Comprehensive History of CSP Genes: Evolution, Phylogenetic Distribution and Functions
Abstract
1. Introduction: The Family of Chemosensory Proteins (CSPs)
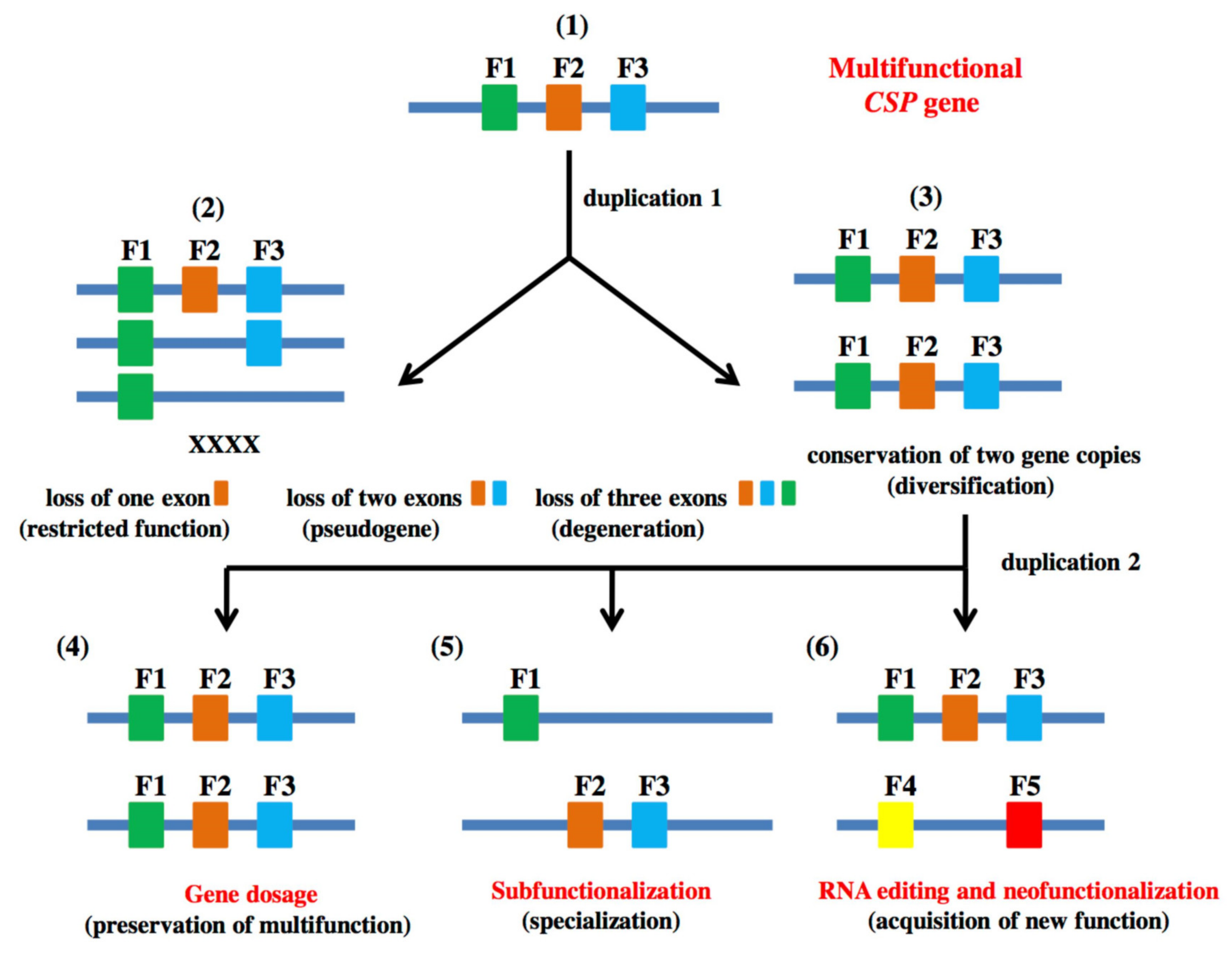
1.1. A Very Ancient Malleable Protein
1.2. CSPs and Cell Evolution
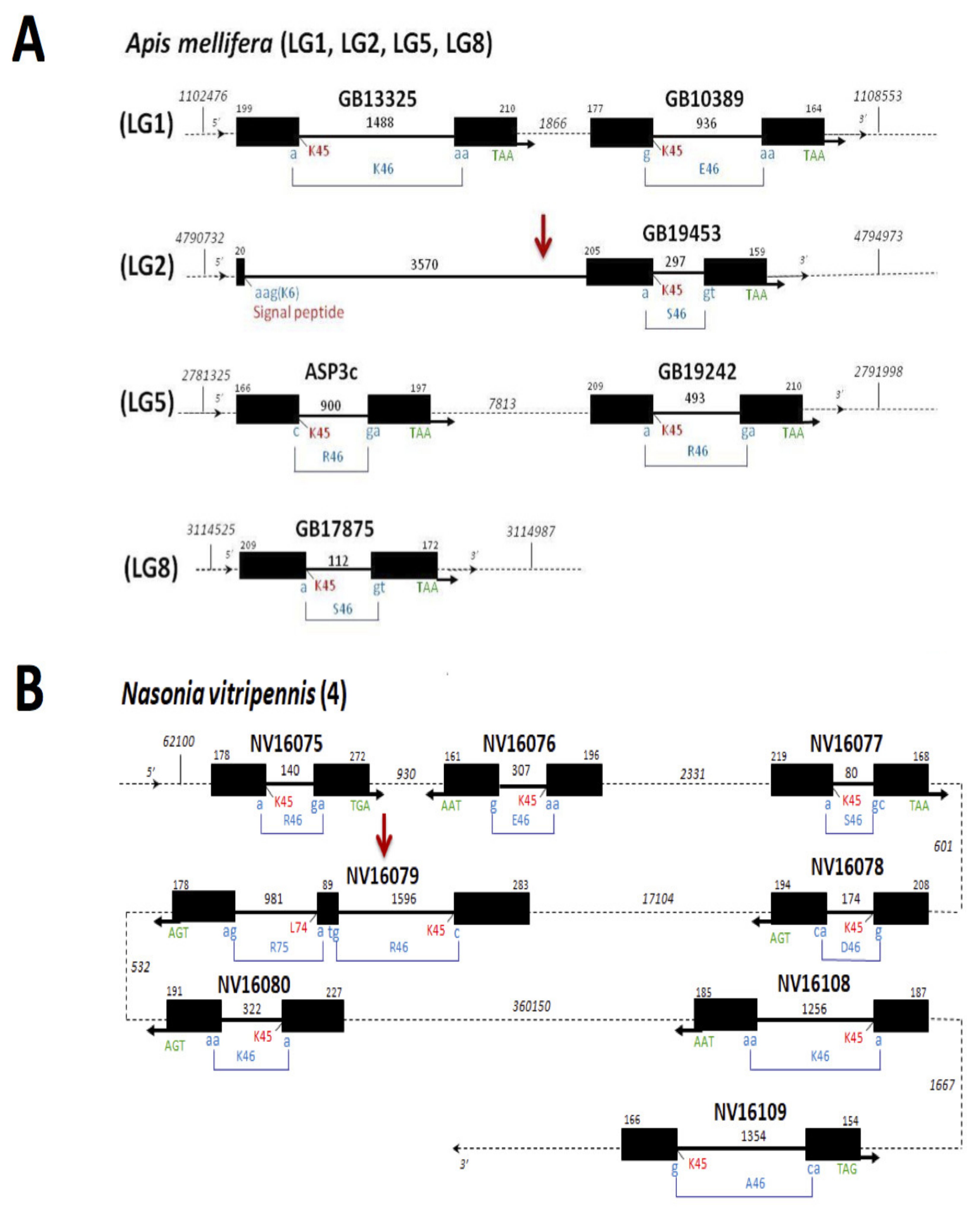
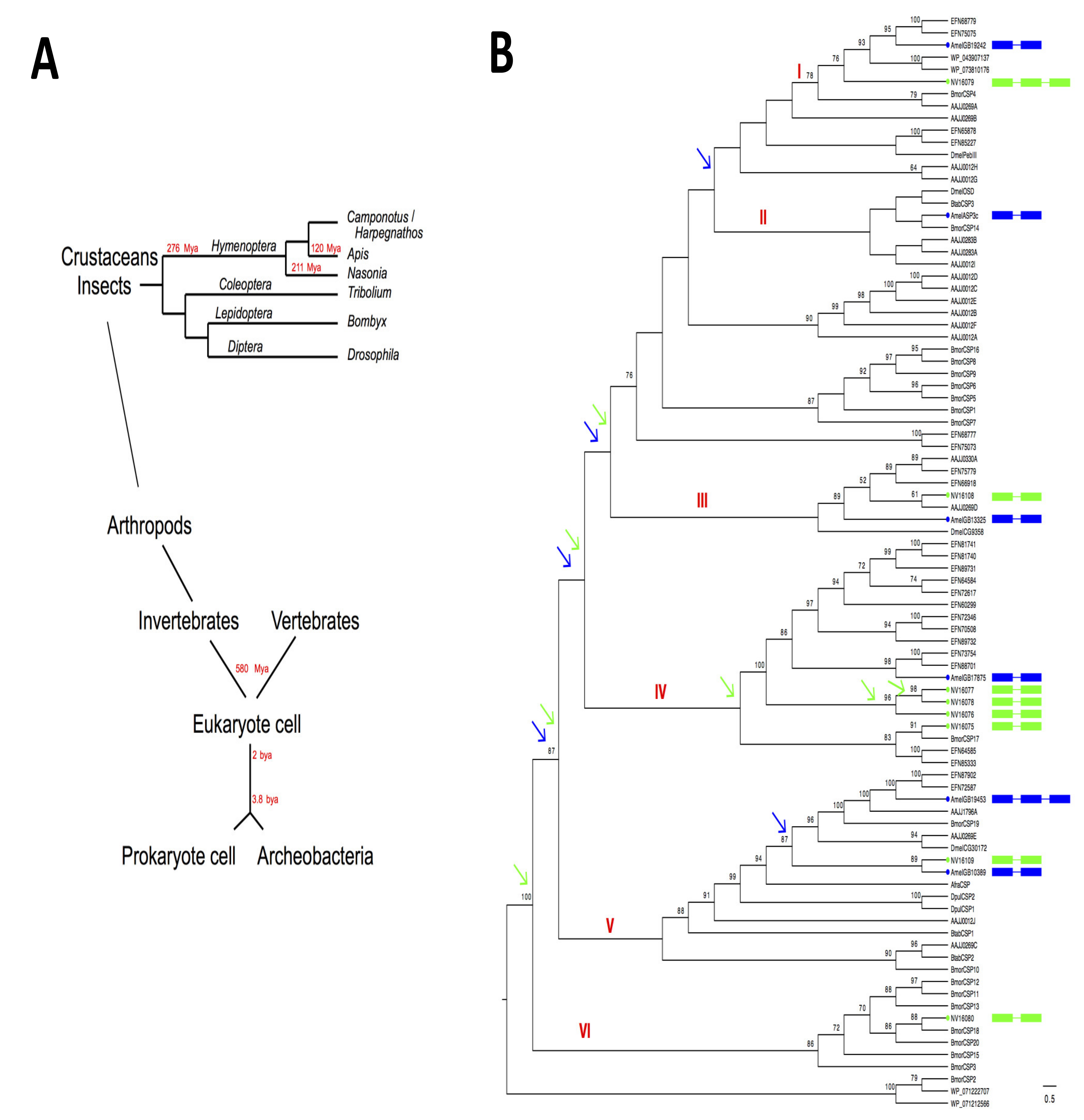
1.3. Genome-Wide Identification, Comparative Genomics and Evolution of CSPs in Hymenoptera
2. Phylogenetic Distribution Analysis in Insects and Bacteria
3. CSP Gene Expression in Response to Environmental Change
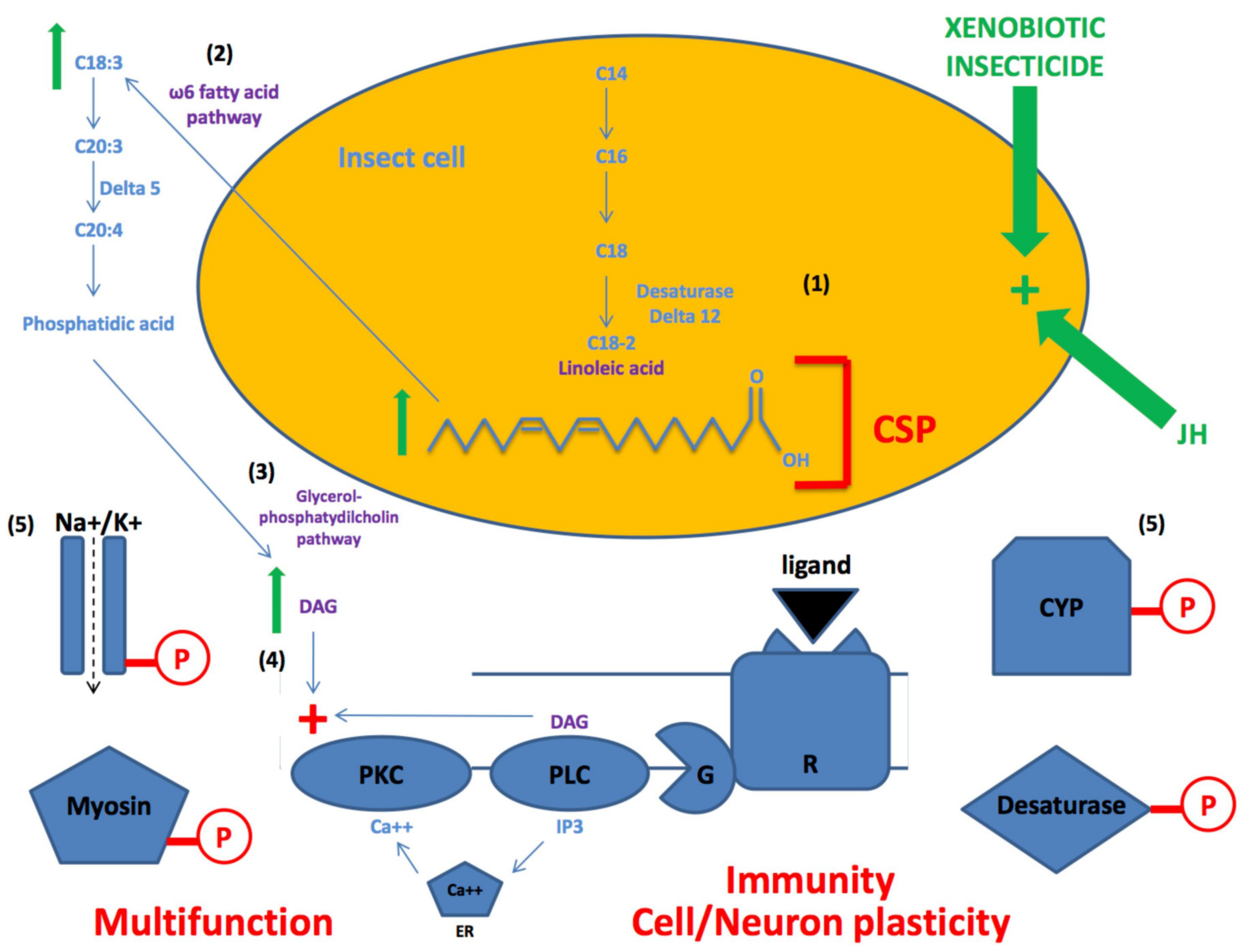
4. CSPs for Lipid- and Fatty Acid- Mediated Pathways
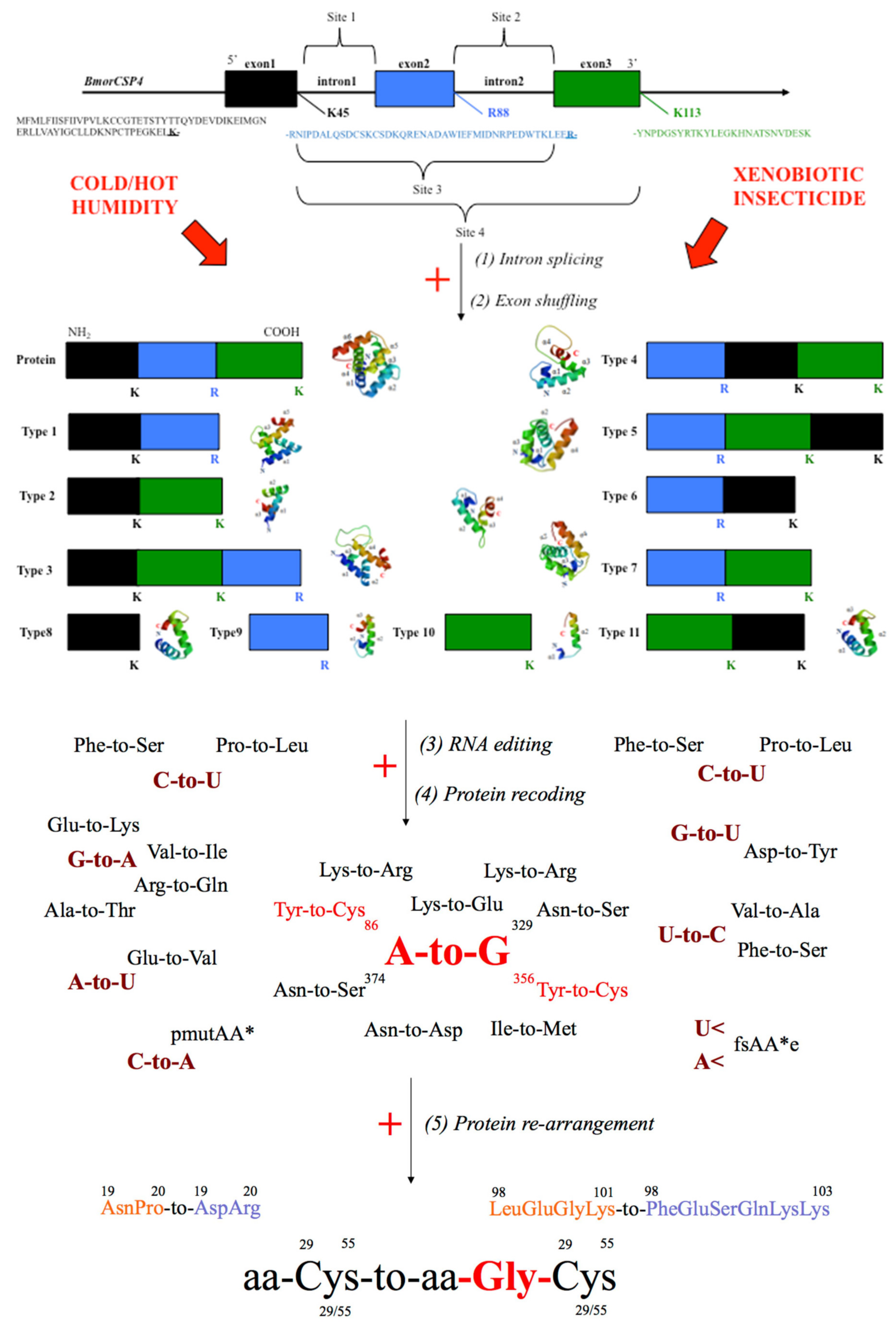
5. Genetic Editing of CSPs for Insecticide Resistance
6. Genetic Plasticity of CSPs for Neuroplasticity
7. Conclusions and Future Research
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nomura, A.; Kawasaki, K.; Kubo, T.; Natori, S. Purification and localization of p10, a novel protein that increases in nymphal regenerating legs of Periplaneta americana (American cockroach). Int. J. Dev. Biol. 1992, 36, 391–398. [Google Scholar] [PubMed]
- Picimbon, J.F.; Leal, W.S. Olfactory soluble proteins of cockroaches. Insect Biochem. Mol. Biol. 1999, 30, 973–978. [Google Scholar] [CrossRef]
- Angeli, S.; Ceron, F.; Scaloni, A.; Monti, M.; Monteforti, G.; Minnocci, A.; Petacchi, R.; Pelosi, P. Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. Eur. J. Biochem. 1999, 262, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Picimbon, J.F.; Dietrich, D.; Breer, H.; Krieger, J. Chemosensory proteins of Locusta migratoria (Orthoptera : Acrididae). Insect Biochem. Mol. Biol. 2000, 30, 233–241. [Google Scholar] [CrossRef]
- Picimbon, J.F.; Dietrich, K.; Angeli, S.; Scaloni, A.; Krieger, J.; Breer, H.; Pelosi, P. Purification and molecular cloning of chemosensory proteins from Bombyx mori. Arch. Insect Biochem. Physiol. 2000, 44, 120–129. [Google Scholar] [CrossRef]
- Picimbon, J.F.; Dietrich, K.; Krieger, J.; Breer, H. Identity and expression pattern of chemosensory proteins in Heliothis virescens (Lepidoptera, Noctuidae). Insect Biochem. Mol. Biol. 2001, 31, 1173–1181. [Google Scholar] [CrossRef]
- Lartigue, A.; Campanacci, V.; Roussel, A.; Larsson, A.M.; Jones, T.A.; Tegoni, M.; Cambillau, C. X-ray structure and ligand binding study of a moth chemosensory protein. J. Biol. Chem. 2002, 277, 32094–32098. [Google Scholar] [CrossRef]
- Campanacci, V.; Lartigue, A.; Hällberg, B.M.; Jones, A.; Giudici-Orticoni, M.T.; Tegoni, M.; Cambillau, C. Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding. Proc. Natl. Acad. Sci. USA 2003, 100, 5069–5074. [Google Scholar] [CrossRef]
- Jansen, S.; Zídek, L.; Löfstedt, C.; Picimbon, J.F.; Sklenar, V. 1H, 13C, and 15N resonance assignment of Bombyx mori chemosensory protein 1 (BmorCSP1). J. Biomol. NMR 2006, 36, 47. [Google Scholar] [CrossRef]
- Jansen, S.; Chmelik, J.; Zídek, L.; Padrta, P.; Novak, P.; Zdrahal, Z.; Picimbon, J.F.; Löfstedt, C.; Sklenar, V. Structure of Bombyx mori Chemosensory Protein 1 in solution. Arch. Insect Biochem. Physiol. 2007, 66, 135–145. [Google Scholar] [CrossRef]
- Tomaselli, S.; Crescenzi, O.; Sanfelice, D.; Ab, E.; Wechselberger, R.; Angeli, S.; Scaloni, A.; Boelens, R.; Tancredi, T.; Pelosi, P.; et al. Solution structure of a chemosensory protein from the desert locust Schistocerca gregaria. Biochemistry 2006, 45, 1606–1613. [Google Scholar] [CrossRef] [PubMed]
- Picimbon, J.F. Biochemistry and Evolution of CSP and OBP Proteins. In Insect Pheromone Biochemistry and Molecular Biology—The Biosynthesis and Detection of Pheromones and Plant Volatiles; Blomquist, J.G., Vogt, R.G., Eds.; Elsevier Academic Press: London, UK; San Diego, CA, USA, 2003; pp. 539–566. [Google Scholar]
- Xuan, N.; Bu, X.; Liu, Y.Y.; Yang, X.; Liu, G.X.; Fan, Z.X.; Bi, Y.P.; Yang, L.Q.; Lou, Q.N.; Rajashekar, B.; et al. Molecular evidence of RNA editing in Bombyx chemosensory protein family. PLoS ONE 2014, 9, e86932. [Google Scholar] [CrossRef] [PubMed]
- Xuan, N.; Guo, X.; Xie, H.Y.; Lou, Q.N.; Bo, L.X.; Liu, G.X.; Picimbon, J.F. Increased expression of CSP and CYP genes in adult silkworm females exposed to avermectins. Insect Sci. 2015, 22, 203–219. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.X.; Ma, H.M.; Xie, H.Y.; Xuan, N.; Xia, G.; Fan, Z.X.; Rajashekar, B.; Arnaud, P.; Offmann, B.; Picimbon, J.F. Biotype characterization, developmental profiling, insecticide response and binding property of Bemisia tabaci chemosensory proteins: Role of CSP in insect defense. PLoS ONE 2016, 11, e0154706. [Google Scholar] [CrossRef] [PubMed]
- Xuan, N.; Rajashekar, B.; Picimbon, J.F. DNA and RNA-dependent polymerization in editing of Bombyx chemosensory protein (CSP) gene family. Agric. Gene 2019, 12, 100087. [Google Scholar] [CrossRef]
- Xuan, X.; Rajashekar, B.; Kasvandik, S.; Picimbon, J.F. Structural components of chemosensory protein mutations in the silkworm moth, Bombyx mori. Agric. Gene 2016, 2, 53–58. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Taurellio, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Picimbon, J.F. A new view of genetic mutations. Australas. Med. J. 2017, 10, 701–715. [Google Scholar] [CrossRef]
- Picimbon, J.F. Evolution of Protein Physical Structures in Insect Chemosensory Systems. In Olfactory Concepts of Insect Control-Alternative to Insecticides; Springer Nature Switzerland: Basel, Switzerland, 2019; Volume 2, pp. 231–263. [Google Scholar]
- Liu, Z.; Song, W.; Dong, K. Persistent tetrodotoxin-sensitive sodium current resulting from U-to-C RNA editing of an insect sodium channel. Proc. Natl. Acad. Sci. USA 2004, 101, 11862–11867. [Google Scholar] [CrossRef]
- Song, W.; Liu, Z.; Tan, J.; Nomura, Y.; Dong, K. RNA editing generates tissue-specific sodium channels with distinct gating properties. J. Biol. Chem. 2004, 279, 32554–32561. [Google Scholar] [CrossRef]
- Black, D.L. Splicing in the inner ear: A familiar tune, but what are the instruments? Neuron 1998, 20, 165–168. [Google Scholar] [CrossRef][Green Version]
- Neves, G.; Zucker, J.; Daly, M.; Chess, A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nat. Genet. 2004, 36, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016, 17, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Wooldridge, L.; Ekeruche-Makinde, J.; van den Berg, H.A.; Skowera, A.; Miles, J.J.; Tan, M.P.; Dolton, G.; Clement, M.; Llewellyn-Lacey, S.; Price, D.A.; et al. A single autoimmune T cell receptor recognizes more than a million different peptides. J. Biol. Chem. 2012, 287, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Bushdid, C.; Magnasco, M.O.; Vosshall, L.B.; Keller, A. Humans can discriminate more than 1 trillion olfactory stimuli. Science 2014, 343, 1370–1372. [Google Scholar] [CrossRef]
- Liu, G.X.; Ma, H.M.; Xie, H.Y.; Xuan, N.; Picimbon, J.F. Sequence variation of Bemisia tabaci Chemosensory protein 2 in cryptic species B and Q: New DNA markers for whitefly recognition. Gene 2016, 576, 284–291. [Google Scholar] [CrossRef]
- Vizueta, J.; Frias-Lopez, C.; Macias-Hernandez, N.; Arnedo, M.A.; Sanchez-Gracia, A.; Rozas, J. Evolution of chemosensory gene families in arthropods: Insight from the first inclusive comparative transcriptome analysis across spider appendages. Genome Biol. Evol. 2017, 9, 178–196. [Google Scholar] [CrossRef]
- Liu, G.X.; Picimbon, J.F. Bacterial origin of chemosensory odor-binding proteins. Gene Transl. Bioinform. 2017, 3, e1548. [Google Scholar] [CrossRef]
- Liu, G.X.; Yue, S.; Rajashekar, B.; Picimbon, J.F. Expression of chemosensory protein (CSP) structures in Pediculus humanus corporis and Acinetobacter baumannii. SOJ Microbiol. Infect. Dis. 2019, 7, 1–17. [Google Scholar]
- Siepel, A.; Bejerano, G.; Pedersen, J.S.; Hinrichs, A.S.; Hou, M.; Rosenbloom, K.; Clawson, H.; Spieth, J.; Hillier, L.D.W.; Richards, S.; et al. Evolutionary conserved elements in vertebrate, insect, worm and yeast genomes. Genome Res. 2005, 15, 1034–1050. [Google Scholar] [CrossRef]
- Isenbarger, T.A.; Carr, C.E.; Johnson, S.S.; Finney, M.; Church, G.M.; Gilbert, W.; Zuber, M.T.; Ruvkun, G. The most conserved genome segments for life detection on earth and other planets. Orig. Life Evol. Biosph. 2008, 38, 517–533. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, G.; Pelosi, P. Plant transcriptomes reveal hidden guests. Biochem. Biophys. Res. Commun. 2016, 474, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Ban, L.; Scaloni, A.; Brandazza, A.; Angeli, S.; Zhang, L.; Pelosi, P. Chemosensory proteins of Locusta migratoria. Insect Mol. Biol. 2003, 12, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Mao, Y.; Zhang, L. Binding properties of four antennae-expressed chemosensory proteins (CSPs) with insecticides indicates the adaptation of Spodoptera litura to environment. Pest Biochem. Physiol. 2018, 146, 43–51. [Google Scholar] [CrossRef]
- Wanner, K.W.; Willis, L.G.; Theilmann, D.A.; Isman, M.B.; Feng, Q.; Plettner, E. Analysis of the insect os-d-like gene family. J. Chem. Ecol. 2004, 30, 889–911. [Google Scholar] [CrossRef]
- Liu, G.X.; Arnaud, P.; Offmann, B.; Picimbon, J.F. Genotyping and bio-sensing chemosensory proteins in insects. Sensors 2017, 17, 1801. [Google Scholar] [CrossRef]
- Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 2006, 443, 931–949. [Google Scholar] [CrossRef]
- Forêt, S.; Wanner, K.W.; Maleszka, R. Chemosensory proteins in the honeybee: Insights from the annotated genome, comparative analysis and expression profiling. Insect Biochem. Mol. Biol. 2007, 37, 19–28. [Google Scholar] [CrossRef]
- Wallberg, A.; Bunikis, I.; Vinnere Pettersson, O.; Mosbech, M.B.; Childers, A.K.; Evans, J.D.; Mikheyev, A.S.; Robertson, H.M.; Robinson, G.E.; Webster, M.T. A hybrid de novo genome assembly of the honeybee, Apis mellifera, with chromosome length scaffold. BMC Genomics 2019, 20, 275. [Google Scholar] [CrossRef]
- Desjardins, C.A.; Oliveira, D.C.S.G.; Edwards, R.M.; Dang, P.M.; Lee, D.; Colbourne, J.K.; Tettelin, H.; Hunter, W.B.; Werren, J.H. Nasonia Expression Libraries from Larvae, Pupae, and Adults. 2007. Available online: NasoniaBase, http://hymenopteragenome.org.
- Werren, J.H.; Richards, S.; Desjardins, C.A.; Niehuis, O.; Gadau, J.; Colbourne, J.; Beukeboom, L.W.; Desplan, C.; Elsik, C.G.; Grimmelikhuijzen, C.J.P.; et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 2010, 327, 343–348. [Google Scholar] [CrossRef]
- Bonasio, R.; Zhang, G.; Ye, C.; Mutti, N.; Fang, X.; Qin, N.; Donahue, G.; Yang, P.; Li, Q.; Li, C.; et al. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 2010, 329, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Kulmuni, J.; Wurm, Y.; Pamilo, P. Comparative genomics and chemosensory protein genes reveals rapid evolution and positive selection in ant-specific duplicates. Heredity 2013, 110, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Sabatier, L.; Jouanguy, E.; Dostert, C.; Zachary, D.; Dimarcq, J.L.; Bulet, P.; Imler, J.L. Pherokine-2 and -3: Two Drosophila molecules related to pheromone/odor-binding proteins induced by viral and bacterial infections. Eur. J. Biol. 2003, 270, 3398–33407. [Google Scholar] [CrossRef] [PubMed]
- Voordekers, K.; Verstrepen, K. Experimental evolution of the model eukaryote Saccharomyces cerevisiae yields insight into the molecular mechanisms underlying adaptation. Curr. Opin. Microbiol. 2015, 28, 1–9. [Google Scholar] [CrossRef]
- Chen, T.; Reith, M.E.; Ross, N.W.; MacRae, T.H. Expressed sequence tag (EST)-based characterization of gene regulation in Artemia larvae. Invert. Rep. Dev. 2003, 44, 33–44. [Google Scholar] [CrossRef]
- Gottfried, J.A.; Wilson, D.A. Smell. In Neurobiology of Sensation and Reward; Gottfried, J.A., Ed.; CRC Press/Taylor & Francis: Boca Raton, FL, USA, 2011. [Google Scholar]
- Celniker, S.E.; Wheeler, D.A.; Kronmiller, B.; Carlson, J.W.; Halpern, A.; Patel, S.; Adams, M.; Champe, M.; Dugan, S.P.; Frise, E.; et al. Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002, 3. [Google Scholar] [CrossRef]
- Stapleton, M.; Carlson, J.; Brokstein, P.; Yu, C.; Champe, M.; George, R.; Guarin, H.; Kronmiller, B.; Pacleb, J.; Park, S.; et al. A Drosophila full-length cDNA resource. Genome Biol. 2002, 3. [Google Scholar] [CrossRef]
- Stapleton, M.; Liao, G.; Brokstein, P.; Hong, L.; Carninci, P.; Shiraki, T.; Hayashizaki, Y.; Champe, M.; Pacleb, J.; Wan, K.; et al. The Drosophila gene collection: Identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 2002, 12, 1294–1300. [Google Scholar] [CrossRef]
- Mita, K.; Morimyo, M.; Okano, K.; Koike, Y.; Nohata, J.; Kawasaki, H.; Kadono-Okuda, K.; Yamamoto, K.; Suzuki, M.G.; Shimada, T.; et al. The construction of an EST database for Bombyx mori and its application. Proc. Natl. Acad. Sci. USA 2003, 104, 14121–14126. [Google Scholar] [CrossRef]
- Ai, H. Sensors and sensory processing for airborne vibrations in silk moths and honeybees. Sensors 2013, 13, 9344–9363. [Google Scholar] [CrossRef]
- Ellison, C.; Brun, Y.V. Mechanosensing: A regulation sensation. Curr. Biol. 2015, 25, R113–R115. [Google Scholar] [CrossRef] [PubMed]
- Topham, M.K.; Prescott, S.M. Handbook of Cell Signaling, 2nd ed.; Elsevier Academic Press: Cambridge, MA, USA, 2009. [Google Scholar]
- Lohr, J.B.; Kuhn-Velten, W.N. Protein phosphorylation changes ligand-binding efficiency of cytochrome P450c17 (CYP17) and accelerates its proteolytic degradation: Putative relevance for hormonal regulation of CYP17 activity. Biochem. Physiol. Res. Commun. 1997, 231, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Helling, S.; Huttermann, M.; Ramzan, R.; Kim, S.H.; Lee, I.; Muller, T.; Langerfeld, E.; Meyer, H.E.; Kadenbach, B.; Vogt, S.; et al. Multiple phosphorylations of cytochrome c oxidase and their functions. Proteomics 2012, 12, 950–959. [Google Scholar] [CrossRef] [PubMed]
- Mansilla, M.C.; de Mendoza, D. The Bacillus subtilis desaturase: A model to understand phospholipid modification and temperature sensing. Arch. Microbiol. 2005, 183, 229–235. [Google Scholar] [CrossRef]
- Tang, G.Q.; Novitzky, W.P.; Griffin, H.C.; Huber, S.C.; Dewey, R.E. Oleate desaturase enzymes of soybean: Evidence of regulation through differential stability and phosphorylation. Plant J. 2005, 44, 433–446. [Google Scholar] [CrossRef]
- Ohnishi, A.; Hull, J.; Kaji, M.; Hashimoto, K.; Lee, J.M.; Tsuneizumi, K.; Suzuki, T.; Dohmae, N.; Matsumoto, S. Hormone signaling linked to silkmoth sex pheromone biosynthesis involves Ca2+/Calmodulin-dependent protein kinase II-mediated phosphorylation of the insect PAT family protein Bombyx mori lipid storage droplet protein-1 (BmLsD1). J. Biol. Chem. 2011, 286, 24101–24112. [Google Scholar] [CrossRef]
- Du, M.; Yin, X.; Zhang, S.; Zhu, B.; Song, Q.; An, S. Identification of lipases involved in PBAN stimulated pheromone production in Bombyx mori using the DGE and RNAi approaches. PLoS ONE 2012, 7, e31045. [Google Scholar] [CrossRef]
- Liu, P.; Peng, H.J.; Zhu, J. Juvenile hormone-activated phospholipase C pathway enhances transcriptional activation by the methoprene-tolerant protein. Proc. Natl. Acad. Sci. USA 2015, 112, E1871–E1879. [Google Scholar] [CrossRef]
- Enan, E.; Matsumura, F. Stimulation of protein phosphorylation in intact rat brain synaptosomes by a pyrethroid insecticide, deltamethrin. Pest Biochem. Physiol. 1991, 39, 182–195. [Google Scholar] [CrossRef]
- Gollamudi, S.; Johri, A.; Callingasan, N.Y.; Yang, L.; Elemento, O.; Beal, M.F. Concordant signaling pathways produced by pesticide exposure in mice correspond to pathways identified in human Parkinson’s disease. PLoS ONE 2012, 7, e36191. [Google Scholar] [CrossRef]
- Yang, J.S.; Symington, S.; Clark, J.M.; Park, Y. Permethrin, a pyrethroid insecticide, regulates ERK1/2 activation through membrane depolarization-mediated pathway in HepG2 hepatocytes. Food Chem. Toxicol. 2018, 121, 387–395. [Google Scholar] [CrossRef]
- Le Goff, G.; Giraudo, M. Effects of Pesticides on the Environment and Insecticide Resistance. In Olfactory Concepts of Insect Control-Alternative to Insecticides; Picimbon, J.F., Ed.; Springer Nature Switzerland: Basel, Switzerland, 2019; Volume 1, pp. 51–78. [Google Scholar]
- Devillard, E.; McIntosh, F.M.; Duncan, S.H.; Wallace, R.J. Metabolism of linoleic acid by human gut bacteria: Different routes for biosynthesis of conjugated linoleic acid. J. Bacteriol. 2007, 189, 2566–2570. [Google Scholar] [CrossRef]
- Takatsuka, J.; Nakai, M.; Shinoda, T. A virus carries a gene encoding juvenile hormone acid methyltransferase, a key regulatory enzyme in insect metamorphosis. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Nates, S.F.; McKenney, C.L., Jr. Growth, lipid class and fatty acid composition in juvenile mud crabs (Rhithropanopeus harrisii) following larval exposure to Fenoxycarb, insect juvenile hormone analog. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2000, 127, 317–325. [Google Scholar] [CrossRef]
- Nagaraju, G.P.C. Reproductive regulators in decapod crustaceans: An overview. J. Exp. Biol. 2011, 214, 3–16. [Google Scholar] [CrossRef]
- Tamone, S.L.; Harrison, J.F. Linking insects with crustacea: Physiology of the Pancrustacea: An introduction to the symposium. Integr. Comp. Physiol. 2015, 55, 765–770. [Google Scholar] [CrossRef]
- Nijhout, H.F. Insect Hormones; Princeton University Press: Princeton, NJ, USA, 1994; p. 280. [Google Scholar]
- Blomquist, G.J.; Dwyer, L.A.; Chu, A.J.; Ryan, R.O.; de Renobales, M. Biosynthesis of linoleic acid in a termite, cockroach and cricket. Insect Biochem. 1982, 12, 349–353. [Google Scholar] [CrossRef]
- Aksenov, V.; Rollo, C.D. Necromone death cues and risk avoidance by the cricket Acheta domesticus: Effects of sex and duration of exposure. J. Insect Behav. 2017, 30, 259–272. [Google Scholar] [CrossRef]
- Blaul, B.; Steinbauer, R.; Merkl, P.; Merkl, R.; Tschochner, H.T.; Ruther, J. Oleic acid is a precursor of linoleic acid and the male sex pheromone in Nasonia vitripennis. Insect Biochem. Mol. Biol. 2014, 51, 33–40. [Google Scholar] [CrossRef]
- Rule, G.; Roelofs, W.L. Biosynthesis of sex pheromone components from linolenic acid in arctiid moths. Arch. Insect Biochem. Physiol. 1989, 12, 89–97. [Google Scholar] [CrossRef]
- Sakai, R.; Fukuzawa, M.; Nakano, R.; Tatsuki, S.; Ishikawa, Y. Alternative suppression of transcription from two desaturase genes is the key for species-specific sex pheromone biosynthesis in two Ostrinia moths. Insect Biochem. Mol. Biol. 2009, 39, 62–67. [Google Scholar] [CrossRef]
- Moto, K.; Suzuki, M.G.; Hull, J.J.; Kurata, R.; Takahashi, S.; Yamamoto, M.; Okano, K.; Imai, K.; Ando, T.; Matsumoto, S. Involvement of a bifunctional fatty-acyl desaturase in the biosynthesis of the silkmoth, Bombyx mori, sex pheromone. Proc. Natl. Acad. Sci. USA 2004, 101, 8631–8636. [Google Scholar] [CrossRef]
- Vukašinović, E.L.; Pond, D.W.; Worland, M.R.; Kojić, D.; Purać, J.; Blagojević, D.P.; Grubor-Lajsić, G. Diapause induces changes in the composition and biophysical properties of lipids in larvae of the European corn borer, Ostrinia nubilalis (Lepidoptera: Crambidae). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2013, 165, 219–225. [Google Scholar] [CrossRef]
- Briand, L.; Swasdipan, N.; Nespoulos, C.; Bézirard, V.; Blon, F.; Huet, J.C.; Ebert, P.; Pernollet, J.C. Characterization of a chemosensory protein (ASP3c) from honeybee (Apis mellifera L.) as a brood pheromone carrier. Eur. J. Biochem. 2002, 269, 4586–4596. [Google Scholar] [CrossRef]
- Li, H.L.; Lou, B.G.; Cheng, J.A.; Gao, Q.K. The Chemosensory protein of Chinese honeybee, Apis cerana cerana: Molecular cloning of cDNA, immunocytochemical localization and expression. Chin. Sci. Bull. 2007, 52, 1355–1364. [Google Scholar] [CrossRef]
- Pikielny, C.W.; Hasan, G.; Rouyer, F.; Rosbach, M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron 1994, 12, 35–49. [Google Scholar] [CrossRef]
- McKenna, M.P.; Hekmat-Scafe, D.S.; Gaines, P.; Carlson, J.R. Putative Drosophila pheromone-binding-proteins expressed in a subregion of the olfactory system. J. Biol. Chem. 1994, 269, 16340–16347. [Google Scholar]
- Claridge-Chang, A.; Wijnen, H.; Naef, F.; Boothroyd, C.; Rajewsky, N.; Young, M.W. Circadian regulation of gene expression systems in the Drosophila head. Neuron 2001, 32, 657–671. [Google Scholar] [CrossRef]
- McDonald, M.J.; Rosbach, M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell 2001, 107, 567–578. [Google Scholar] [CrossRef]
- Stathopoulos, A.; Van Drenth, M.; Erives, A.; Markstein, M.; Levine, M. Whole genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell 2002, 111, 687–701. [Google Scholar] [CrossRef]
- Malcicka, M.; Visser, B.; Ellers, J. An evolutionary perspective on linoleic acid synthesis in animals. Evol. Biol. 2018, 45, 15–26. [Google Scholar] [CrossRef]
- Guo, W.; Wang, X.; Ma, Z.; Xue, L.; Han, J.; Yu, D.; Kang, L. CSP and Takeout genes modulate the switch between attraction and repulsion during behavioral phase change in the migratory locust. PLoS Genet. 2011, 7, e1001291. [Google Scholar] [CrossRef]
- Martín-Blázquez, R.; Chen, B.; Kang, L.; Bakkali, M. Evolution, expression and association of the chemosensory protein genes with the outbreak phase of two main pest locusts. Sci. Rep. 2017, 7, 6653. [Google Scholar] [CrossRef]
- Whitfield, C.W.; Band, M.R.; Bonaldo, M.F.; Kumar, C.G.; Liu, L.; Pardinas, J.R.; Robertson, H.M.; Bento Soares, M.; Robinson, G.E. Annotated Expressed Sequence Tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Res. 2002, 12, 555–566. [Google Scholar] [CrossRef]
- Porath, H.T.; Hazan, E.; Shpigler, H.; Cohen, M.; Band, M.; Ben-Shahar, Y.; Levanon, E.Y.; Eisenberg, E.; Bloch, G. RNA editing is abundant and correlates with task performance in a social bumblebee. Nat. Commun. 2019, 10, 1605. [Google Scholar] [CrossRef]
- Duan, Y.; Dou, S.; Huang, J.; Eisenberg, E.; Lu, J. Evolutionary forces on A-to-I RNA editing revealed by sequencing individual honeybee drones. bioRXiv 2020. [Google Scholar] [CrossRef]
- Alyokhin, A.; Baker, M.B.; Mota-Sanchez, D.; Dively, G.; Grafius, E. Colorado potato beetle resistance to insecticide. Am. J. Potato Res. 2008, 85, 395–413. [Google Scholar] [CrossRef]
- Boyer, S.; Zhang, H.Y.; Lemperiere, G. A review of control methods and resistance mechanisms in stored-product insects. Bull. Entomol. Res. 2011, 102, 213–229. [Google Scholar] [CrossRef]
- Mota-Sanchez, D.; Wise, J.C.; Vander Poppen, R.; Gut, L.J.; Hollingworth, R.M. Resistance of codling moth, Cydia pomonella (L.) (Lepidoptera: Tortricidae), larvae in Michigan to different insecticides with different modes of action and the impact on field residual activity. Pest Manag. Sci. 2010, 64, 881–890. [Google Scholar] [CrossRef]
- Troczka, B.J.; Williamson, M.S.; Field, L.M.; Davies, T.G.E. Rapid selection for resistance to diamide insecticides in Plutella xylostella via specific amino acid polymorphisms in the ryanodine receptor. Neurotoxicology 2017, 60, 224–233. [Google Scholar] [CrossRef]
- Luo, C.; Jones, C.M.; Devine, G.; Zhang, F.; Denholm, I.; Gorman, K. Insecticide resistance in Bemisia tabaci biotype Q (Hemiptera: Aleyrodidae) from China. Crop Prot. 2010, 29, 429–434. [Google Scholar] [CrossRef]
- Liu, G.X.; Xuan, N.; Chu, D.; Xie, H.Y.; Fan, Z.X.; Bi, Y.P.; Picimbon, J.F.; Qin, Y.C.; Zhong, S.T.; Li, Y.F.; et al. Biotype expression and insecticide response of Bemisia tabaci chemosensory protein-1. Arch. Insect Biochem. Physiol. 2014, 85, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Wells, J.; Senan, A.; Bermudez, I.; Jones, A.K. Species specific RNA A-to-I editing of mosquito RDL modulates GABA potency and influences agonistic, potentiating and antagonistic actions of ivermectin. Insect Biochem. Mol. Biol. 2018, 93, 1–11. [Google Scholar] [CrossRef]
- Dong, K. Insect sodium channels and insecticide resistance. Invert. Neurosci. 2007, 7, 17–30. [Google Scholar] [CrossRef]
- Chang, C.; Shen, W.K.; Wang, T.T.; Lin, Y.H.; Hsu, E.L.; Dai, S.M. A novel amino acid substitution in a voltage-gated sodium channel is associated with knockdown resistance to permethrin in Aedes aegypti. Insect Biochem. Mol. Biol. 2009, 39, 272–278. [Google Scholar] [CrossRef]
- Claudianos, C.; Ranson, H.; Johnson, R.M.; Biswas, S.; Schuler, M.A.; Berenbaum, M.R.; Feyereisen, R.; Oakeshott, J.G. A deficit of detoxification enzymes: Pesticide sensitivity and environmental response in the honeybee. Insect Mol. Biol. 2006, 15, 615–636. [Google Scholar] [CrossRef]
- Maleszka, J.; Forêt, S.; Saint, R.; Maleszka, R. RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev. Genes Evol. 2007, 217, 189–196. [Google Scholar] [CrossRef]
- Zimmerlin, L.; Park, T.S.; Zambidis, E.T. Capturing human naïve pluripotency in the embryo and in the dish. Stem Cells Dev. 2017, 26, 1141–1161. [Google Scholar] [CrossRef]
- Graf, T.; Stadtfeld, M. Heterogeneity of embryonic and adult stem cells. Cell Stem Cell 2008, 3, 480–483. [Google Scholar] [CrossRef]
- Tweedel, K.S. The adaptability of somatic stem cells: A review. J. Stem Cell Regen. Med. 2017, 13, 3–13. [Google Scholar]
- Pyza, E.M. Plasticity in invertebrate sensory systems. Front. Physiol. 2013, 4, 226. [Google Scholar] [CrossRef] [PubMed]
- Fahrbach, S.E.; Van Nest, B.N. Synapsin-based approaches to brain plasticity in adult social insects. Curr. Opin. Insect Sci. 2016, 18, 27–34. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Anton, S.; Gadenne, C. Effect of juvenile hormone on the central nervous processing of sex pheromone in an insect. Proc. Natl. Acad. Sci. USA 1999, 96, 5764–5767. [Google Scholar] [CrossRef]
- Lechelt, J.L.; Evenden, M.L. Peripheral and behavioral plasticity of pheromone response and its hormonal control in a long-lived moth. J. Exp. Biol. 2009, 212, 2000–2006. [Google Scholar]
- Barrozo, R.B.; Gadenne, C.; Anton, S. Switching attraction to inhibition: Mating-induced reversed role of sex pheromone in an insect. J. Exp. Biol. 2010, 213, 2933–2939. [Google Scholar] [CrossRef]
- Barrozo, R.B.; Jarriault, D.; Deisig, N.; Gemeno, C.; Monsempes, C.; Lucas, P.; Gadenne, C.; Anton, S. Mating-induced differential coding of plant odour and sex pheromone in a male moth. Eur. J. Neurosci. 2011, 33, 1841–1850. [Google Scholar] [CrossRef]
- Cheng, S.S.; Liu, J.Y.; Tsai, K.H.; Chen, W.J.; Chang, S.T. Chemical composition and mosquito larvicidal activity of essential oils from leaves of different Cinnamomum osmophloeum provenances. J. Agric. Food Chem. 2004, 52, 4395–4400. [Google Scholar] [CrossRef]
- Baumann, K. Stem cells A key to totipotency. Nat. Rev. Mol. Cell Biol. 2017, 18, 137. [Google Scholar] [CrossRef]
- Brooks, A.N.; Turkarslan, S.; Beer, K.D.; Lo, F.Y.; Baliga, N.S. Adaptation of cells to new environments. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011, 3, 544–561. [Google Scholar] [CrossRef]
- Picimbon, J.F. RNA/peptide editing in small soluble binding proteins: A new theory for the origin of life on earth’s crust. Preprints 2020, 2020010357. [Google Scholar]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, G.; Xuan, N.; Rajashekar, B.; Arnaud, P.; Offmann, B.; Picimbon, J.-F. Comprehensive History of CSP Genes: Evolution, Phylogenetic Distribution and Functions. Genes 2020, 11, 413. https://doi.org/10.3390/genes11040413
Liu G, Xuan N, Rajashekar B, Arnaud P, Offmann B, Picimbon J-F. Comprehensive History of CSP Genes: Evolution, Phylogenetic Distribution and Functions. Genes. 2020; 11(4):413. https://doi.org/10.3390/genes11040413
Chicago/Turabian StyleLiu, Guoxia, Ning Xuan, Balaji Rajashekar, Philippe Arnaud, Bernard Offmann, and Jean-François Picimbon. 2020. "Comprehensive History of CSP Genes: Evolution, Phylogenetic Distribution and Functions" Genes 11, no. 4: 413. https://doi.org/10.3390/genes11040413
APA StyleLiu, G., Xuan, N., Rajashekar, B., Arnaud, P., Offmann, B., & Picimbon, J.-F. (2020). Comprehensive History of CSP Genes: Evolution, Phylogenetic Distribution and Functions. Genes, 11(4), 413. https://doi.org/10.3390/genes11040413





