Validation of the Reference Genes for the Gene Expression Studies in Chicken DT40 Cell Line
Abstract
1. Introduction
2. Materials and Methods
2.1. Reference Genes Selection
2.2. Sample Collection for Analysis
2.3. RT-qPCR
2.4. Statistical Tools to Select the Appropriate Reference Genes
2.5. Target Genes
2.6. Relative Gene Expression and Statistical Analysis
3. Results
3.1. Reference Genes Selection
3.2. Amplification Efficiencies in the qPCR Analysis
3.3. Target Genes Selection
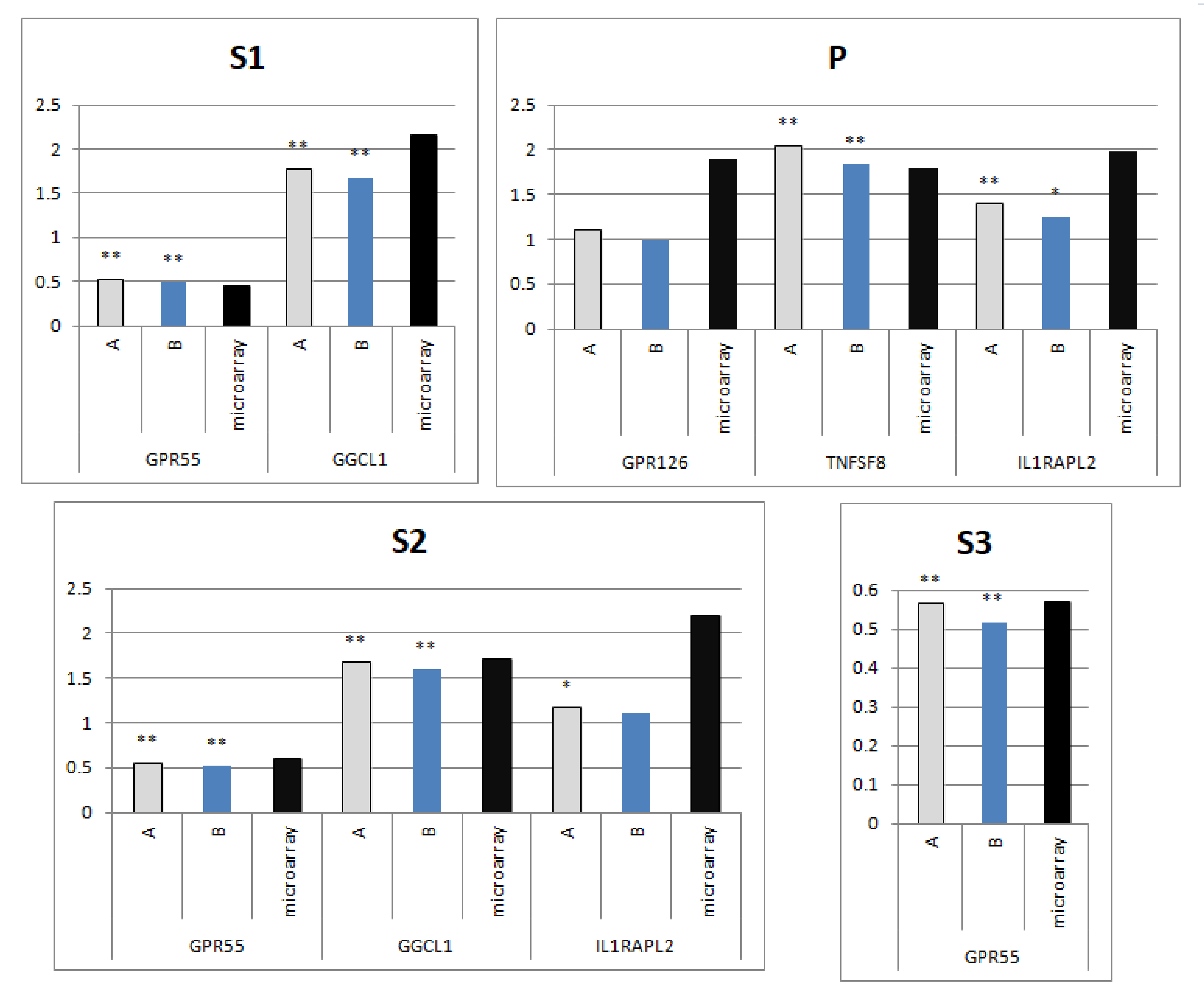
3.4. Comparison qPCR Results with Microarrays Data Based on the Selected Reference Genes
4. Discussion
4.1. Reference Genes Selection
4.2. Comparison qPCR Results with Microarrays Data Based on Selected Reference Genes
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes, revisited. Trends Genet. 2013, 29, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Zakrajsek, B.A. Effect of experimental treatment on housekeeping gene expression: Validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods 2000, 46, 69–81. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]
- Molnár, J.; Póti, Á.; Pipek, O.; Krzystanek, M.; Kanu, N.; Swanton, C.; Tusnády, G.E.; Szallasi, Z.; Csabai, I.; Szüts, D. The genome of the chicken DT40 bursal lymphoma cell line. G3 Genes Genomes Genet. 2014, 4, 2231–2240. [Google Scholar] [CrossRef]
- Chang, H.; Delany, M.E. Karyotype stability of the DT40 chicken B cell line: Macrochromosome variation and cytogenetic mosaicism. Chromosom. Res. 2004, 12, 299–307. [Google Scholar] [CrossRef]
- Dunislawska, A.; Slawinska, A.; Siwek, M. Expression of FOXJ1 and ITGB4 is Activated upon KLH and LTA Stimulation in the DT40 Cell Line. Folia Biol. (Krakow) 2017, 65, 9–18. [Google Scholar] [CrossRef]
- Sevane, N.; Bialade, F.; Velasco, S.; Rebolé, A.; Rodríguez, M.L.; Ortiz, L.T.; Cañón, J.; Dunner, S. Dietary inulin supplementation modifies significantly the liver transcriptomic profile of broiler chickens. PLoS One 2014, 9. [Google Scholar] [CrossRef]
- De Boever, S.; Vangestel, C.; De Backer, P.; Croubels, S.; Sys, S.U. Identification and validation of housekeeping genes as internal control for gene expression in an intravenous LPS inflammation model in chickens. Vet. Immunol. Immunopathol. 2008, 122, 312–317. [Google Scholar] [CrossRef]
- Cinar, M.U.; Islam, M.A.; Pröll, M.; Kocamis, H.; Tholen, E.; Tesfaye, D.; Looft, C.; Schellander, K.; Uddin, M.J. Evaluation of suitable reference genes for gene expression studies in porcine PBMCs in response to LPS and LTA. BMC Res. Notes 2013, 6, 56. [Google Scholar] [CrossRef]
- Yang, F.; Lei, X.; Rodriguez-Palacios, A.; Tang, C.; Yue, H. Selection of reference genes for quantitative real-time PCR analysis in chick embryo fibroblasts infected with avian leukosis virus subgroup J. BMC Res Notes. 2013, 402. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant. Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Dhar, A.K.; Bowers, R.M.; Licon, K.S.; Veazey, G.; Read, B. Validation of reference genes for quantitative measurement of immune gene expression in shrimp. Mol. Immunol. 2009, 46, 1688–1695. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Winding, P.; Berchtold, M.W. The chicken B cell line DT40: A novel tool for gene disruption experiments. J. Immunol. Methods 2001, 249, 1–16. [Google Scholar] [CrossRef]
- Katarzyńska-Banasik, D.; Grzesiak, M.; Sechman, A. Selection of reference genes for quantitative real-time PCR analysis in chicken ovary following silver nanoparticle treatment. Environ. Toxicol. Pharmacol. 2017, 56, 186–190. [Google Scholar] [CrossRef]
- Simon, Á.; Jávor, A.; Bai, P.; Oláh, J.; Czeglédi, L. Reference gene selection for reverse transcription quantitative polymerase chain reaction in chicken hypothalamus under different feeding status. J. Anim. Physiol. Anim. Nutr. (Berl.) 2018, 102, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gao, Y.Y.; Huang, Y.Q.; Fan, Q.; Lu, X.T.; Wang, C.K. Selection of housekeeping genes for quantitative gene expression analysis in yellow-feathered broilers. Ital. J. Anim. Sci. 2018, 17, 540–546. [Google Scholar] [CrossRef]
- Dunislawska, A.; Slawinska, A.; Bednarczyk, M.; Siwek, M. Transcriptome modulation by in ovo delivered Lactobacillus synbiotics in a range of chicken tissues. Gene 2019, 698, 27–33. [Google Scholar] [CrossRef] [PubMed]

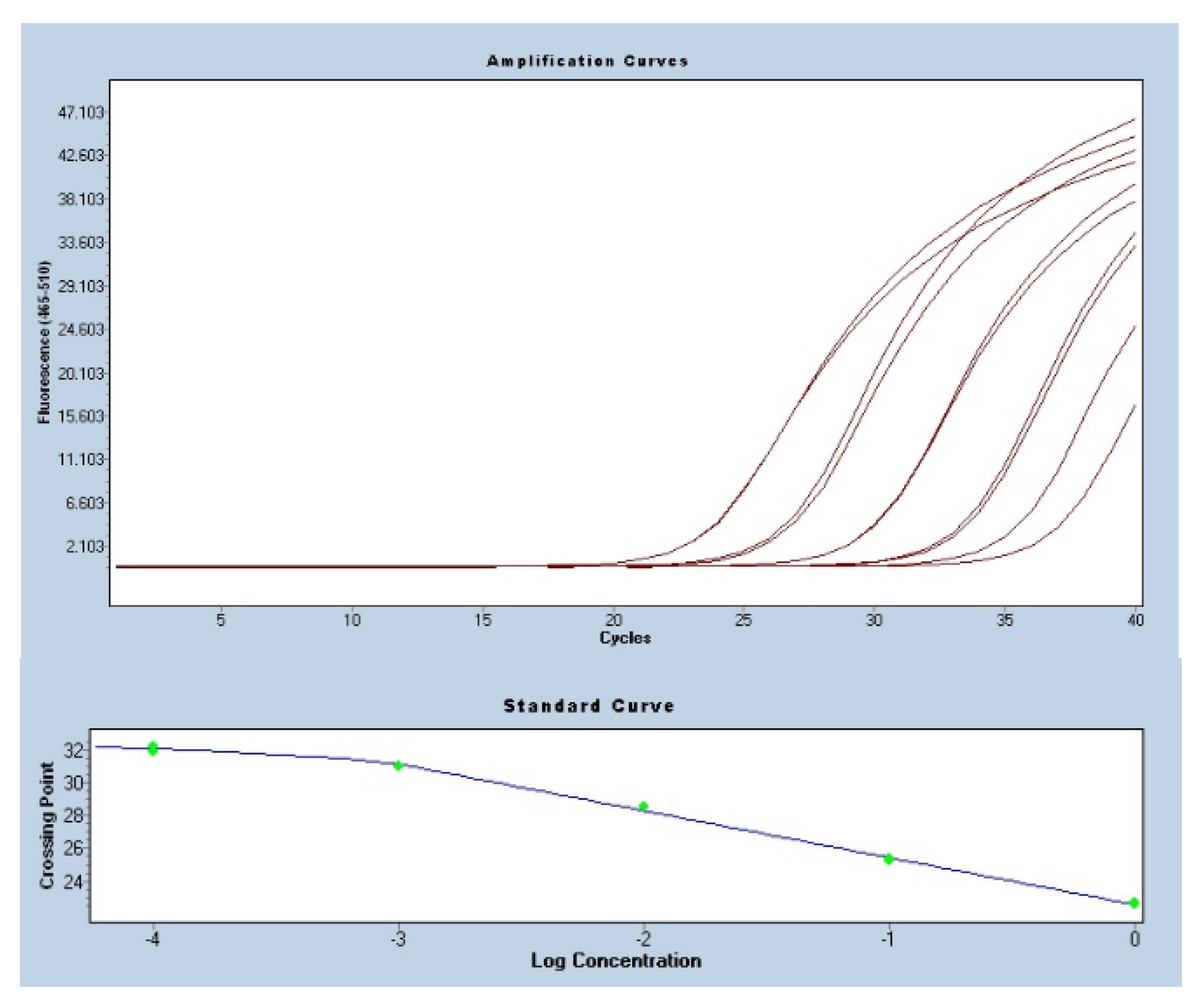
| Gene | Gene name | Primer sequence (5′-3′) | Reference |
|---|---|---|---|
| G6PDH | glucose-6-phosphate dehydrogenase | F: CGGGAACCAAATGCACTTCGT R: GGCTGCCGTAGAGGTATGGGA | [8] |
| HPRT | hypoxanthine phophoribosyl-transferase | F: CCCAAACATTATGCAGACGA R: TGTCCTGTCCATGATGAGC | [9] |
| SDHA | succinate dehydro-gense complex, subunit A | F: AGAGCCTCAAGTTCGGGAAG R: CAGGAGATCCAAGGCAAAAT | [10] |
| B2M | beta-2-microglobulin | F: ACTTTTCACACCGCTCCAGT R: CGGATGGAACCCAGATACAT | [10] |
| RPL4 | ribosomal protein L4 | F: AGGAGGCTGTTCTGCTTCTG R: TCCAGGGATGTTTCTGAAGG | [10] |
| RPL30 | ribosomal protein L30 | F: GAGTCACCTGGGTCAATAA R: CCAACAACTGTCCTGCTTT | [11] |
| UB | Ubiquitin C | F: GGGATGCAGATCTTCGTGAAA R: CTTGCCAGCAAAGATCAACCTT | [9] |
| Reference Gene | Delta Ct | BestKeeper | NormFinder | geNorm |
|---|---|---|---|---|
| Average of STED | Standard Deviation SD | Stability Value | Stability Value | |
| G6PDH | 0.146 | 0.184 | 0.091 | 0.093 |
| HPRT | 0.166 | 0.231 | 0.126 | 0.11 |
| SDHA | 0.132 | 0.197 | 0.063 | 0.079 |
| B2M | 0.193 | 0.241 | 0.162 | 0.16 |
| RPL4 | 0.126 | 0.194 | 0.033 | 0.079 |
| RPL30 | 0.184 | 0.264 | 0.15 | 0.146 |
| UB | 0.17 | 0.245 | 0.131 | 0.13 |
| Gene | Gene Name | Primer Sequence (5´-3´) | Gene ID |
|---|---|---|---|
| GPR55 | G protein-coupled receptor 55 | F: GGAGTCACCCGCTGGAGAA R: TCTCCCCCTGCTGAGATGTT | 770641 |
| GPR126 | G protein-coupled receptor 126 | F: TGTGCGAATTGCCGTGTCT R: CACTGCTGCTCTTGGTTGCTT | 421673 |
| TNFSF8 | tumor necrosis factor (ligand) superfamily, member 8 | F: GCAAAGGGAACACCTCTGAAGA R: TGAGTTTGACACTCTCATGTATGCA | 378930 |
| GGCL1 | chemokine-like ligand 1 | F: TGTATGAGTACACGGGAAGCAGAT R: ACAGCACTTCTTCCCCTCAAAG | 417464 |
| IL1RAPL2 | interleukin 1 receptor accessory protein-like 2 | F: GATCTGGCTAACTATACGTGTCATGTG R: ATCAAATCTTTCTTGCGCAGAAG | 422379 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dunislawska, A.; Slawinska, A.; Siwek, M. Validation of the Reference Genes for the Gene Expression Studies in Chicken DT40 Cell Line. Genes 2020, 11, 372. https://doi.org/10.3390/genes11040372
Dunislawska A, Slawinska A, Siwek M. Validation of the Reference Genes for the Gene Expression Studies in Chicken DT40 Cell Line. Genes. 2020; 11(4):372. https://doi.org/10.3390/genes11040372
Chicago/Turabian StyleDunislawska, Aleksandra, Anna Slawinska, and Maria Siwek. 2020. "Validation of the Reference Genes for the Gene Expression Studies in Chicken DT40 Cell Line" Genes 11, no. 4: 372. https://doi.org/10.3390/genes11040372
APA StyleDunislawska, A., Slawinska, A., & Siwek, M. (2020). Validation of the Reference Genes for the Gene Expression Studies in Chicken DT40 Cell Line. Genes, 11(4), 372. https://doi.org/10.3390/genes11040372






