N.; Bian, X.W.; Shen, H.Y.; Li, C.; et al. Promoting oligodendroglial-oriented differentiation of glioma stem cell: A repurposing of quetiapine for the treatment of malignant glioma. Oncotarget 2017, 8, 37511–37524. [Google Scholar] [CrossRef]
- Choi, K.H.; Elashoff, M.; Higgs, B.W.; Song, J.; Kim, S.; Sabunciyan, S.; Diglisic, S.; Yolken, R.H.; Knable, M.B.; Fuller, E.F.; et al. Putative psychosis genes in the prefrontal cortex: Combined analysis of gene expression microarrays. BMC Psychiatry 2008, 8, 87. [Google Scholar] [CrossRef]
- Ebadi, M.; Iversen, P.L.; Hao, R.; Cerutis, D.R.; Rojas, P.; Happe, H.K.; Murrin, L.C.; Pfeiffer, R.F. Expression and regulation of brain metallothionein. Neurochem. Int. 1995, 27, 1–22. [Google Scholar] [CrossRef]
- Roy, S.; Dasgupta, A.; Banerjee, U.; Chowdhury, P.; Mukhopadhyay, A.; Saha, G.; Singh, O. Role of membrane cholesterol and lipid peroxidation in regulating the Na+/K+-ATPase activity in schizophrenia. Indian J. Psychiatry 2016, 58, 317–325. [Google Scholar]
- Kurauchi, Y.; Hisatsune, A.; Seki, T.; Katsuki, H. Na+, K+-ATPase dysfunction causes cerebrovascular endothelial cell degeneration in rat prefrontal cortex slice cultures. Brain Res. 2016, 1644, 249–257. [Google Scholar] [CrossRef]
- Corti, C.; Xuereb, J.H.; Crepaldi, L.; Corsi, M.; Michielin, F.; Ferraguti, F. Altered levels of glutamatergic receptors and Na+/K+ ATPase-α1 in the prefrontal cortex of subjects with schizophrenia. Schizophr. Res. 2011, 128, 7–14. [Google Scholar] [CrossRef]
- O’Donovan, S.M.; Sullivan, C.; Koene, R.; Devine, E.; Hasselfeld, K.; Moody, C.L.; McCullumsmith, R.E. Cell-subtype-specific changes in adenosine pathways in schizophrenia. Neuropsychopharmacology 2018, 43, 1667–1674. [Google Scholar] [CrossRef] [PubMed]
- Venkataramaiah, C. Modulations in the ATPases during ketamine-induced schizophrenia and regulatory effect of “3-(3, 4-dimethoxy phenyl) -1- (4-methoxyphenyl) prop-2-en-1-one”: An in vivo and in silico studies. J. Recept. Signal Transduct. 2020, 40, 148–156. [Google Scholar] [CrossRef]
- Hodes, A.; Rosen, H.; Cohen-Ben Ami, H.; Lichtstein, D. Na+, K+-ATPase α3 isoform in frontal cortex GABAergic neurons in psychiatric diseases. J. Psychiatr. Res. 2019, 115, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Toker, L.; Mancarci, B.O.; Tripathy, S.; Pavlidis, P. Transcriptomic evidence for alterations in astrocytes and parvalbumin interneurons in subjects with bipolar disorder and schizophrenia. Biol. Psychiatry 2018, 84, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Trépanier, M.O.; Hopperton, K.E.; Mizrahi, R.; Mechawar, N.; Bazinet, R.P. Postmortem evidence of cerebral inflammation in schizophrenia: A systematic review. Mol. Psychiatry 2016, 21, 1009–1026. [Google Scholar] [CrossRef] [PubMed]
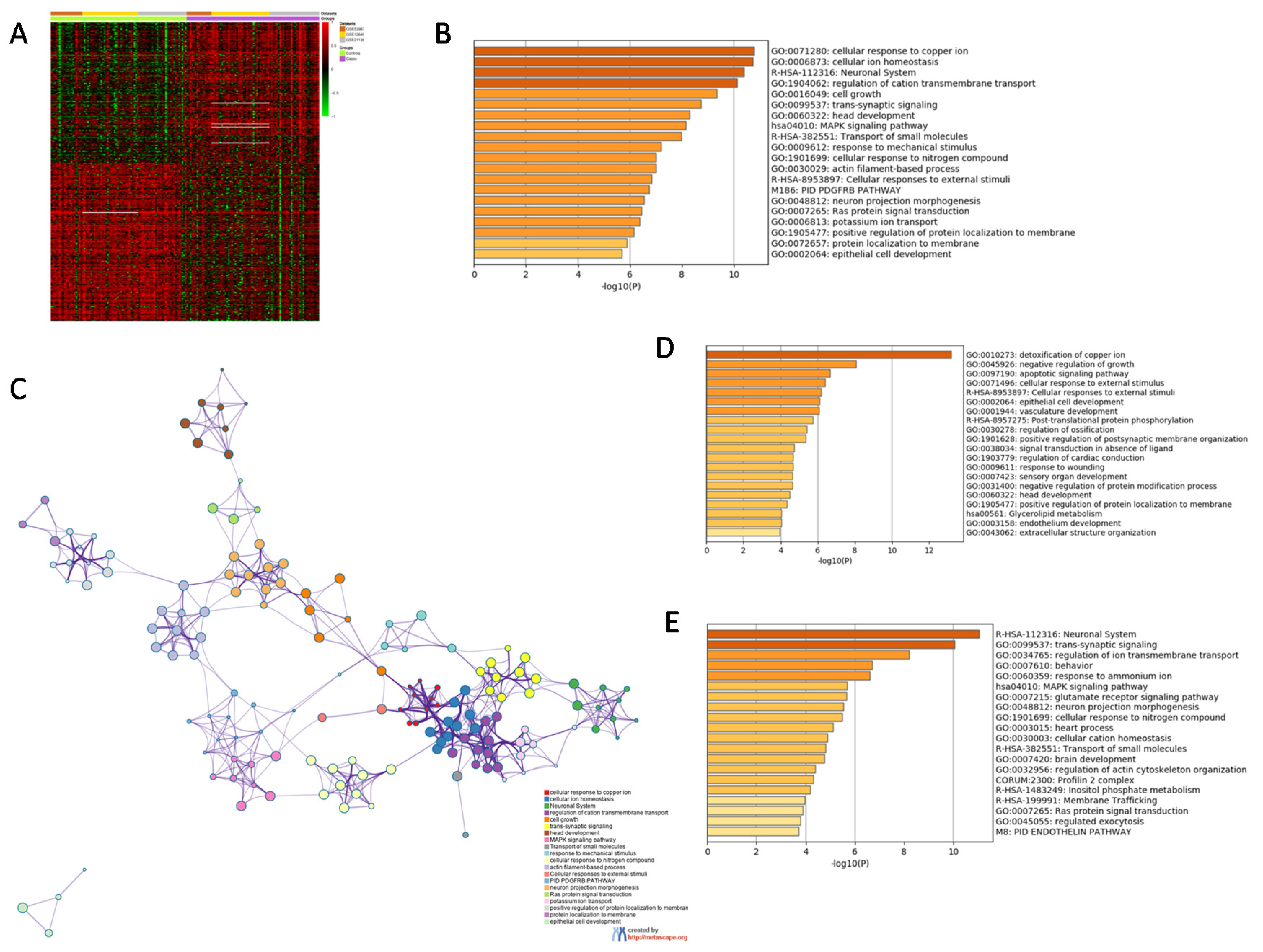


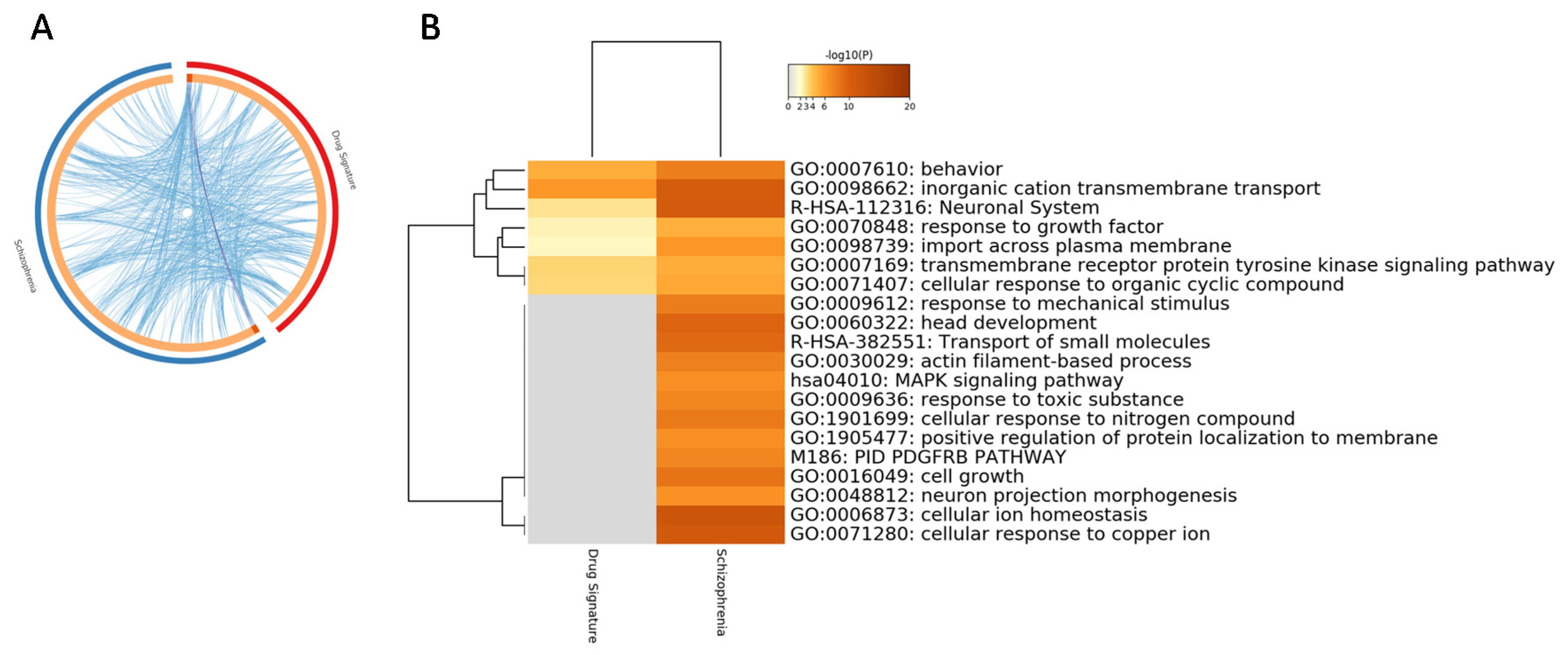
| GO | Description | Count | % | Log10 (p-Value) | Log10 (adj. p-Value) |
|---|---|---|---|---|---|
| GO:0010959 | regulation of metal ion transport | 10 | 15.15 | −6.95 | −2.79 |
| GO:0044282 | small molecule catabolic process | 10 | 15.15 | −6.44 | −2.75 |
| GO:0009612 | response to mechanical stimulus | 7 | 10.61 | −5.77 | −2.62 |
| GO:0051047 | positive regulation of secretion | 9 | 13.64 | −5.61 | −2.54 |
| GO:0019932 | second-messenger-mediated signaling | 9 | 13.64 | −5.52 | −2.51 |
| R-HSA-382551 | Transport of small molecules | 11 | 16.67 | −5.39 | −2.41 |
| GO:0007423 | sensory organ development | 9 | 13.64 | −4.76 | −2.03 |
| GO:0034368 | protein-lipid complex remodeling | 3 | 4.55 | −4.17 | −1.65 |
| GO:0050954 | sensory perception of mechanical stimulus | 5 | 7.58 | −4.05 | −1.58 |
| GO:0009636 | response to toxic substance | 8 | 12.12 | −4.05 | −1.58 |
| GO | Description | Count | % | Log10 (p-Value) | Log10 (adj. p-Value) |
|---|---|---|---|---|---|
| GO:0007268 | chemical synaptic transmission | 14 | 35.00 | −11.47 | −7.64 |
| R-HSA-112316 | Neuronal System | 10 | 25.00 | −8.99 | −5.37 |
| GO:0098662 | inorganic cation transmembrane transport | 12 | 30.00 | −8.72 | −5.18 |
| GO:0043269 | regulation of ion transport | 11 | 27.50 | −7.91 | −4.50 |
| GO:0007610 | behavior | 10 | 25.00 | −7.47 | −4.11 |
| GO:0072347 | response to anesthetic | 4 | 10.00 | −5.07 | −2.22 |
| R-HSA-422356 | Regulation of insulin secretion | 4 | 10.00 | −5.07 | −2.22 |
| GO:0035637 | multicellular organismal signaling | 5 | 12.50 | −4.66 | −1.88 |
| hsa04726 | Serotonergic synapse | 4 | 10.00 | −4.44 | −1.70 |
| GO:0001508 | action potential | 4 | 10.00 | −4.16 | −1.49 |
| ID | Adj. p-Value | p-Value | z-Val | Gene_Name |
|---|---|---|---|---|
| GPR15 | 0.00754 | 1.00E−06 | 4.891597 | G protein-coupled receptor 15 |
| ICAM2 | 0.00754 | 5.99E−07 | −4.99165 | intercellular adhesion molecule 2 |
| KLHL21 | 0.013174 | 4.37E−06 | −4.59301 | kelch like family member 21 |
| RANGAP1 | 0.013174 | 3.01E−06 | −4.67032 | Ran GTPase activating protein 1 |
| UBA3 | 0.013174 | 4.11E−06 | 4.605957 | ubiquitin like modifier activating enzyme 3 |
| MAPK7 | 0.013234 | 6.14E−06 | −4.52136 | mitogen-activated protein kinase 7 |
| SCAP | 0.013234 | 5.60E−06 | −4.54101 | SREBF chaperone |
| EAF2 | 0.018421 | 1.10E−05 | 4.396598 | ELL associated factor 2 |
| MAML1 | 0.018421 | 1.02E−05 | −4.41356 | mastermind like transcriptional coactivator 1 |
| CCDC107 | 0.019182 | 1.50E−05 | −4.3293 | coiled-coil domain containing 107 |
| FAM193A | 0.019182 | 1.94E−05 | −4.27156 | family with sequence similarity 193 member A |
| KIF21B | 0.019182 | 1.53E−05 | −4.32392 | kinesin family member 21B |
| RNF44 | 0.019182 | 1.93E−05 | −4.27328 | ring finger protein 44 |
| SELPLG | 0.019182 | 1.58E−05 | −4.31701 | selectin P ligand |
| TCL1B | 0.019182 | 1.80E−05 | 4.288751 | T cell leukemia/lymphoma 1B |
| ZNF76 | 0.019182 | 2.04E−05 | −4.26095 | zinc finger protein 76 |
| ABHD15 | 0.019623 | 2.50E−05 | −4.21456 | abhydrolase domain containing 15 |
| MEF2D | 0.019623 | 2.67E−05 | −4.19974 | myocyte enhancer factor 2D |
| SEC24C | 0.019623 | 2.73E−05 | −4.19463 | SEC24 homolog C, COPII coat complex component |
| TMEM123 | 0.019623 | 2.33E−05 | 4.230499 | transmembrane protein 123 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petralia, M.C.; Ciurleo, R.; Saraceno, A.; Pennisi, M.; Basile, M.S.; Fagone, P.; Bramanti, P.; Nicoletti, F.; Cavalli, E. Meta-Analysis of Transcriptomic Data of Dorsolateral Prefrontal Cortex and of Peripheral Blood Mononuclear Cells Identifies Altered Pathways in Schizophrenia. Genes 2020, 11, 390. https://doi.org/10.3390/genes11040390
Petralia MC, Ciurleo R, Saraceno A, Pennisi M, Basile MS, Fagone P, Bramanti P, Nicoletti F, Cavalli E. Meta-Analysis of Transcriptomic Data of Dorsolateral Prefrontal Cortex and of Peripheral Blood Mononuclear Cells Identifies Altered Pathways in Schizophrenia. Genes. 2020; 11(4):390. https://doi.org/10.3390/genes11040390
Chicago/Turabian StylePetralia, Maria Cristina, Rosella Ciurleo, Andrea Saraceno, Manuela Pennisi, Maria Sofia Basile, Paolo Fagone, Placido Bramanti, Ferdinando Nicoletti, and Eugenio Cavalli. 2020. "Meta-Analysis of Transcriptomic Data of Dorsolateral Prefrontal Cortex and of Peripheral Blood Mononuclear Cells Identifies Altered Pathways in Schizophrenia" Genes 11, no. 4: 390. https://doi.org/10.3390/genes11040390
APA StylePetralia, M. C., Ciurleo, R., Saraceno, A., Pennisi, M., Basile, M. S., Fagone, P., Bramanti, P., Nicoletti, F., & Cavalli, E. (2020). Meta-Analysis of Transcriptomic Data of Dorsolateral Prefrontal Cortex and of Peripheral Blood Mononuclear Cells Identifies Altered Pathways in Schizophrenia. Genes, 11(4), 390. https://doi.org/10.3390/genes11040390







