A Whole Genome Re-Sequencing Based GWA Analysis Reveals Candidate Genes Associated with Ivermectin Resistance in Haemonchus contortus
Abstract
1. Introduction
2. Materials and Methods
2.1. Background of Strains, Worm Collection, and Identification
2.2. DNA Isolation, Library Construction, and Whole Genome Re-Sequencing
2.3. Mapping Reads and Variant Calling
2.4. Characterization of Genetic Variability and Tracing Selection
2.5. Egg Extraction and Culturing Eggs into Larvae
2.6. RNAi-Based Silencing of Candidate Genes
2.7. Larval Feeding Inhibition Assay (LFIA)
2.8. RNA Extraction and qPCR
2.9. Statistical Analysis
2.10. Data Availability
3. Results
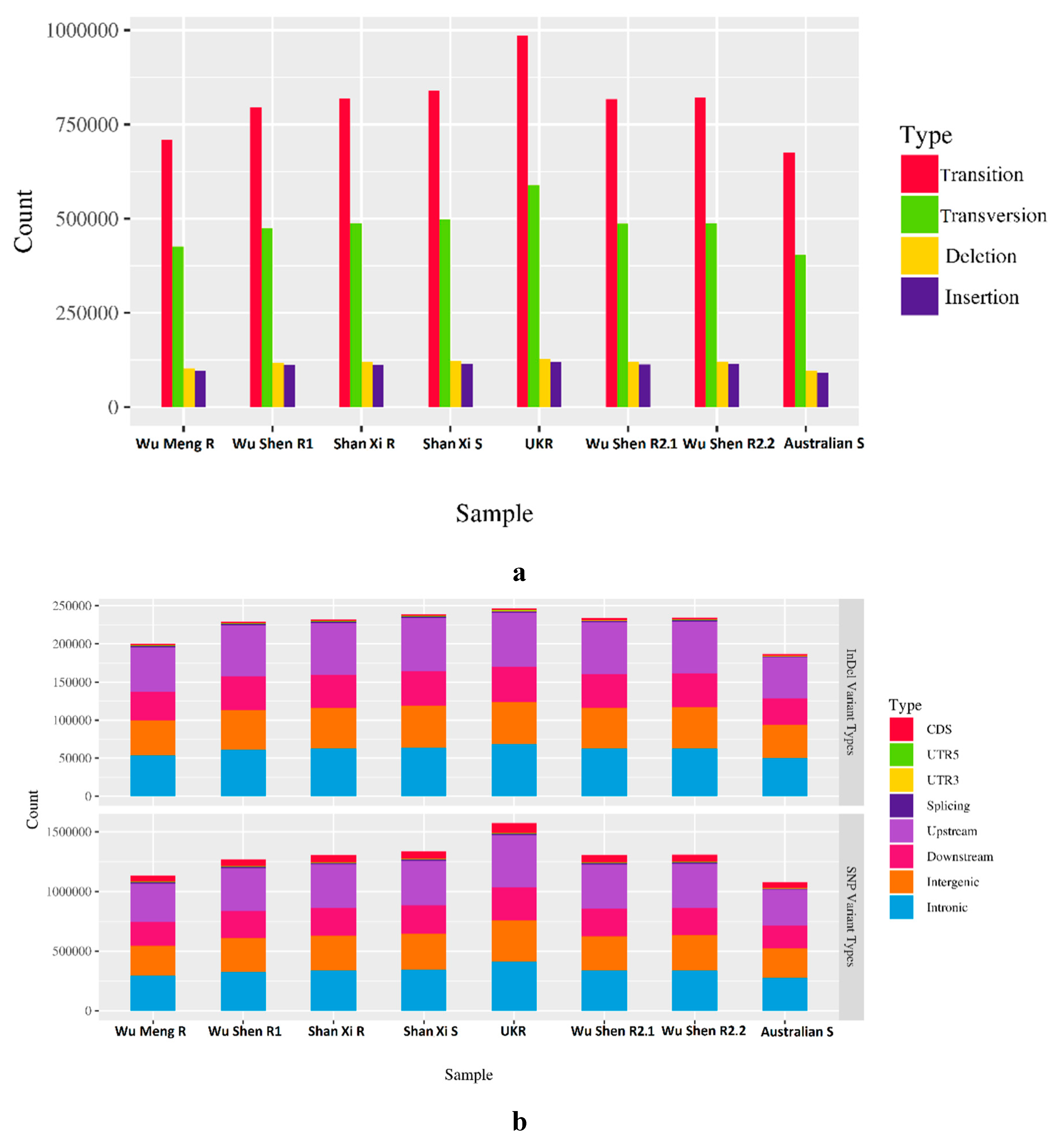
3.1. Results of Whole Genome Re-Sequencing
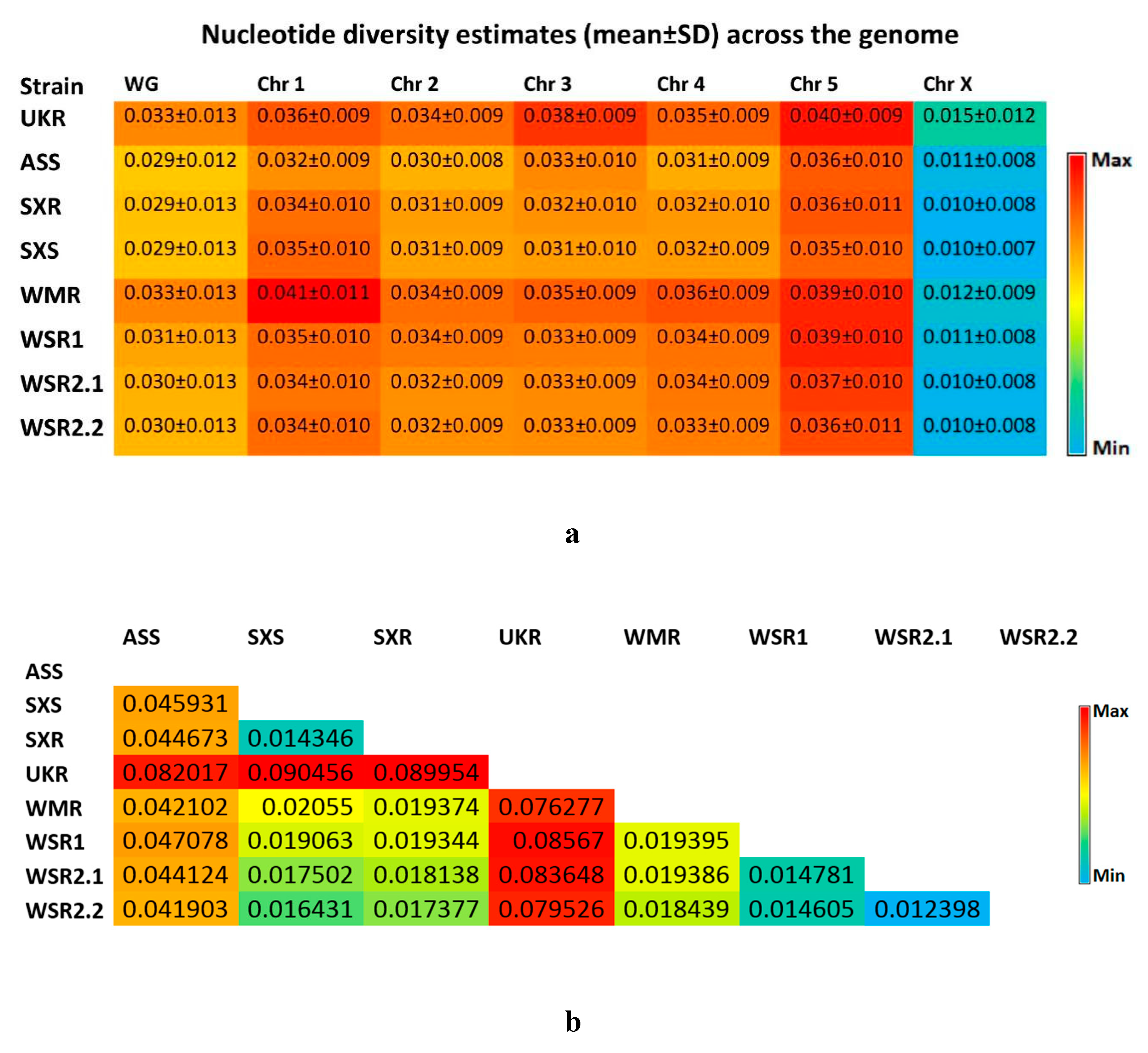
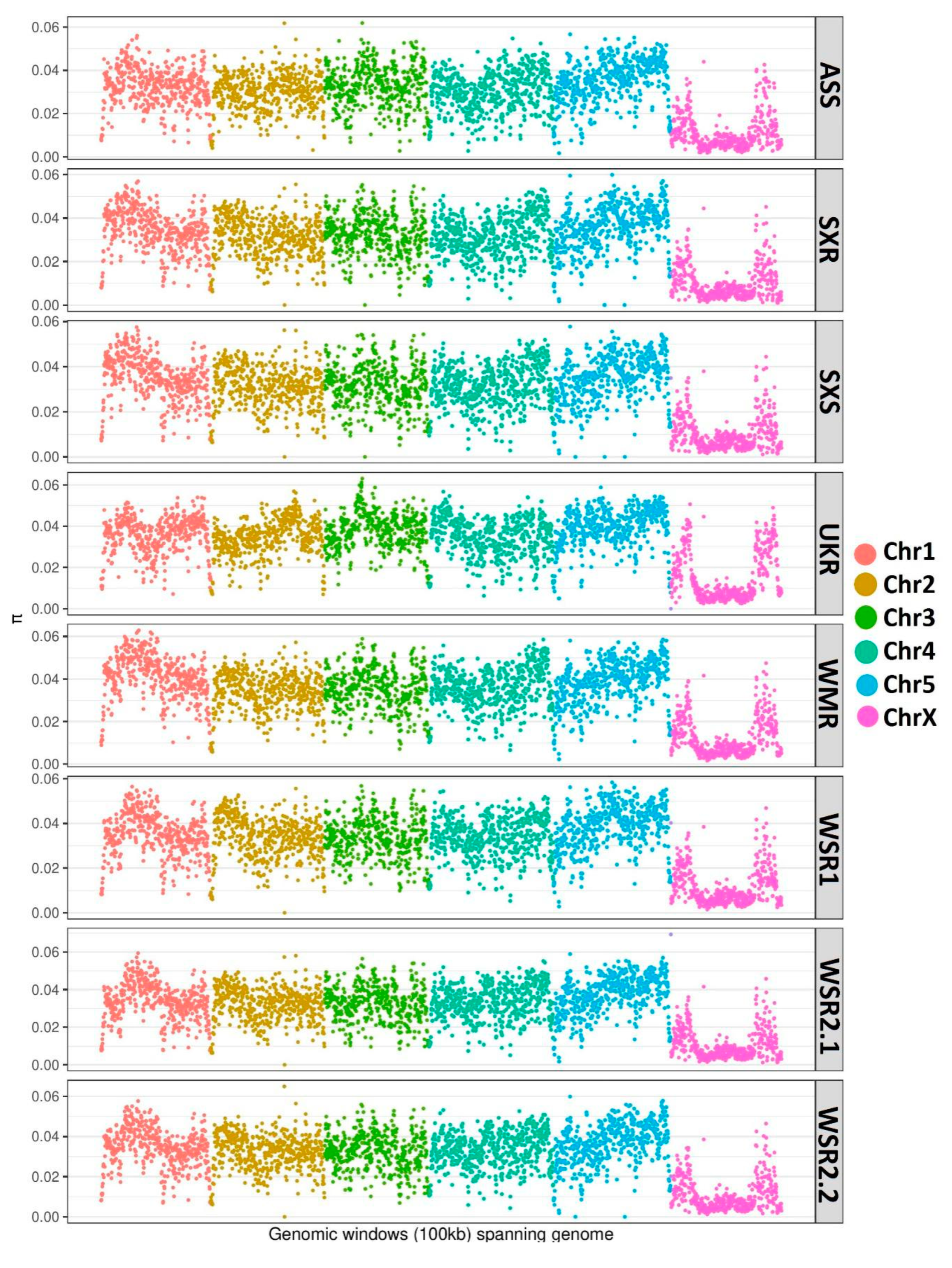
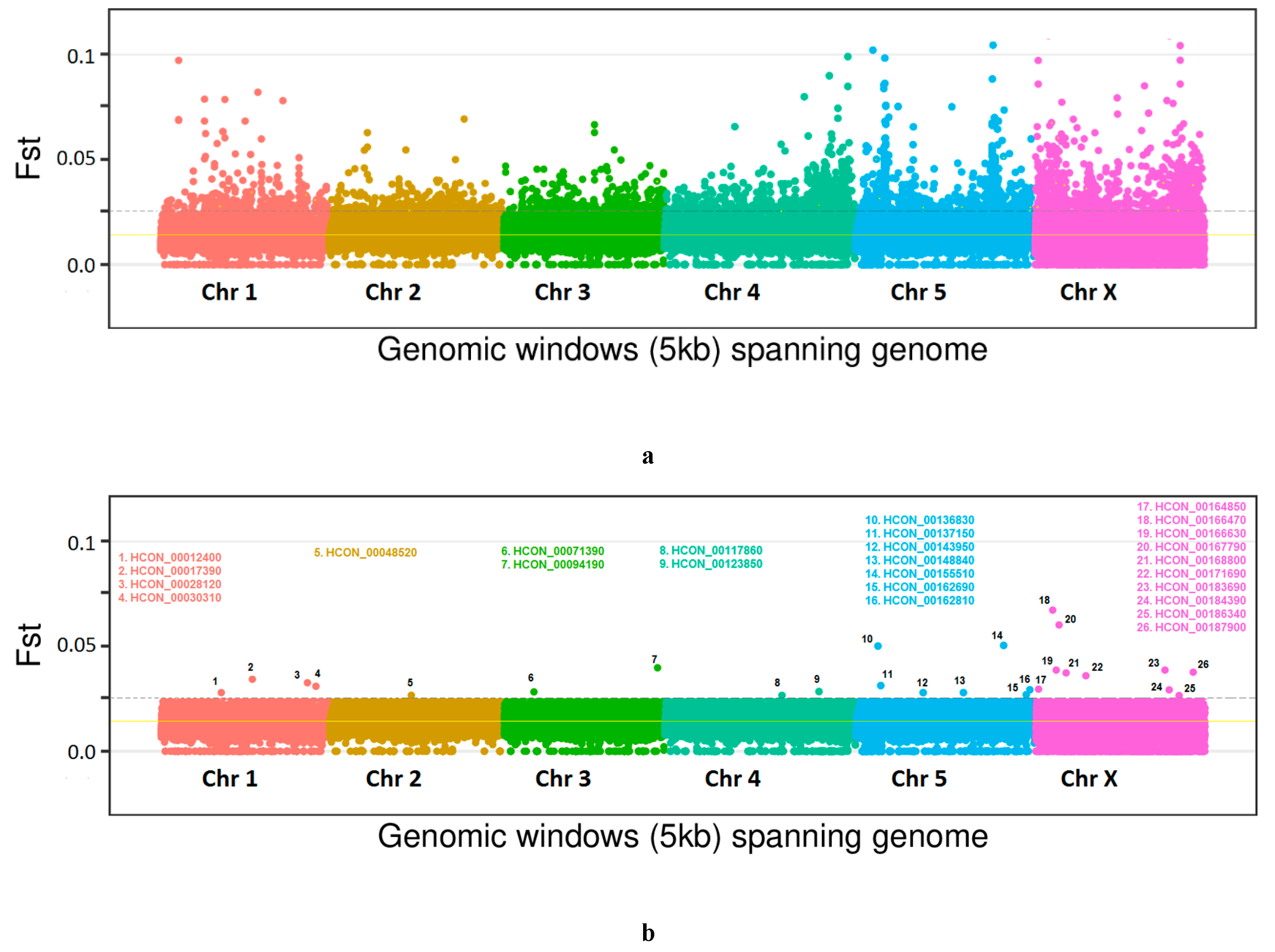
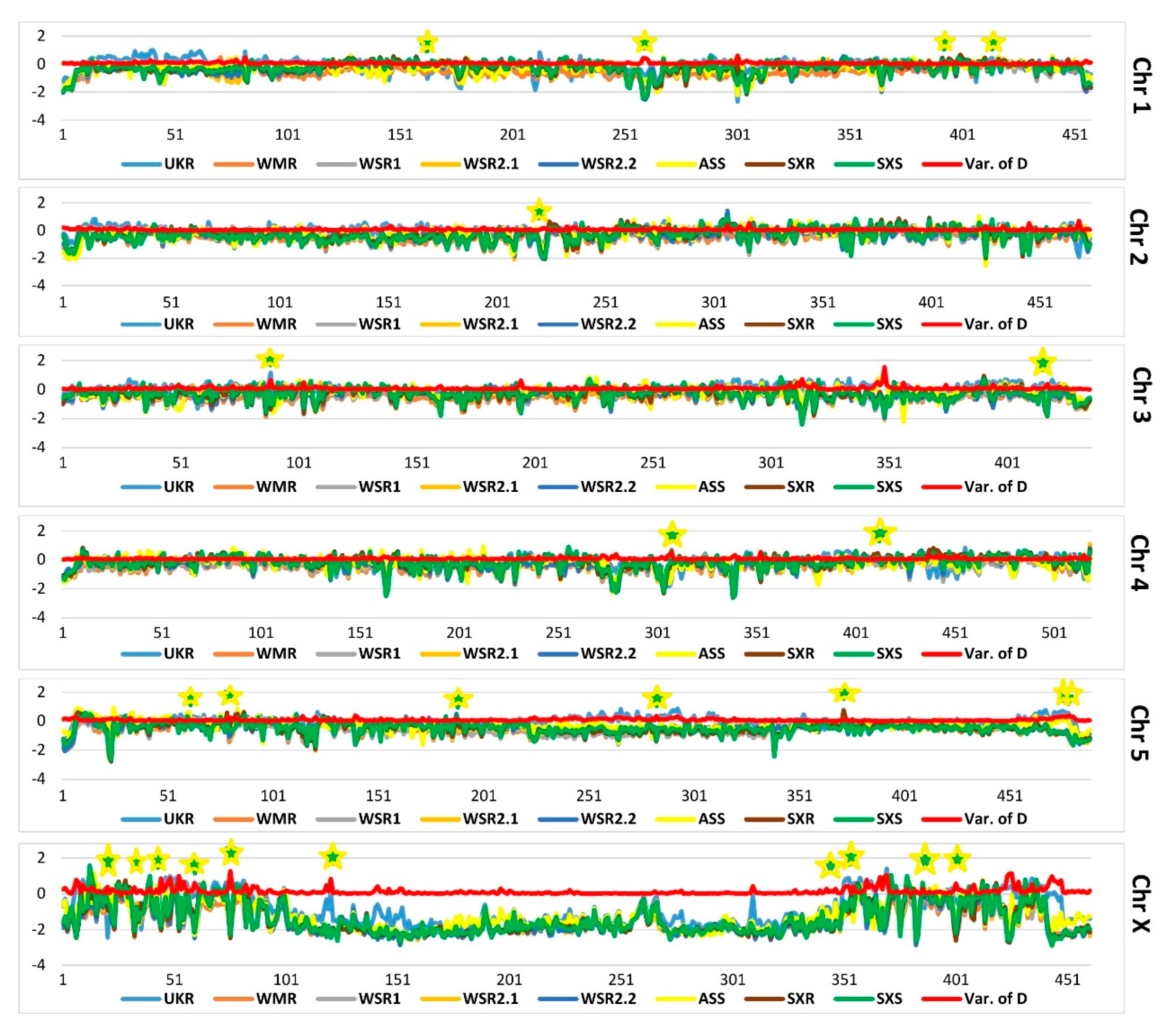
3.2. Genome-wide Analysis of Genetic Diversity and Tracing Selection
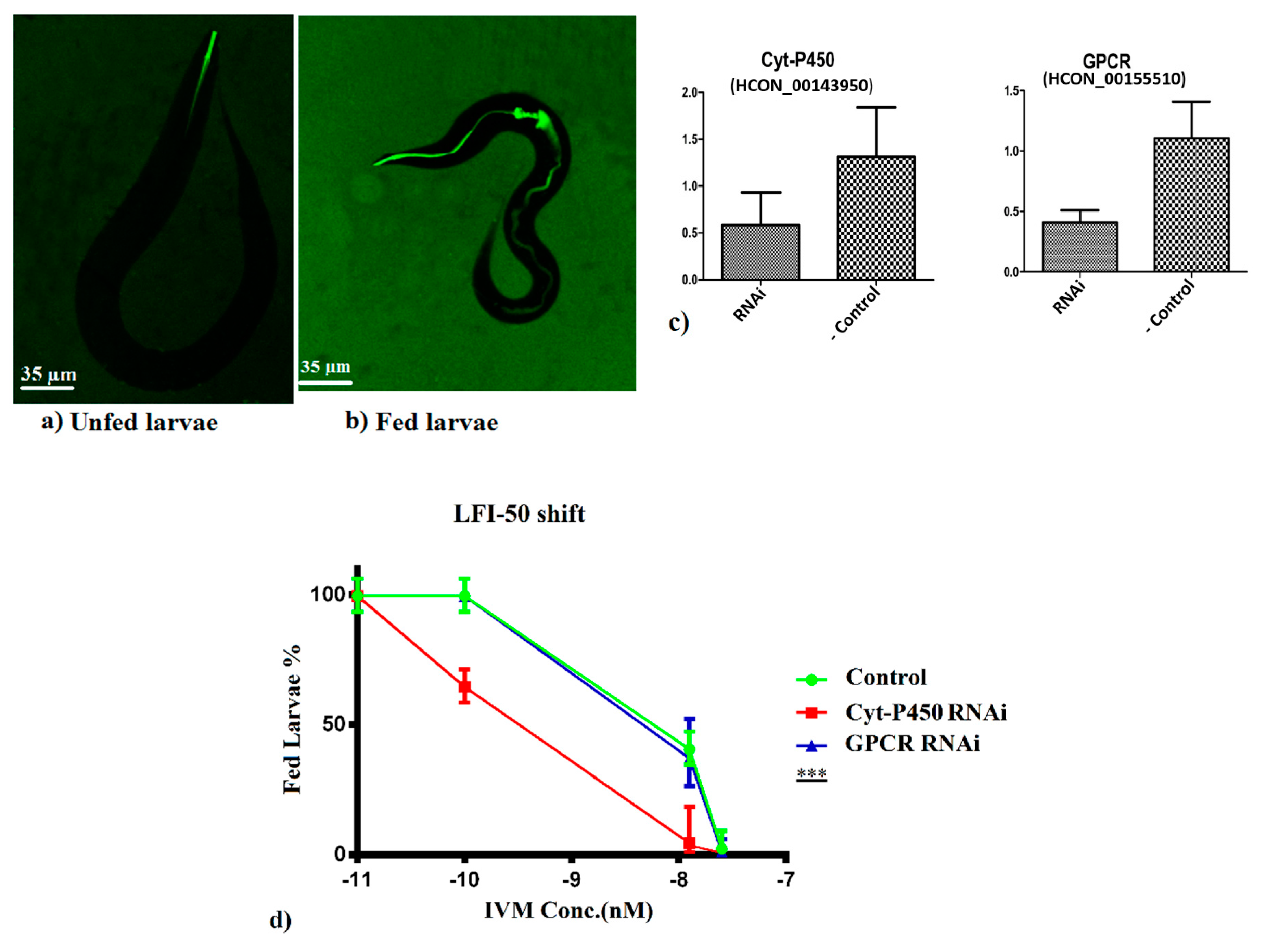
3.3. RNAi Assays for Functional Validations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ménez, C.; Alberich, M.; Courtot, E.; Guegnard, F.; Blanchard, A.; Aguilaniu, H.; Lespine, A. The transcription factor NHR-8: A new target to increase ivermectin efficacy in nematodes. PLoS Pathog. 2019, 15, e1007598. [Google Scholar] [CrossRef] [PubMed]
- Hoberg, E.P.; Zarlenga, D.S. Evolution and Biogeography of Haemonchus contortus: Linking Faunal Dynamics in Space and Time. Adv. Parasitol. 2016, 93, 1–30. [Google Scholar] [PubMed]
- Emery, D.L.; Hunt, P.W.; Le Jambre, L.F. Haemonchus contortus: the then and now, and where to from here? Int. J. Parasitol. 2016, 46, 755–769. [Google Scholar] [CrossRef] [PubMed]
- Roeber, F.; Jex, A.R.; Gasser, R.B. Impact of gastrointestinal parasitic nematodes of sheep, and the role of advanced molecular tools for exploring epidemiology and drug resistance - an Australian perspective. Parasit. Vectors 2013, 6, 153. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Luo, X.; Yuan, C.; Zhao, X.; Nisar, A.; Li, J.; Yang, X.; Zhang, J.; Feng, X. Microsatellite analysis reveals extensive gene flow, and lack of population structure in the farm populations of Haemonchus contortus in northern China. Parasitol. Int. 2019, 73, 101959. [Google Scholar] [CrossRef] [PubMed]
- Omura, S.; Crump, A. The life and times of ivermectin - a success story. Nat. Rev. Microbiol. 2004, 2, 984–989. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, R.M.; Vidyashankar, A.N. An inconvenient truth: global worming and anthelmintic resistance. Vet. Parasitol. 2012, 186, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Kotze, A.; Prichard, R. Anthelmintic Resistance in Haemonchus contortus: History, Mechanisms and Diagnosis. Adv. Parasitol. 2016, 93, 397–428. [Google Scholar] [PubMed]
- Rose, H.; Rinaldi, L.; Bosco, A.; Mavrot, F.; de Waal, T.; Skuce, P.; Charlier, J.; Torgerson, P.R.; Hertzberg, H.; Hendrickx, G.; et al. Widespread anthelmintic resistance in European farmed ruminants: a systematic review. Vet. Rec. 2015, 176, 546. [Google Scholar] [CrossRef] [PubMed]
- Osei-Atweneboana, M.Y.; Awadzi, K.; Attah, S.K.; Boakye, D.A.; Gyapong, J.O.; Prichard, R.K. Phenotypic evidence of emerging ivermectin resistance in Onchocerca volvulus. PLoS Negl. Trop. Dis. 2011, 5, e998. [Google Scholar] [CrossRef]
- Doyle, S.R.; Bourguinat, C.; Nana-Djeunga, H.C.; Kengne-Ouafo, J.A.; Pion, S.D.S.; Bopda, J.; Kamgno, J.; Wanji, S.; Che, H.; Kuesel, A.C.; et al. Genome-wide analysis of ivermectin response by Onchocerca volvulus reveals that genetic drift and soft selective sweeps contribute to loss of drug sensitivity. PLoS Negl. Trop. Dis. 2017, 11, e0005816. [Google Scholar] [CrossRef] [PubMed]
- Geerts, S.; Gryseels, B. Drug resistance in human helminths: current situation and lessons from livestock. Clin. Microbiol. Rev. 2000, 13, 207–222. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, R.M. Drug resistance in nematodes of veterinary importance: a status report. Trends Parasitol. 2004, 20, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Horsberg, T.E. Avermectin use in aquaculture. Curr. Pharm. Biotechnol. 2012, 13, 1095–1102. [Google Scholar] [CrossRef]
- Ghosh, R.; Andersen, E.C.; Shapiro, J.A.; Gerke, J.P.; Kruglyak, L. Natural variation in a chloride channel subunit confers avermectin resistance in C. elegans. Science 2012, 335, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, I.A.; Leathwick, D.M. Anthelmintic resistance in nematode parasites of cattle: a global issue? Trends Parasitol. 2011, 27, 176–181. [Google Scholar] [CrossRef]
- Harhay, M.O.; Horton, J.; Olliaro, P.L. Epidemiology and control of human gastrointestinal parasites in children. Expert Rev. Anti Infect. Ther. 2010, 8, 219–234. [Google Scholar] [CrossRef]
- Molento, M.B. Parasite control in the age of drug resistance and changing agricultural practices. Vet. Parasitol. 2009, 163, 229–234. [Google Scholar] [CrossRef]
- Besier, R.B.; Kahn, L.P.; Sargison, N.D.; Van Wyk, J.A. Chapter Four - The Pathophysiology, Ecology and Epidemiology of Haemonchus contortus Infection in Small Ruminants. In Advances in parasitology; Gasser, R.B., Samson-Himmelstjerna, G.V., Eds.; Academic Press: Cambridge, MA, USA, 2016; Volume 93, pp. 95–143. [Google Scholar]
- James, C.E.; Hudson, A.L.; Davey, M.W. Drug resistance mechanisms in helminths: is it survival of the fittest? Trends Parasitol. 2009, 25, 328–335. [Google Scholar] [CrossRef]
- Kotze, A.C.; Hunt, P.W.; Skuce, P.; von Samson-Himmelstjerna, G.; Martin, R.J.; Sager, H.; Krücken, J.; Hodgkinson, J.; Lespine, A.; Jex, A.R. Recent advances in candidate-gene and whole-genome approaches to the discovery of anthelmintic resistance markers and the description of drug/receptor interactions. Int. J. Parasitol. Drugs Drug Resist. 2014, 4, 164–184. [Google Scholar] [CrossRef]
- Lespine, A.; Ménez, C.; Bourguinat, C.; Prichard, R.K. P-glycoproteins and other multidrug resistance transporters in the pharmacology of anthelmintics: Prospects for reversing transport-dependent anthelmintic resistance. Int. J. Parasitol. Drugs Drug Resist. 2012, 2, 58–75. [Google Scholar] [CrossRef] [PubMed]
- Redman, E.; Sargison, N.; Whitelaw, F.; Jackson, F.; Morrison, A.; Bartley, D.J.; Gilleard, J.S. Introgression of ivermectin resistance genes into a susceptible Haemonchus contortus strain by multiple backcrossing. PLoS Pathog. 2012, 8, e1002534. [Google Scholar] [CrossRef] [PubMed]
- Blackhall, W.J.; Pouliot, J.F.; Prichard, R.K.; Beech, R.N. Haemonchus contortus: selection at a glutamate-gated chloride channel gene in ivermectin- and moxidectin-selected strains. Exp. Parasitol. 1998, 90, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Eng, J.K.; Blackhall, W.J.; Osei-Atweneboana, M.Y.; Bourguinat, C.; Galazzo, D.; Beech, R.N.; Unnasch, T.R.; Awadzi, K.; Lubega, G.W.; Prichard, R.K. Ivermectin selection on beta-tubulin: evidence in Onchocerca volvulus and Haemonchus contortus. Mol. Biochem. Parasitol. 2006, 150, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Shi, X.; Yuan, C.; Ai, M.; Ge, C.; Hu, M.; Feng, X.; Yang, X. Genome-wide SNP analysis using 2b-RAD sequencing identifies the candidate genes putatively associated with resistance to ivermectin in Haemonchus contortus. Parasit. Vectors 2017, 10, 31. [Google Scholar] [CrossRef]
- Blackhall, W.J.; Prichard, R.K.; Beech, R.N. Selection at a gamma-aminobutyric acid receptor gene in Haemonchus contortus resistant to avermectins/milbemycins. Mol. Biochem. Parasitol. 2003, 131, 137–145. [Google Scholar] [CrossRef]
- Urdaneta-Marquez, L.; Bae, S.H.; Janukavicius, P.; Beech, R.; Dent, J.; Prichard, R. A dyf-7 haplotype causes sensory neuron defects and is associated with macrocyclic lactone resistance worldwide in the nematode parasite Haemonchus contortus. Int. J. Parasitol. 2014, 44, 1063–1071. [Google Scholar] [CrossRef]
- Raza, A.; Kopp, S.R.; Bagnall, N.H.; Jabbar, A.; Kotze, A.C. Effects of in vitro exposure to ivermectin and levamisole on the expression patterns of ABC transporters in Haemonchus contortus larvae. Int. J. Parasitol. Drugs Drug Resist. 2016, 6, 103–115. [Google Scholar] [CrossRef]
- Lloberas, M.; Alvarez, L.; Entrocasso, C.; Virkel, G.; Ballent, M.; Mate, L.; Lanusse, C.; Lifschitz, A. Comparative tissue pharmacokinetics and efficacy of moxidectin, abamectin and ivermectin in lambs infected with resistant nematodes: Impact of drug treatments on parasite P-glycoprotein expression. Int. J. Parasitol. Drugs Drug Resist. 2013, 3, 20–27. [Google Scholar] [CrossRef]
- Laing, R.; Maitland, K.; Lecová, L.; Skuce, P.J.; Tait, A.; Devaney, E. Analysis of putative resistance gene loci in UK field populations of Haemonchus contortus after 6 years of macrocyclic lactone use. Int. J. Parasitol. 2016, 46, 621–630. [Google Scholar] [CrossRef]
- Rezansoff, A.M.; Laing, R.; Gilleard, J.S. Evidence from two independent backcross experiments supports genetic linkage of microsatellite Hcms8a20, but not other candidate loci, to a major ivermectin resistance locus in Haemonchus contortus. Int. J. Parasitol. 2016, 46, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Prichard, R. Genetic variability following selection of Haemonchus contortus with anthelmintics. Trends Parasitol. 2001, 17, 445–453. [Google Scholar] [CrossRef]
- Doyle, S.R.; Cotton, J.A. Genome-wide Approaches to Investigate Anthelmintic Resistance. Trends Parasitol. 2019, 35, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Beech, R.N.; Skuce, P.; Bartley, D.J.; Martin, R.J.; Prichard, R.K.; Gilleard, J.S. Anthelmintic resistance: markers for resistance, or susceptibility? Parasitology 2011, 138, 160–174. [Google Scholar] [CrossRef]
- Laing, R.; Kikuchi, T.; Martinelli, A.; Tsai, I.J.; Beech, R.N.; Redman, E.; Holroyd, N.; Bartley, D.J.; Beasley, H.; Britton, C.; et al. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. 2013, 14, R88. [Google Scholar] [CrossRef]
- Schwarz, E.M.; Korhonen, P.K.; Campbell, B.E.; Young, N.D.; Jex, A.R.; Jabbar, A.; Hall, R.S.; Mondal, A.; Howe, A.C.; Pell, J.; et al. The genome and developmental transcriptome of the strongylid nematode Haemonchus contortus. Genome Biol. 2013, 14, R89. [Google Scholar] [CrossRef]
- Harris, T.W.; Antoshechkin, I.; Bieri, T.; Blasiar, D.; Chan, J.; Chen, W.J.; De La Cruz, N.; Davis, P.; Duesbury, M.; Fang, R.; et al. WormBase: A comprehensive resource for nematode research. Nucleic Acids Res. 2010, 38, D463–D467. [Google Scholar] [CrossRef]
- Harris, T.W.; Arnaboldi, V.; Cain, S.; Chan, J.; Chen, W.J.; Cho, J.; Davis, P.; Gao, S.; Grove, C.A.; Kishore, R.; et al. WormBase: A modern Model Organism Information Resource. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef]
- Doyle, S.R.; Illingworth, C.J.R.; Laing, R.; Bartley, D.J.; Redman, E.; Martinelli, A.; Holroyd, N.; Morrison, A.A.; Rezansoff, A.; Tracey, A.; et al. Population genomic and evolutionary modelling analyses reveal a single major QTL for ivermectin drug resistance in the pathogenic nematode, Haemonchus contortus. BMC Genom. 2019, 20, 218. [Google Scholar] [CrossRef]
- Doyle, S.R.; Laing, R.; Bartley, D.J.; Britton, C.; Chaudhry, U.; Gilleard, J.S.; Holroyd, N.; Mable, B.K.; Maitland, K.; Morrison, A.A.; et al. A Genome Resequencing-Based Genetic Map Reveals the Recombination Landscape of an Outbred Parasitic Nematode in the Presence of Polyploidy and Polyandry. Genome. Biol. Evol. 2018, 10, 396–409. [Google Scholar] [CrossRef]
- Sargison, N.D.; Redman, E.; Morrison, A.A.; Bartley, D.J.; Jackson, F.; Naghra-van Gijzel, H.; Holroyd, N.; Berriman, M.; Cotton, J.A.; Gilleard, J.S. A method for single pair mating in an obligate parasitic nematode. Int. J. Parasitol. 2018, 48, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Sallé, G.; Doyle, S.R.; Cortet, J.; Cabaret, J.; Berriman, M.; Holroyd, N.; Cotton, J.A. The global diversity of Haemonchus contortus is shaped by human intervention and climate. Nat. Commun. 2019, 10, 4811. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Zhao, X.; Hou, Y.; Yuan, C.; Li, Y.; Luo, X.; Liu, J.; Feng, X. Analysis of genome-wide SNPs based on 2b-RAD sequencing of pooled samples reveals signature of selection in different populations of Haemonchus contortus. J. Biosci. 2019, 44, 97. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, L.A.; Chilton, N.B.; Gasser, R.B. Differentiation of Haemonchus placei from H. contortus (Nematoda: Trichostrongylidae) by the ribosomal DNA second internal transcribed spacer. Int. J. Parasitol. 1995, 25, 483–488. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang le, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Doyle, S.R.; Tracey, A.; Laing, R.; Holroyd, N.; Bartley, D.; Bazant, W.; Beasley, H.; Beech, R.; Britton, C.; Brooks, K.; et al. Extensive genomic and transcriptomic variation defines the chromosome-scale assembly of Haemonchus contortus, a model gastrointestinal worm. 2020. Available online: https://www.biorxiv.org/content/10.1101/2020.02.18.945246v1.article-info (accessed on 20 February 2020).
- Howe, K.L.; Bolt, B.J.; Shafie, M.; Kersey, P.; Berriman, M. WormBase ParaSite − a comprehensive resource for helminth genomics. Mol. Biochem. Parasitol. 2017, 215, 2–10. [Google Scholar] [CrossRef]
- Ferretti, L.; Ramos-Onsins, S.E.; Perez-Enciso, M. Population genomics from pool sequencing. Mol. Ecol. 2013, 22, 5561–5576. [Google Scholar] [CrossRef] [PubMed]
- Kofler, R.; Pandey, R.V.; Schlotterer, C. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 2011, 27, 3435–3436. [Google Scholar] [CrossRef] [PubMed]
- Coles, G.C.; Jackson, F.; Pomroy, W.E.; Prichard, R.K.; von Samson-Himmelstjerna, G.; Silvestre, A.; Taylor, M.A.; Vercruysse, J. The detection of anthelmintic resistance in nematodes of veterinary importance. Vet. Parasitol. 2006, 136, 167–185. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, A.; Guégnard, F.; Charvet, C.L.; Crisford, A.; Courtot, E.; Sauvé, C.; Harmache, A.; Duguet, T.; O’Connor, V.; Castagnone-Sereno, P.; et al. Deciphering the molecular determinants of cholinergic anthelmintic sensitivity in nematodes: When novel functional validation approaches highlight major differences between the model Caenorhabditis elegans and parasitic species. PLoS Pathog. 2018, 14, e1006996. [Google Scholar] [CrossRef] [PubMed]
- Bartley, D.J.; McAllister, H.; Bartley, Y.; Dupuy, J.; Menez, C.; Alvinerie, M.; Jackson, F.; Lespine, A. P-glycoprotein interfering agents potentiate ivermectin susceptibility in ivermectin sensitive and resistant isolates of Teladorsagia circumcincta and Haemonchus contortus. Parasitology 2009, 136, 1081–1088. [Google Scholar] [CrossRef]
- Geary, T.G.; Sims, S.M.; Thomas, E.M.; Vanover, L.; Davis, J.P.; Winterrowd, C.A.; Klein, R.D.; Ho, N.F.; Thompson, D.P. Haemonchus contortus: ivermectin-induced paralysis of the pharynx. Exp. Parasitol. 1993, 77, 88–96. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yin, F.; Gasser, R.B.; Li, F.; Bao, M.; Huang, W.; Zou, F.; Zhao, G.; Wang, C.; Yang, X.; Zhou, Y.; et al. Genetic variability within and among Haemonchus contortus isolates from goats and sheep in China. Parasit. Vectors 2013, 6, 279. [Google Scholar] [CrossRef]
- Yin, F.; Gasser, R.B.; Li, F.; Bao, M.; Huang, W.; Zou, F.; Zhao, G.; Wang, C.; Yang, X.; Zhou, Y.; et al. Population structure of Haemonchus contortus from seven geographical regions in China, determined on the basis of microsatellite markers. Parasit. Vectors 2016, 9, 586. [Google Scholar] [CrossRef]
- Redman, E.; Packard, E.; Grillo, V.; Smith, J.; Jackson, F.; Gilleard, J.S. Microsatellite analysis reveals marked genetic differentiation between Haemonchus contortus laboratory isolates and provides a rapid system of genetic fingerprinting. Int. J. Parasitol. 2008, 38, 111–122. [Google Scholar] [CrossRef]
- Silvestre, A.; Sauve, C.; Cortet, J.; Cabaret, J. Contrasting genetic structures of two parasitic nematodes, determined on the basis of neutral microsatellite markers and selected anthelmintic resistance markers. Mol. Ecol. 2009, 18, 5086–5100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Gasser, R.B.; Yang, X.; Yin, F.; Zhao, G.; Bao, M.; Pan, B.; Huang, W.; Wang, C.; Zou, F.; et al. Two benzimidazole resistance-associated SNPs in the isotype-1 beta-tubulin gene predominate in Haemonchus contortus populations from eight regions in China. Int. J. Parasitol. Drugs Drug Resist. 2016, 6, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Deitsch, K.W.; Lukehart, S.A.; Stringer, J.R. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat. Rev. Microbiol. 2009, 7, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Gilleard, J.; Redman, E. Genetic Diversity and Population Structure of Haemonchus contortus. Adv. Parasitol. 2016, 93, 31–68. [Google Scholar]
- Troell, K.; Engstrom, A.; Morrison, D.A.; Mattsson, J.G.; Hoglund, J. Global patterns reveal strong population structure in Haemonchus contortus, a nematode parasite of domesticated ruminants. Int. J. Parasitol. 2006, 36, 1305–1316. [Google Scholar] [CrossRef]
- Williamson, S.M.; Storey, B.; Howell, S.; Harper, K.M.; Kaplan, R.M.; Wolstenholme, A.J. Candidate anthelmintic resistance-associated gene expression and sequence polymorphisms in a triple-resistant field isolate of Haemonchus contortus. Mol. Biochem. Parasitol. 2011, 180, 99–105. [Google Scholar] [CrossRef]
- Atif, M.; Estrada-Mondragon, A.; Nguyen, B.; Lynch, J.W.; Keramidas, A. Effects of glutamate and ivermectin on single glutamate-gated chloride channels of the parasitic nematode H. contortus. PLoS Pathog. 2017, 13, e1006663. [Google Scholar] [CrossRef]
- Atif, M.; Smith, J.J.; Estrada-Mondragon, A.; Xiao, X.; Salim, A.A.; Capon, R.J.; Lynch, J.W.; Keramidas, A. GluClR-mediated inhibitory postsynaptic currents reveal targets for ivermectin and potential mechanisms of ivermectin resistance. PLoS Pathog. 2019, 15, e1007570. [Google Scholar] [CrossRef]
- Yates, D.M.; Portillo, V.; Wolstenholme, A.J. The avermectin receptors of Haemonchus contortus and Caenorhabditis elegans. Int. J. Parasitol. 2003, 33, 1183–1193. [Google Scholar] [CrossRef]
- Cully, D.F.; Vassilatis, D.K.; Liu, K.K.; Paress, P.S.; Van der Ploeg, L.H.; Schaeffer, J.M.; Arena, J.P. Cloning of an avermectin-sensitive glutamate-gated chloride channel from Caenorhabditis elegans. Nature 1994, 371, 707–711. [Google Scholar] [CrossRef]
- Wolstenholme, A.J.; Rogers, A.T. Glutamate-gated chloride channels and the mode of action of the avermectin/milbemycin anthelmintics. Parasitology 2005, 13, S85–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Puinean, A.M.; O´Reilly, A.O.; Williamson, M.S.; Smelt, C.L.C.; Millar, N.S.; Wu, Y. Mutations on M3 helix of Plutella xylostella glutamate-gated chloride channel confer unequal resistance to abamectin by two different mechanisms. Insect Biochem. Mol. Biol. 2017, 86, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.H.; Yoon, K.S.; Clark, J.M.; Lee, S.H. A point mutation in a glutamate-gated chloride channel confers abamectin resistance in the two-spotted spider mite, Tetranychus urticae Koch. Insect Mol. Biol. 2010, 19, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Dermauw, W.; Ilias, A.; Riga, M.; Tsagkarakou, A.; Grbic, M.; Tirry, L.; Van Leeuwen, T.; Vontas, J. The cys-loop ligand-gated ion channel gene family of Tetranychus urticae: implications for acaricide toxicology and a novel mutation associated with abamectin resistance. Insect Biochem. Mol. Biol. 2012, 42, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Menez, C.; Alberich, M.; Kansoh, D.; Blanchard, A.; Lespine, A. Acquired Tolerance to Ivermectin and Moxidectin after Drug Selection Pressure in the Nematode Caenorhabditis elegans. Antimicrob. Agents Chemother. 2016, 60, 4809–4819. [Google Scholar] [CrossRef] [PubMed]
- Cvilink, V.; Lamka, J.; Skalova, L. Xenobiotic metabolizing enzymes and metabolism of anthelminthics in helminths. Drug Metab. Rev. 2009, 41, 8–26. [Google Scholar] [CrossRef]
- Alvarez, L.I.; Solana, H.D.; Mottier, M.L.; Virkel, G.L.; Fairweather, I.; Lanusse, C.E. Altered drug influx/efflux and enhanced metabolic activity in triclabendazole-resistant liver flukes. Parasitology 2005, 131, 501–510. [Google Scholar] [CrossRef]
- Rochat, B. Role of cytochrome P450 activity in the fate of anticancer agents and in drug resistance: focus on tamoxifen, paclitaxel and imatinib metabolism. Clin. Pharm. 2005, 44, 349–366. [Google Scholar] [CrossRef]
- David, J.P.; Ismail, H.M.; Chandor-Proust, A.; Paine, M.J. Role of cytochrome P450s in insecticide resistance: impact on the control of mosquito-borne diseases and use of insecticides on Earth. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120429. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, T.; Wang, J.F.; Wei, D.Q. Advances in human cytochrome p450 and personalized medicine. Curr. Drug Metab. 2011, 12, 436–444. [Google Scholar] [CrossRef]
- Barrett, J. Cytochrome P450 in parasitic protozoa and helminths. Comp. Biochem. Physiol. C Pharm. Toxicol. Endocrinol. 1998, 121, 181–183. [Google Scholar] [CrossRef]
- Precious, W.Y.; Barrett, J. The possible absence of cytochrome P-450 linked xenobiotic metabolism in helminths. Biochim. Biophys. Acta 1989, 992, 215–222. [Google Scholar] [CrossRef]
- Menzel, R.; Bogaert, T.; Achazi, R. A systematic gene expression screen of Caenorhabditis elegans cytochrome P450 genes reveals CYP35 as strongly xenobiotic inducible. Arch. Biochem. Biophys. 2001, 395, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Lindblom, T.H.; Dodd, A.K. Xenobiotic detoxification in the nematode Caenorhabditis elegans. J. Exp. Zool. A Comp. Exp. Biol. 2006, 305, 720–730. [Google Scholar] [CrossRef] [PubMed]
- Menzel, R.; Rodel, M.; Kulas, J.; Steinberg, C.E. CYP35: xenobiotically induced gene expression in the nematode Caenorhabditis elegans. Arch. Biochem. Biophys. 2005, 438, 93–102. [Google Scholar] [CrossRef]
- Reichert, K.; Menzel, R. Expression profiling of five different xenobiotics using a Caenorhabditis elegans whole genome microarray. Chemosphere 2005, 61, 229–237. [Google Scholar] [CrossRef]
- Roh, J.Y.; Jung, I.H.; Lee, J.Y.; Choi, J. Toxic effects of di(2-ethylhexyl)phthalate on mortality, growth, reproduction and stress-related gene expression in the soil nematode Caenorhabditis elegans. Toxicology 2007, 237, 126–133. [Google Scholar] [CrossRef]
- Roh, J.Y.; Lee, J.; Choi, J. Assessment of stress-related gene expression in the heavy metal-exposed nematode Caenorhabditis elegans: a potential biomarker for metal-induced toxicity monitoring and environmental risk assessment. Env. Toxicol. Chem. 2006, 25, 2946–2956. [Google Scholar] [CrossRef]
- Laing, R.; Bartley, D.J.; Morrison, A.A.; Rezansoff, A.; Martinelli, A.; Laing, S.T.; Gilleard, J.S. The cytochrome P450 family in the parasitic nematode Haemonchus contortus. Int. J. Parasitol. 2015, 45, 243–251. [Google Scholar] [CrossRef]
- Yilmaz, E.; Ramunke, S.; Demeler, J.; Krucken, J. Comparison of constitutive and thiabendazole-induced expression of five cytochrome P450 genes in fourth-stage larvae of Haemonchus contortus isolates with different drug susceptibility identifies one gene with high constitutive expression in a multi-resistant isolate. Int. J. Parasitol. Drugs Drug Resist. 2017, 7, 362–369. [Google Scholar]
- Higgins, C.F.; Linton, K.J. ABC transporters, an introduction and overview. In ABC Proteins; Holland, I.B., Ed.; Academic Press: London, UK, 2003; pp. 317–335. [Google Scholar]
- Alvarez, A.I.; Merino, G.; Molina, A.J.; Pulido, M.M.; McKellar, Q.A.; Prieto, J.G. Role of ABC transporters in veterinary drug research and parasite resistance. Curr. Drug Deliv. 2006, 3, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Lincke, C.R.; The, I.; van Groenigen, M.; Borst, P. The P-glycoprotein gene family of Caenorhabditis elegans. Cloning and characterization of genomic and complementary DNA sequences. J. Mol. Biol. 1992, 228, 701–711. [Google Scholar] [CrossRef]
- Kerboeuf, D.; Blackhall, W.; Kaminsky, R.; von Samson-Himmelstjerna, G. P-glycoprotein in helminths: function and perspectives for anthelmintic treatment and reversal of resistance. Int. J. Antimicrob. Agents 2003, 22, 332–346. [Google Scholar] [CrossRef]
- Smith, J.M.; Prichard, R.K. Localization of p-glycoprotein mRNA in the tissues of Haemonchus contortus adult worms and its relative abundance in drug-selected and susceptible strains. J. Parasitol. 2002, 88, 612–620. [Google Scholar] [CrossRef]
- Xu, M.; Molento, M.; Blackhall, W.; Ribeiro, P.; Beech, R.; Prichard, R. Ivermectin resistance in nematodes may be caused by alteration of P-glycoprotein homolog. Mol. Biochem. Parasitol. 1998, 91, 327–335. [Google Scholar] [CrossRef]
- Blackhall, W.J.; Liu, H.Y.; Xu, M.; Prichard, R.K.; Beech, R.N. Selection at a P-glycoprotein gene in ivermectin- and moxidectin-selected strains of Haemonchus contortus. Mol. Biochem. Parasitol. 1998, 95, 193–201. [Google Scholar] [CrossRef]
- Alberich, M.; Menez, C.; Sutra, J.F.; Lespine, A. Ivermectin exposure leads to up-regulation of detoxification genes in vitro and in vivo in mice. Eur. J. Pharm. 2014, 740, 428–435. [Google Scholar] [CrossRef]
- Omiecinski, C.J.; Vanden Heuvel, J.P.; Perdew, G.H.; Peters, J.M. Xenobiotic metabolism, disposition, and regulation by receptors: From biochemical phenomenon to predictors of major toxicities. Toxicol. Sci. 2011, 120 (Suppl. 1), S49–S75. [Google Scholar] [CrossRef]
- Chai, X.; Zeng, S.; Xie, W. Nuclear receptors PXR and CAR: implications for drug metabolism regulation, pharmacogenomics and beyond. Expert Opin. Drug Metab. Toxicol. 2013, 9, 253–266. [Google Scholar] [CrossRef]
- Wu, B.; Li, S.; Dong, D. 3D structures and ligand specificities of nuclear xenobiotic receptors CAR, PXR and VDR. Drug Discov. Today 2013, 18, 574–581. [Google Scholar] [CrossRef]
- Lindblom, T.H.; Pierce, G.J.; Sluder, A.E. A C. elegans orphan nuclear receptor contributes to xenobiotic resistance. Curr. Biol. 2001, 11, 864–868. [Google Scholar] [CrossRef]
- King-Jones, K.; Horner, M.A.; Lam, G.; Thummel, C.S. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab. 2006, 4, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Wolstenholme, A.J. Ion channels and receptor as targets for the control of parasitic nematodes. Int. J. Parasitol. Drugs Drug Resist. 2011, 1, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, E.; Bouhelal, R.; Gerspacher, M.; Seuwen, K. The 7 TM G-protein-coupled receptor target family. ChemMedChem 2006, 1, 761–782. [Google Scholar] [CrossRef] [PubMed]
- Lagerstrom, M.C.; Schioth, H.B. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008, 7, 339–357. [Google Scholar] [CrossRef] [PubMed]
- McVeigh, P.; Atkinson, L.; Marks, N.J.; Mousley, A.; Dalzell, J.J.; Sluder, A.; Hammerland, L.; Maule, A.G. Parasite neuropeptide biology: Seeding rational drug target selection? Int. J. Parasitol. Drugs Drug Resist. 2012, 2, 76–91. [Google Scholar] [CrossRef]
- Saeger, B.; Schmitt-Wrede, H.P.; Dehnhardt, M.; Benten, W.P.; Krucken, J.; Harder, A.; Von Samson-Himmelstjerna, G.; Wiegand, H.; Wunderlich, F. Latrophilin-like receptor from the parasitic nematode Haemonchus contortus as target for the anthelmintic depsipeptide PF1022A. Faseb J. 2001, 15, 1332–1334. [Google Scholar] [CrossRef]
- Chen, I.S.; Tateyama, M.; Fukata, Y.; Uesugi, M.; Kubo, Y. Ivermectin activates GIRK channels in a PIP2 -dependent, Gbetagamma -independent manner and an amino acid residue at the slide helix governs the activation. J. Physiol. 2017, 595, 5895–5912. [Google Scholar] [CrossRef]
- Komuniecki, R.W.; Hobson, R.J.; Rex, E.B.; Hapiak, V.M.; Komuniecki, P.R. Biogenic amine receptors in parasitic nematodes: what can be learned from Caenorhabditis elegans? Mol. Biochem. Parasitol. 2004, 137, 1–11. [Google Scholar] [CrossRef]
- Chase, D.L.; Koelle, M.R. Biogenic amine neurotransmitters in C. elegans. In WormBook: The Online Review of C. elegans Biology; The Celegans Research Community, Ed.; WormBook: Pasadena, CA, USA, 2007. [Google Scholar]
- Dittman, J.S.; Kaplan, J.M. Behavioral impact of neurotransmitter-activated G-protein-coupled receptors: muscarinic and GABAB receptors regulate Caenorhabditis elegans locomotion. J. Neurosci. 2008, 28, 7104–7112. [Google Scholar] [CrossRef]
- Laing, S.T.; Ivens, A.; Laing, R.; Ravikumar, S.; Butler, V.; Woods, D.J.; Gilleard, J.S. Characterization of the xenobiotic response of Caenorhabditis elegans to the anthelmintic drug albendazole and the identification of novel drug glucoside metabolites. Biochem. J. 2010, 432, 505–514. [Google Scholar] [CrossRef]
- Jones, L.M.; Rayson, S.J.; Flemming, A.J.; Urwin, P.E. Adaptive and specialised transcriptional responses to xenobiotic stress in Caenorhabditis elegans are regulated by nuclear hormone receptors. PLoS ONE 2013, 8, e69956. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J. Forty years of helminth biochemistry. Parasitology 2009, 136, 1633–1642. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.M.; Flemming, A.J.; Urwin, P.E. NHR-176 regulates cyp-35d1 to control hydroxylation-dependent metabolism of thiabendazole in Caenorhabditis elegans. Biochem. J. 2015, 466, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Kotze, A.C. Cytochrome P450 monooxygenase activity in Haemonchus contortus (Nematoda). Int. J. Parasitol. 1997, 27, 33–40. [Google Scholar] [CrossRef]
- Kotze, A.C. Oxidase activities in macrocyclic-resistant and -susceptible Haemonchus contortus. J. Parasitol. 2000, 86, 873–876. [Google Scholar] [CrossRef]
- Kotze, A.C.; Dobson, R.J.; Chandler, D. Synergism of rotenone by piperonyl butoxide in Haemonchus contortus and Trichostrongylus colubriformis in vitro: potential for drug-synergism through inhibition of nematode oxidative detoxification pathways. Vet. Parasitol. 2006, 136, 275–282. [Google Scholar] [CrossRef]
- Kotze, A.C. Peroxide-supported in-vitro cytochrome P450 activities in Haemonchus contortus. Int. J. Parasitol. 1999, 29, 389–396. [Google Scholar] [CrossRef]
| Pool | Strain Name | Code | IVM-Phenotype | Pool Size |
|---|---|---|---|---|
| 1 | Wu Meng-R | WMR | Resistant | 50 |
| 2 | Wu Shen-R1 | WSR1 | Resistant | 50 |
| 3 | Shan Xi-R | SXR | Resistant | 50 |
| 4 | Shan Xi-S | SXS | Sensitive | 50 |
| 5 | UK-R | UKR | Resistant | 50 |
| 6 | Wu Shen-R2.1 | WSR2.1 | Resistant | 50 |
| 7 | Wu Shen-R2.2 | WSR2.2 | Resistant | 50 |
| 8 | Australian-S | ASS | Sensitive | 50 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, S.; Nisar, A.; Yuan, J.; Luo, X.; Dou, X.; Liu, F.; Zhao, X.; Li, J.; Ahmad, H.; Mehmood, S.A.; et al. A Whole Genome Re-Sequencing Based GWA Analysis Reveals Candidate Genes Associated with Ivermectin Resistance in Haemonchus contortus. Genes 2020, 11, 367. https://doi.org/10.3390/genes11040367
Khan S, Nisar A, Yuan J, Luo X, Dou X, Liu F, Zhao X, Li J, Ahmad H, Mehmood SA, et al. A Whole Genome Re-Sequencing Based GWA Analysis Reveals Candidate Genes Associated with Ivermectin Resistance in Haemonchus contortus. Genes. 2020; 11(4):367. https://doi.org/10.3390/genes11040367
Chicago/Turabian StyleKhan, Sawar, Ayesha Nisar, Jianqi Yuan, Xiaoping Luo, Xueqin Dou, Fei Liu, Xiaochao Zhao, Junyan Li, Habib Ahmad, Sardar Azhar Mehmood, and et al. 2020. "A Whole Genome Re-Sequencing Based GWA Analysis Reveals Candidate Genes Associated with Ivermectin Resistance in Haemonchus contortus" Genes 11, no. 4: 367. https://doi.org/10.3390/genes11040367
APA StyleKhan, S., Nisar, A., Yuan, J., Luo, X., Dou, X., Liu, F., Zhao, X., Li, J., Ahmad, H., Mehmood, S. A., & Feng, X. (2020). A Whole Genome Re-Sequencing Based GWA Analysis Reveals Candidate Genes Associated with Ivermectin Resistance in Haemonchus contortus. Genes, 11(4), 367. https://doi.org/10.3390/genes11040367





