Gallop Racing Shifts Mature mRNA towards Introns: Does Exercise-Induced Stress Enhance Genome Plasticity?
Abstract
1. Introduction
2. Materials and Methods
2.1. Training
2.2. Sampling
2.3. RNA Extraction
2.4. Sequencing
2.5. Bioinformatic Analyses
2.5.1. Annotations Retrieval and Count Matrices
2.5.2. Differential Expression Analyses: Genes
2.5.3. Differential Expression Analyses: Isoforms
2.5.4. Differential Expression Analyses: Repetitive Elements
- (1)
- Genome-wide differential expression analysis
- (2)
- Differential expression analysis of repeats classes
- (3)
- Differential expression analysis of long interspersed nuclear elements subclass 1 (LINE1) only.
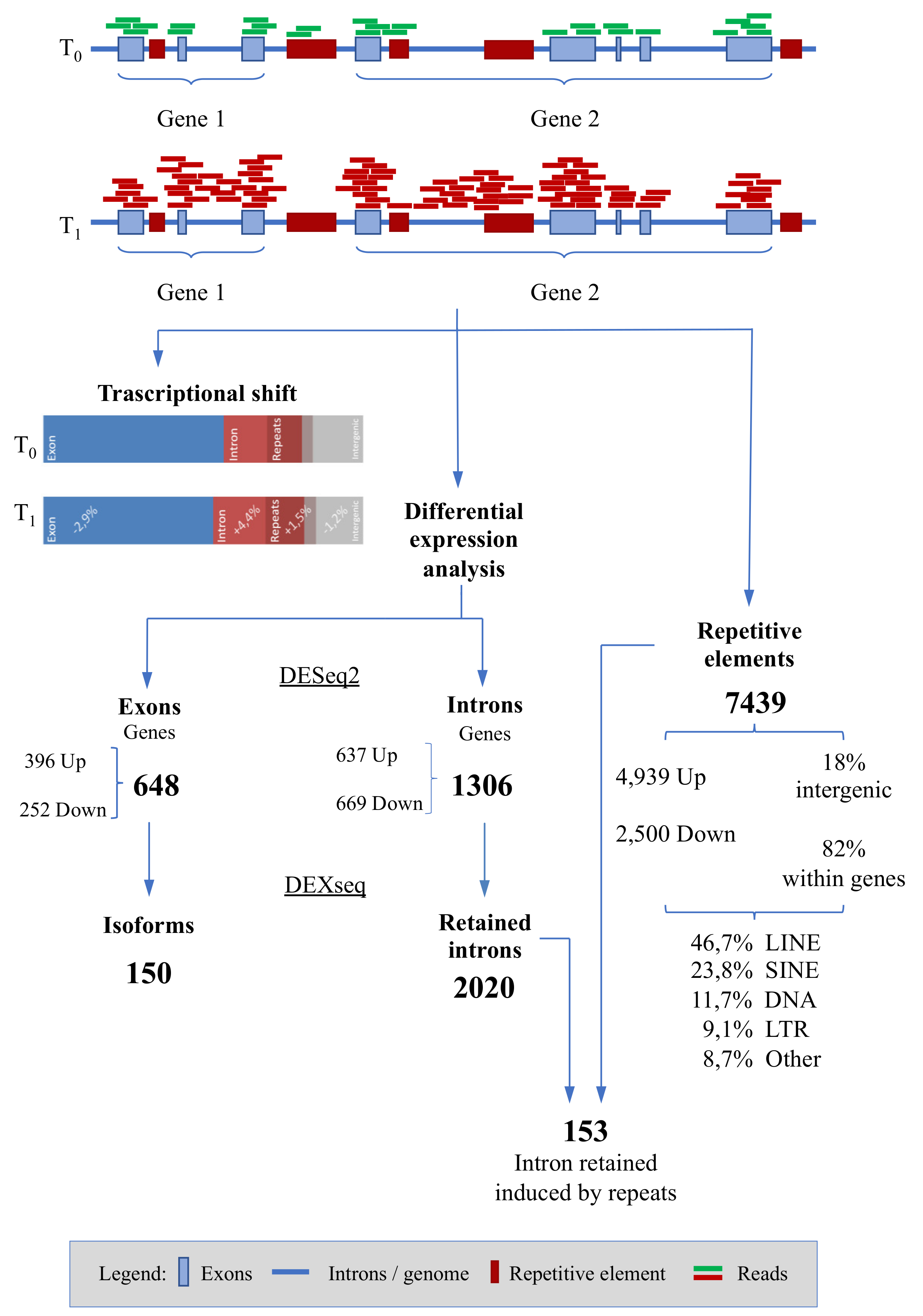
3. Results
3.1. Sequencing Statistics
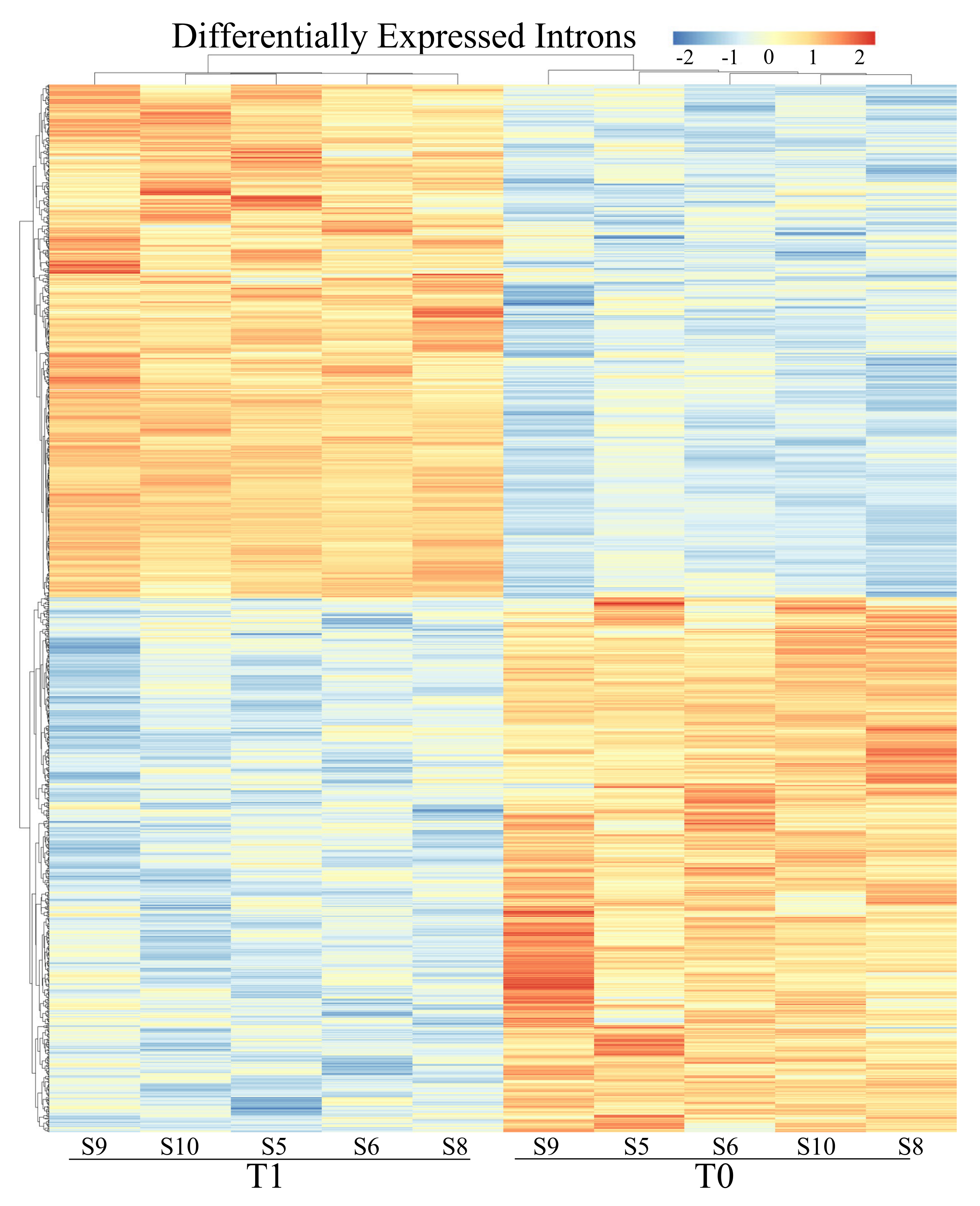
3.2. Differential Expression Analyses: Genes
3.3. Differential Expression Analyses: Isoforms
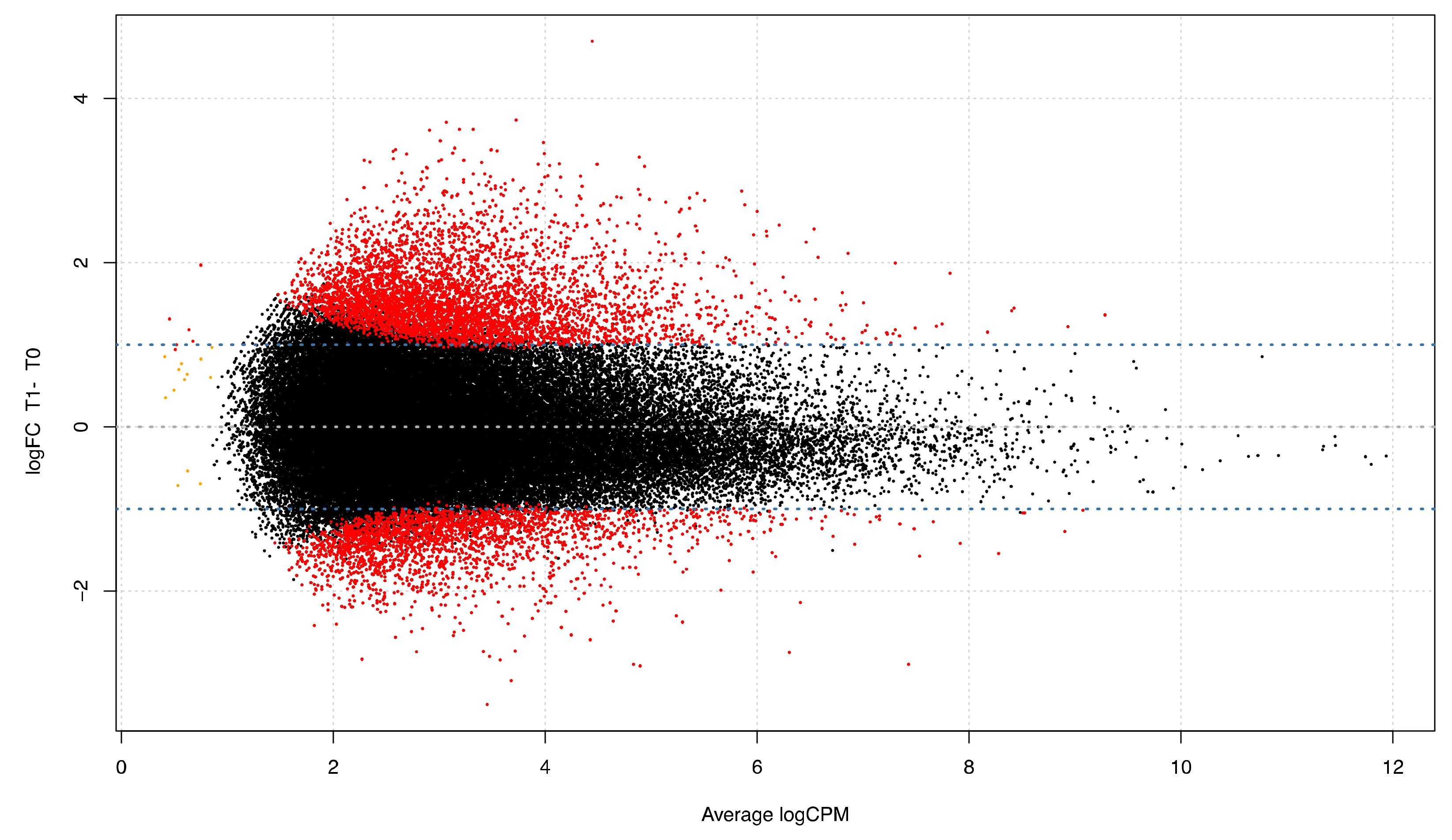
3.4. Differential Expression Analyses: Repetitive Elements
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Giuffra, E.; Tuggle, C.K. and Functional Annotation of Animal Genomes (FAANG): Current Achievements and Roadmap. Annu. Rev. Anim. Biosci. 2019, 7, 65–88. [Google Scholar] [CrossRef] [PubMed]
- De Hoon, M.; Shin, J.W.; Carninci, P. Paradigm shifts in genomics through the FANTOM projects. Mamm. Genome 2015, 26, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Kline, H.; Foreman, J. Heart and spleen weights as a function of breed and somatotype. Equine Exerc Physiol. 1991, 3, 17–21. [Google Scholar]
- Barrey, E.; Galloux, P.; Valette, J.P.; Auvinet, B.; Wolter, R. Stride Characteristics of Overground versus Treadmill Locomotion in the Saddle Horse. Cells Tissues Organs 1993, 146, 90–94. [Google Scholar] [CrossRef]
- Hargreaves, B.J.; Kronfeld, D.S.; Naylor, J.R.J. Ambient temperature and relative humidity influenced packed cell volume, total plasma protein and other variables in horses during an incremental submaximal field exercise test. Equine Vet. J. 1999, 31, 314–318. [Google Scholar] [CrossRef]
- Cappelli, K.; Felicetti, M.; Capomaccio, S.; Nocelli, C.; Silvestrelli, M.; Verini-Supplizi, A. Effect of training status on immune defence related gene expression in Thoroughbred: Are genes ready for the sprint? Vet. J. 2013, 195, 373–376. [Google Scholar] [CrossRef]
- Ohmura, H.; Mukai, K.; Takahashi, T.; Aida, H.; Jones, J.H. Cardiorespiratory function in Thoroughbreds during locomotion on a treadmill at an incline or decline. Am. J. Vet. Res. 2017, 78, 340–349. [Google Scholar] [CrossRef]
- Rivero, J.L.L.; van Breda, E.; Rogers, C.W.; Lindner, A.; Sloet van Oldruitenborgh-Oosterbaan, M.M. Unexplained underperformance syndrome in sport horses: Classification, potential causes and recognition. Equine Vet. J. 2008, 40, 611–618. [Google Scholar] [CrossRef]
- Cappelli, K.; Supplizi, A.V.; Capomaccio, S.; Albertini, E.; Silvestrelli, M. Analysis Of Peripheral Blood Mononuclear Cells Gene Expression in Endurance Horses; Town & Country Convention Center: San Diego, CA, USA, 2005. [Google Scholar]
- Cappelli, K.; Verini-Supplizi, A.; Capomaccio, S.; Silvestrelli, M. Analysis of peripheral blood mononuclear cells gene expression in endurance horses by cDNA-AFLP technique. Res. Vet. Sci. 2007, 82, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Cappelli, K.; Felicetti, M.; Capomaccio, S.; Spinsanti, G.; Silvestrelli, M.; Verini Supplizi, A. Exercise induced stress in horses: Selection of the most stable reference genes for quantitative RT-PCR normalization. BMC Mol. Biol. 2008, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Cappelli, K.; Felicetti, M.; Capomaccio, S.; Pieramati, C.; Silvestrelli, M.; Verini-Supplizi, A. Exercise-induced up-regulation of MMP-1 and IL-8 genes in endurance horses. BMC Physiol. 2009, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Capomaccio, S.; Cappelli, K.; Barrey, E.; Felicetti, M.; Silvestrelli, M.; Verini-Supplizi, A. Microarray analysis after strenuous exercise in peripheral blood mononuclear cells of endurance horses. Anim. Genet. 2010, 41, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Capomaccio, S.; Cappelli, K.; Spinsanti, G.; Mencarelli, M.; Muscettola, M.; Felicetti, M.; Supplizi, A.; Bonifazi, M. Athletic humans and horses: Comparative analysis of interleukin-6 (IL-6) and IL-6 receptor (IL-6R) expression in peripheral blood mononuclear cells in trained and untrained subjects at rest. BMC Physiol. 2011, 11, 3. [Google Scholar] [CrossRef]
- Capomaccio, S.; Vitulo, N.; Verini-Supplizi, A.; Barcaccia, G.; Albiero, A.; D’Angelo, M.; Campagna, D.; Valle, G.; Felicetti, M.; Silvestrelli, M.; et al. RNA Sequencing of the Exercise Transcriptome in Equine Athletes. PLoS ONE 2013, 8, e83504. [Google Scholar]
- Jacob, A.G.; Smith, C.W.J. Intron retention as a component of regulated gene expression programs. Hum. Genet. 2017, 136, 1043–1057. [Google Scholar] [CrossRef]
- Braunschweig, U.; Barbosa-Morais, N.L.; Pan, Q.; Nachman, E.N.; Alipanahi, B.; Gonatopoulos-Pournatzis, T.; Frey, B.; Irimia, M.; Blencowe, B.J. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014, 24, 1774–1786. [Google Scholar] [CrossRef]
- Elbarbary, R.A.; Lucas, B.A.; Maquat, L.E. Retrotransposons as regulators of gene expression. Science 2016, 351, aac7247. [Google Scholar] [CrossRef]
- Capomaccio, S.; Verini-Supplizi, A.; Galla, G.; Vitulo, N.; Barcaccia, G.; Felicetti, M.; Silvestrelli, M.; Cappelli, K. Transcription of LINE-derived sequences in exercise-induced stress in horses. Anim. Genet. 2010, 41, 23–27. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Kalbfleisch, T.S.; Rice, E.S.; DePriest, M.S.; Walenz, B.P.; Hestand, M.S.; Vermeesch, J.R.; O′Connell, B.L.; Fiddes, I.T.; Vershinina, A.O.; Saremi, N.F.; et al. Improved reference genome for the domestic horse increases assembly contiguity and composition. Commun. Biol. 2018, 1, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Anders, S.; Reyes, A.; Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012, 22, 2008–2017. [Google Scholar] [CrossRef]
- Karolchik, D.; Hinrichs, A.S.; Furey, T.S.; Roskin, K.M.; Sugnet, C.W.; Haussler, D.; Kent, W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004, 32, D493–D496. [Google Scholar] [CrossRef]
- Kuhn, R.M.; Haussler, D.; Kent, W.J. The UCSC genome browser and associated tools. Brief. Bioinform. 2013, 14, 144–161. [Google Scholar] [CrossRef]
- Vilborg, A.; Steitz, J.A. Readthrough transcription: How are DoGs made and what do they do? RNA Biol. 2016, 14, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.C.R.; Acuña, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Non-Coding RNA 2019, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Pecinka, A.; Mittelsten Scheid, O. Stress-Induced Chromatin Changes: A Critical View on Their Heritability. Plant Cell Physiol. 2012, 53, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Biamonti, G.; Caceres, J.F. Cellular stress and RNA splicing. Trends Biochem. Sci. 2009, 34, 146–153. [Google Scholar] [CrossRef]
- Kaer, K.; Branovets, J.; Hallikma, A.; Nigumann, P.; Speek, M. Intronic L1 Retrotransposons and Nested Genes Cause Transcriptional Interference by Inducing Intron Retention, Exonization and Cryptic Polyadenylation. PLoS ONE 2011, 6, e26099. [Google Scholar] [CrossRef]
- Sznajder, Ł.J.; Thomas, J.D.; Carrell, E.M.; Reid, T.; McFarland, K.N.; Cleary, J.D.; Oliveira, R.; Nutter, C.A.; Bhatt, K.; Sobczak, K.; et al. Intron retention induced by microsatellite expansions as a disease biomarker. Proc. Natl. Acad. Sci. USA 2018, 115, 4234–4239. [Google Scholar] [CrossRef]
- Muotri, A.R.; Marchetto, M.C.N.; Coufal, N.G.; Gage, F.H. The necessary junk: New functions for transposable elements. Hum. Mol. Genet. 2007, 16, R159–R167. [Google Scholar] [CrossRef]
- Barash, Y.; Calarco, J.A.; Gao, W.; Pan, Q.; Wang, X.; Shai, O.; Blencowe, B.J.; Frey, B.J. Deciphering the splicing code. Nature 2010, 465, 53–59. [Google Scholar] [CrossRef]
- Long, J.C.; Caceres, J.F. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2009, 417, 15–27. [Google Scholar] [CrossRef]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef]
- Dvinge, H. Regulation of alternative mRNA splicing: Old players and new perspectives. FEBS Lett. 2018, 592, 2987–3006. [Google Scholar] [CrossRef] [PubMed]
- Herzel, L.; Neugebauer, K.M. Quantification of co-transcriptional splicing from RNA-Seq data. Methods 2015, 85, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Blencowe, B.J. Alternative Splicing: New Insights from Global Analyses. Cell 2006, 126, 37–47. [Google Scholar] [CrossRef] [PubMed]
- ZHENG, C.L.; FU, X.-D.; GRIBSKOV, M. Characteristics and regulatory elements defining constitutive splicing and different modes of alternative splicing in human and mouse. RNA 2005, 11, 1777–1787. [Google Scholar] [CrossRef] [PubMed]
- Garg, K.; Green, P. Differing patterns of selection in alternative and constitutive splice sites. Genome Res. 2007, 17, 1015–1022. [Google Scholar] [CrossRef] [PubMed]
- Shalgi, R.; Hurt, J.A.; Lindquist, S.; Burge, C.B. Widespread Inhibition of Posttranscriptional Splicing Shapes the Cellular Transcriptome following Heat Shock. Cell Rep. 2014, 7, 1362–1370. [Google Scholar] [CrossRef]
- Ninomiya, K.; Adachi, S.; Natsume, T.; Iwakiri, J.; Terai, G.; Asai, K.; Hirose, T. LncRNA-dependent nuclear stress bodies promote intron retention through SR protein phosphorylation. EMBO J. 2019, 39, e102729. [Google Scholar]
- Anufrieva, K.S.; Shender, V.О.; Arapidi, G.P.; Pavlyukov, M.S.; Shakhparonov, M.I.; Shnaider, P.V.; Butenko, I.O.; Lagarkova, M.A.; Govorun, V.M. Therapy-induced stress response is associated with downregulation of pre-mRNA splicing in cancer cells. Genome Med. 2018, 10, 49. [Google Scholar] [CrossRef]
- Brady, L.K.; Wang, H.; Radens, C.M.; Bi, Y.; Radovich, M.; Maity, A.; Ivan, C.; Ivan, M.; Barash, Y.; Koumenis, C. Transcriptome analysis of hypoxic cancer cells uncovers intron retention in EIF2B5 as a mechanism to inhibit translation. PLoS Biol. 2017, 15, e2002623. [Google Scholar] [CrossRef]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [CrossRef]
- Wade, C.M.; Giulotto, E.; Sigurdsson, S.; Zoli, M.; Gnerre, S.; Imsland, F.; Lear, T.L.; Adelson, D.L.; Bailey, E.; Bellone, R.R.; et al. Genome Sequence, Comparative Analysis, and Population Genetics of the Domestic Horse. Science 2009, 326, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Crichton, J.H.; Dunican, D.S.; MacLennan, M.; Meehan, R.R.; Adams, I.R. Defending the genome from the enemy within: Mechanisms of retrotransposon suppression in the mouse germline. Cell. Mol. Life Sci. 2014, 71, 1581–1605. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.R.; Doucet, A.J.; Kopera, H.C.; Moldovan, J.B.; Garcia-Pérez, J.L.; Moran, J.V. The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes. Microbiol. Spectr. 2015, 3, 1165–1208. [Google Scholar] [CrossRef] [PubMed]
- Aporntewan, C.; Phokaew, C.; Piriyapongsa, J.; Ngamphiw, C.; Ittiwut, C.; Tongsima, S.; Mutirangura, A. Hypomethylation of Intragenic LINE-1 Represses Transcription in Cancer Cells through AGO2. PLoS ONE 2011, 6, e17934. [Google Scholar] [CrossRef]
- Wongpaiboonwattana, W.; Tosukhowong, P.; Dissayabutra, T.; Mutirangura, A.; Boonla, C. Oxidative Stress Induces Hypomethylation of LINE-1 and Hypermethylation of the RUNX3 Promoter in a Bladder Cancer Cell Line. Asian Pac. J. Cancer Prev. 2013, 14, 3773–3778. [Google Scholar] [CrossRef]
- Miousse, I.R.; Koturbash, I. The Fine LINE: Methylation Drawing the Cancer Landscape. Available online: https://www.hindawi.com/journals/bmri/2015/131547/ (accessed on 5 November 2019).
- Scott, E.; Devine, S. The Role of Somatic L1 Retrotransposition in Human Cancers. Viruses 2017, 9, 131. [Google Scholar] [CrossRef]
- Kroutter, E.N.; Belancio, V.P.; Wagstaff, B.J.; Roy-Engel, A.M. The RNA Polymerase Dictates ORF1 Requirement and Timing of LINE and SINE Retrotransposition. PLoS Genet 2009, 5, e1000458. [Google Scholar] [CrossRef]
- Ostertag, E.M.; Kazazian, H.H., Jr. Biology of Mammalian L1 Retrotransposons. Annu. Rev. Genet. 2001, 35, 501–538. [Google Scholar] [CrossRef]
- Pizarro, J.G.; Cristofari, G. Post-Transcriptional Control of LINE-1 Retrotransposition by Cellular Host Factors in Somatic Cells. Front. Cell Dev. Biol. 2016, 4. [Google Scholar] [CrossRef]
- Mackinnon, L.T. Chronic exercise training effects on immune function. Med. Sci. Sports Exerc. 2000, 32, S369. [Google Scholar] [CrossRef]

| Sample | Reads before Trimming | Reads after Trimming | Quality Check Passed Rate (%) | Uniquely Mapped Reads | Alignment Rate (%) |
|---|---|---|---|---|---|
| S5_T0 | 18,986,960 | 18,391,294 | 96.9 | 7,930,946 | 86.2 |
| S5_T1 | 19,209,286 | 18,644,550 | 97.1 | 8,112,987 | 87.0 |
| S6_T0 | 19,143,702 | 18,542,082 | 96.9 | 7,833,923 | 84.4 |
| S6_T1 | 15,681,066 | 15,223,494 | 97.1 | 6,619,839 | 87.0 |
| S8_T0 | 20,252,456 | 19,654,560 | 97.0 | 8,542,631 | 86.9 |
| S8_T1 | 31,767,674 | 30,848,206 | 97.1 | 13,349,560 | 86.6 |
| S9_T0 | 15,724,850 | 15,241,064 | 96.9 | 6,598,277 | 86.6 |
| S9_T1 | 25,053,756 | 24,340,766 | 97.2 | 10,589,195 | 86.7 |
| S10_T0 | 20,330,330 | 19,746,518 | 97.1 | 8,631,228 | 87.4 |
| S10_T1 | 33,503,070 | 32,465,956 | 96.9 | 14,217,209 | 87.4 |
| Average | 21,965,315 | 21,309,849 | 97.0 | 9,242,580 | 86.60 |
| Sample | Total Alignments | Successfully Assigned Alignments EXONS | % | Successfully Assigned Alignments INTRONS | % | Successfully Assigned Alignments REPEATS | % |
|---|---|---|---|---|---|---|---|
| S10_T0 | 9,418,426 | 5,220,406 | 55.4 | 2,282,112 | 24.2 | 1,397,171 | 14.8 |
| S5_T0 | 8,731,754 | 4,697,224 | 53.8 | 2,233,626 | 25.6 | 1,342,363 | 15.4 |
| S6_T0 | 8,600,396 | 4,899,846 | 57 | 1,863,096 | 21.7 | 1,145,141 | 13.3 |
| S8_T0 | 9,313,192 | 5,146,610 | 55.3 | 2,268,959 | 24.4 | 1,423,386 | 15.3 |
| S9_T0 | 7,236,941 | 4,332,077 | 59.9 | 1,442,060 | 19.9 | 921,749 | 12.7 |
| Average | 8,660,141.80 | 4,859,232.60 | 56.28 | 2,017,970.60 | 23.16 | 1,245,962.00 | 14.30 |
| S10_T1 | 15,479,241 | 8,015,665 | 51.8 | 4,616,059 | 29.8 | 2,603,246 | 16.8 |
| S5_T1 | 8,858,376 | 4,813,955 | 54.3 | 2,389,440 | 27 | 1,377,956 | 15.6 |
| S6_T1 | 7,230,807 | 3,883,746 | 53.7 | 1,976,927 | 27.3 | 1,138,745 | 15.7 |
| S8_T1 | 14,650,606 | 7,835,416 | 53.5 | 3,893,566 | 26.6 | 2,256,959 | 15.4 |
| S9_T1 | 11,642,249 | 6,259,752 | 53.8 | 3,167,804 | 27.2 | 1,806,643 | 15.5 |
| Average | 11,572,255.80 | 6,161,706.80 | 53.42 | 3,208,759.20 | 27.58 | 1,836,709.80 | 15.80 |
| Race VS. Basal | −2.86 | +4.42 | +1.50 | ||||
| t-Test | 0.029 | 0.008 | 0.028 |
| Genes | Introns | |||
|---|---|---|---|---|
| ID | log2Fold Change | ID | log2Fold Change | |
| Upregulated | ENSECAG00000030595 | 10.53 | ENSECAG00000018841 | 6.29 |
| ENSECAG00000019352 | 7.58 | ENSECAG00000039315 | 6.09 | |
| ENSECAG00000001516 | 6.98 | ENSECAG00000020003 | 5.39 | |
| ENSECAG00000038063 | 6.24 | ENSECAG00000017073 | 4.79 | |
| ENSECAG00000023163 | 6.24 | ENSECAG00000011929 | 4.49 | |
| ENSECAG00000009129 | 5.97 | ENSECAG00000004515 | 4.33 | |
| ENSECAG00000009755 | 5.96 | ENSECAG00000002619 | 4.30 | |
| ENSECAG00000021383 | 5.75 | ENSECAG00000010669 | 4.00 | |
| ENSECAG00000011929 | 5.57 | ENSECAG00000005905 | 3.95 | |
| ENSECAG00000020402 | 5.54 | ENSECAG00000030110 | 3.91 | |
| ENSECAG00000034297 | 5.48 | ENSECAG00000003573 | 3.86 | |
| ENSECAG00000039315 | 5.45 | ENSECAG00000010860 | 3.77 | |
| ENSECAG00000033016 | 5.34 | ENSECAG00000016321 | 3.68 | |
| ENSECAG00000015766 | 4.67 | ENSECAG00000013594 | 3.66 | |
| ENSECAG00000020003 | 4.53 | ENSECAG00000000051 | 3.66 | |
| ENSECAG00000040244 | 4.51 | ENSECAG00000039959 | 3.57 | |
| ENSECAG00000002234 | 4.36 | ENSECAG00000014979 | 3.56 | |
| ENSECAG00000010860 | 4.31 | ENSECAG00000023173 | 3.47 | |
| ENSECAG00000033856 | 4.31 | ENSECAG00000009215 | 3.43 | |
| ENSECAG00000015992 | 4.00 | ENSECAG00000015318 | 3.37 | |
| Downregulated | ENSECAG00000032106 | −23.04 | ENSECAG00000040402 | −4.91 |
| ENSECAG00000034632 | −6.08 | ENSECAG00000023475 | −4.33 | |
| ENSECAG00000007460 | −5.64 | ENSECAG00000028489 | −4.14 | |
| ENSECAG00000021087 | −4.55 | ENSECAG00000011895 | −3.99 | |
| ENSECAG00000010281 | −4.17 | ENSECAG00000031371 | −3.62 | |
| ENSECAG00000009895 | −4.07 | ENSECAG00000032756 | −3.58 | |
| ENSECAG00000035315 | −3.54 | ENSECAG00000036032 | −3.56 | |
| ENSECAG00000008274 | −3.40 | ENSECAG00000036704 | −3.48 | |
| ENSECAG00000009869 | −3.22 | ENSECAG00000000386 | −3.45 | |
| ENSECAG00000032544 | −3.09 | ENSECAG00000017676 | −3.43 | |
| ENSECAG00000032503 | −3.04 | ENSECAG00000025038 | −3.34 | |
| ENSECAG00000030934 | −2.90 | ENSECAG00000022510 | −3.34 | |
| ENSECAG00000034974 | −2.85 | ENSECAG00000006114 | −3.28 | |
| ENSECAG00000009625 | −2.78 | ENSECAG00000027794 | −3.26 | |
| ENSECAG00000034925 | −2.72 | ENSECAG00000011660 | −3.18 | |
| ENSECAG00000010326 | −2.68 | ENSECAG00000014338 | −3.12 | |
| ENSECAG00000036608 | −2.63 | ENSECAG00000033795 | −3.10 | |
| ENSECAG00000037661 | −2.55 | ENSECAG00000033519 | −2.99 | |
| ENSECAG00000014338 | −2.53 | ENSECAG00000011723 | −2.97 | |
| ENSECAG00000015083 | −2.48 | ENSECAG00000036290 | −2.85 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cappelli, K.; Mecocci, S.; Gioiosa, S.; Giontella, A.; Silvestrelli, M.; Cherchi, R.; Valentini, A.; Chillemi, G.; Capomaccio, S. Gallop Racing Shifts Mature mRNA towards Introns: Does Exercise-Induced Stress Enhance Genome Plasticity? Genes 2020, 11, 410. https://doi.org/10.3390/genes11040410
Cappelli K, Mecocci S, Gioiosa S, Giontella A, Silvestrelli M, Cherchi R, Valentini A, Chillemi G, Capomaccio S. Gallop Racing Shifts Mature mRNA towards Introns: Does Exercise-Induced Stress Enhance Genome Plasticity? Genes. 2020; 11(4):410. https://doi.org/10.3390/genes11040410
Chicago/Turabian StyleCappelli, Katia, Samanta Mecocci, Silvia Gioiosa, Andrea Giontella, Maurizio Silvestrelli, Raffaele Cherchi, Alessio Valentini, Giovanni Chillemi, and Stefano Capomaccio. 2020. "Gallop Racing Shifts Mature mRNA towards Introns: Does Exercise-Induced Stress Enhance Genome Plasticity?" Genes 11, no. 4: 410. https://doi.org/10.3390/genes11040410
APA StyleCappelli, K., Mecocci, S., Gioiosa, S., Giontella, A., Silvestrelli, M., Cherchi, R., Valentini, A., Chillemi, G., & Capomaccio, S. (2020). Gallop Racing Shifts Mature mRNA towards Introns: Does Exercise-Induced Stress Enhance Genome Plasticity? Genes, 11(4), 410. https://doi.org/10.3390/genes11040410








