Cigarette and Cannabis Smoking Effects on GPR15+ Helper T Cell Levels in Peripheral Blood: Relationships with Epigenetic Biomarkers
Abstract
1. Introduction
- First, we examine whether the proportion of GPR15-expressing CD3+CD4+ Th cells is associated with (a) tobacco and cannabis smoking patterns and (b) confounding variables that could impact immune function such as age, sex, race, perceived stress, depressive symptomatology, and use of over-the-counter non-steroidal anti-inflammatory drugs (NSAIDs).
- Second, we examine whether the addition of cg19859270 and cg05575921 can fully account for the variance in the level of GPR15-expressing CD3+CD4+ Th cells due to smoking patterns and other immunological variables.
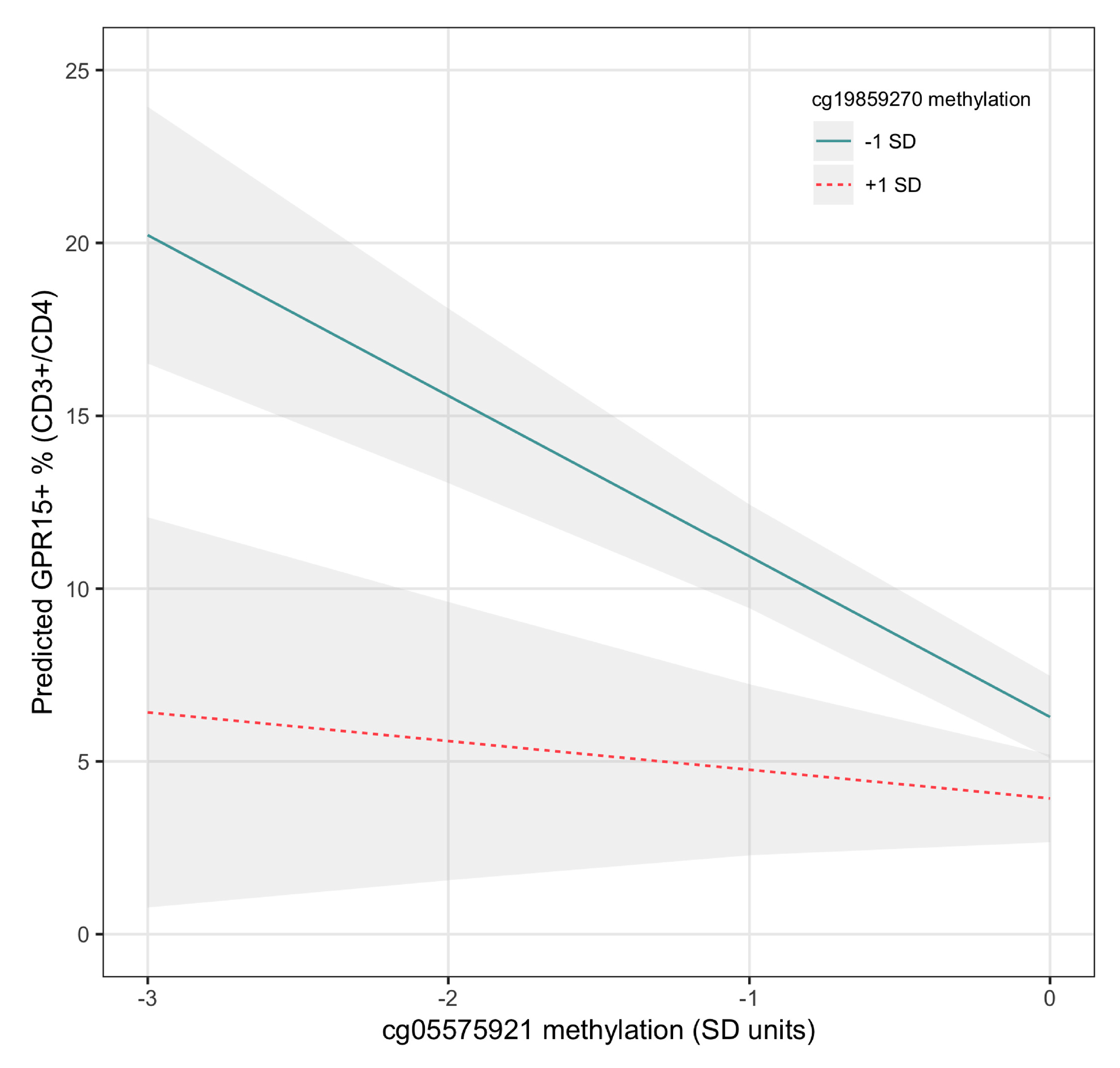
- Third, we examine whether interaction effects between cg19859270 and cg05575921 influence the level of GPR15-expressing CD3+CD4+ Th cells.
2. Materials and Methods
2.1. Sample
2.2. Subject Procedures
2.3. Biomaterials and Assays
2.3.1. ELISAs
2.3.2. ddPCR Assays
2.3.3. PBMC Isolation and Cryopreservation
2.3.4. Flow Cytometry
2.4. Statistical Analysis
2.4.1. Coding
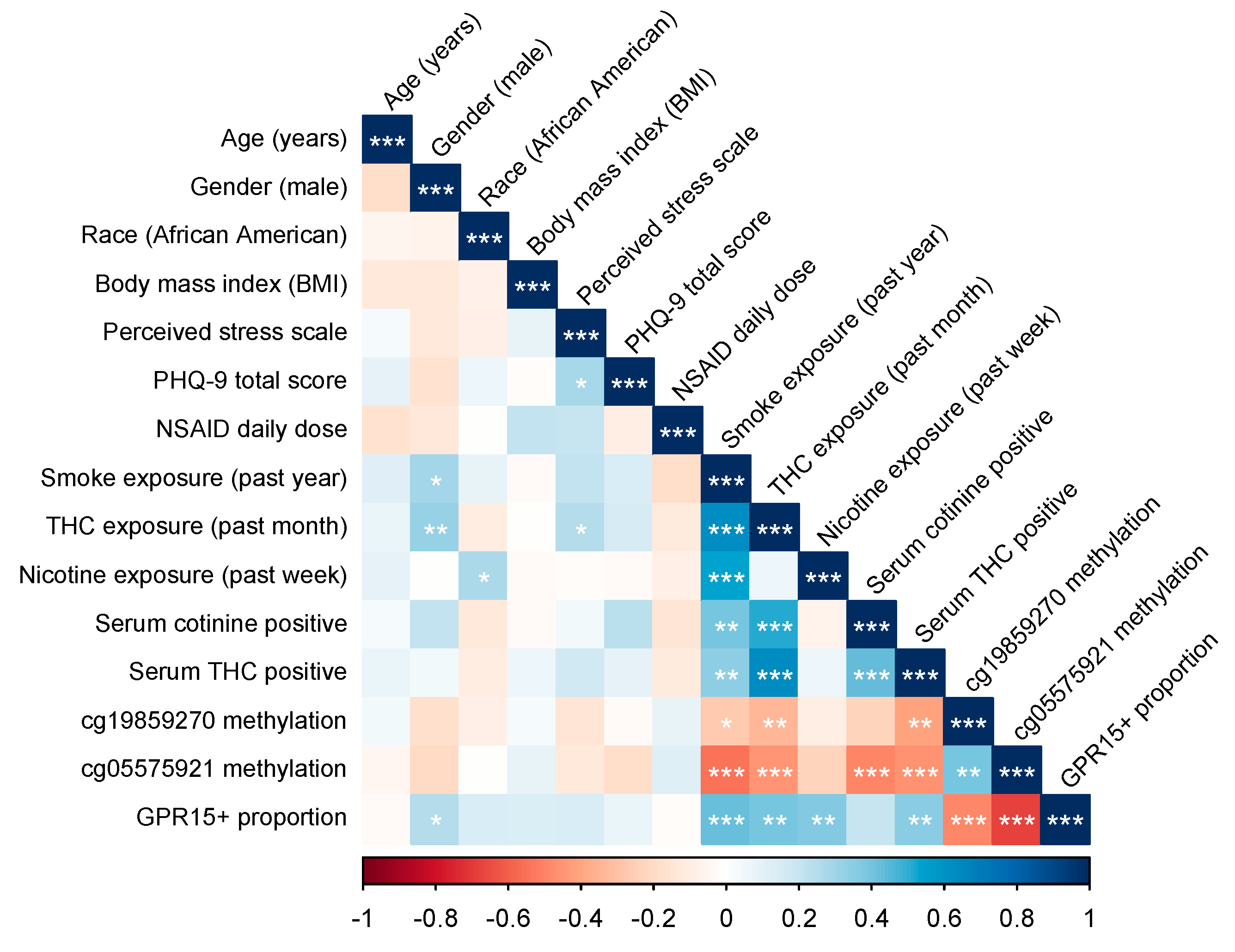
2.4.2. Correlational Analyses
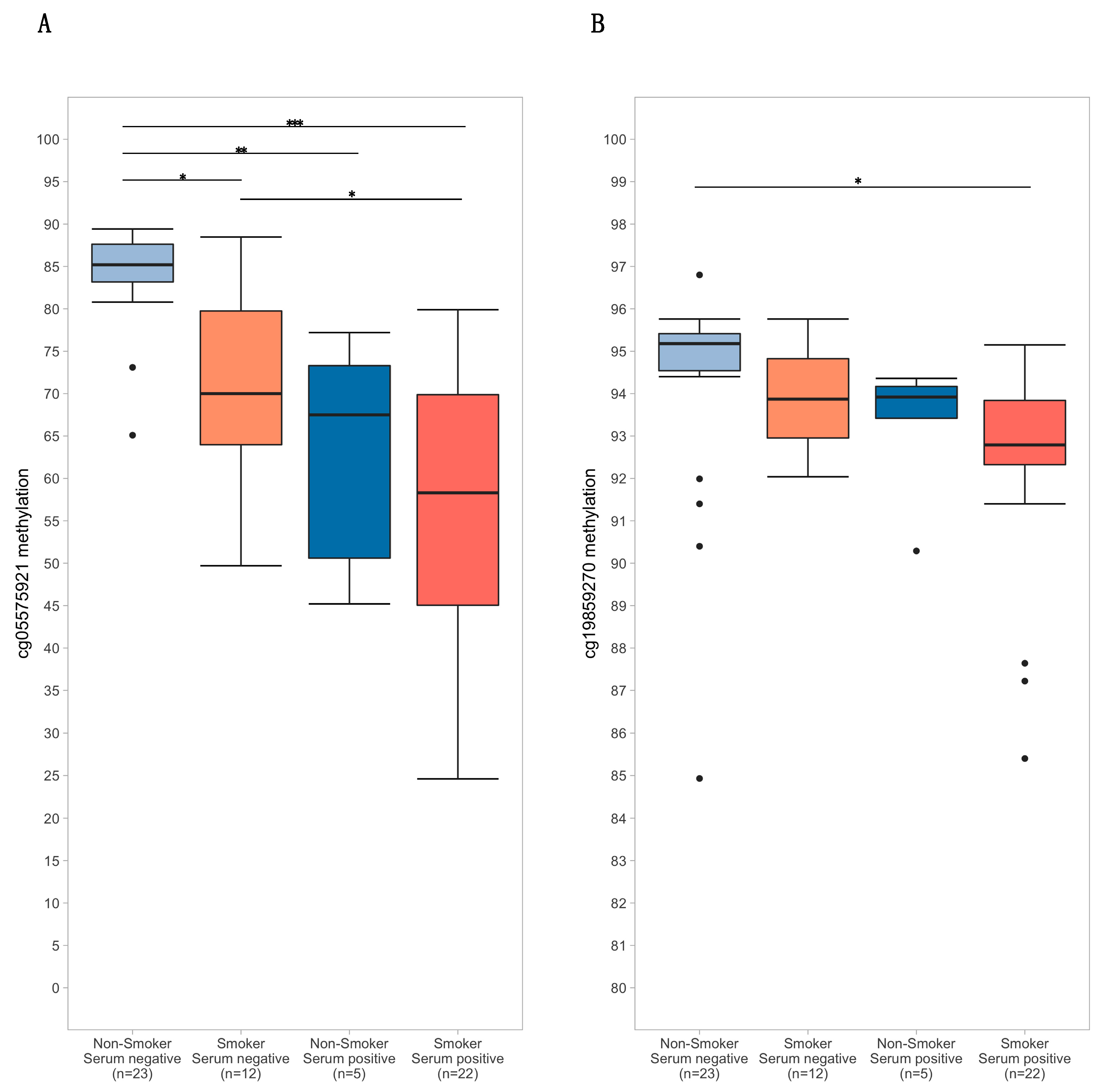
2.4.3. ANOVAs
2.4.4. Regression Analyses
3. Results
3.1. Subject Characteristics and Assay ResuLts
3.2. Study Variable Correlations
3.3. Regression Analysis Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- DHHS. The Health Consequences of Smoking—50 Years of Progress: A Report of the Surgeon General; National Center for Chronic Disease Prevention and Health Promotion Office on Smoking and Health: Atlanta, GA, USA, 2014; Available online: https://aahb.org/Resources/Pictures/Meetings/2014-Charleston/PPT%20Presentations/Sunday%20Welcome/Abrams.AAHB.3.13.v1.o.pdf (accessed on 27 January 2020).
- Jamal, A.; Phillips, E.; Gentzke, A.S.; Homa, D.M.; Babb, S.D.; King, B.A.; Linda, J.N. Current cigarette smoking among adults—United States, 2016. Morb. Mortal. Wkly. Rep. 2018, 67, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Fantini, D.; Seiler, R.; Meeks, J.J. Molecular footprints of muscle-invasive bladder cancer in smoking and nonsmoking patients. In Urologic Oncology: Seminars and Original Investigations; Elsevier: Amsterdam, The Netherlands, 2018; pp. 818–825. [Google Scholar]
- Mokdad, A.H.; Marks, J.S.; Stroup, D.F.; Gerberding, J.L. Actual causes of death in the United States, 2000. Jama 2004, 291, 1238–1245. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C.; Bernstein, C.N.; Vatn, M.H.; Lakatos, P.L.; Loftus, E.V.; Tysk, C.; O’Morain, C.; Moum, B.; Colombel, J.F. Geographical variability and environmental risk factors in inflammatory bowel disease. Gut 2013, 62, 630–649. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.; Yang, S.M.; Kim, S.H.; Han, K.H.; Park, S.J.; Shin, J.I. Smoking and rheumatoid arthritis. Int. J. Mol. Sci. 2014, 15, 22279–22295. [Google Scholar] [CrossRef] [PubMed]
- Riise, T.; Nortvedt, M.W.; Ascherio, A. Smoking is a risk factor for muLtiple sclerosis. Neurology 2003, 61, 1122–1124. [Google Scholar] [CrossRef]
- Freiman, A.; Bird, G.; Metelitsa, A.I.; Barankin, B.; Lauzon, G.J. Cutaneous effects of smoking. J. Cutan. Med. Surg. 2004, 8, 415–423. [Google Scholar] [CrossRef]
- Mahid, S.S.; Minor, K.S.; Soto, R.E.; Hornung, C.A.; Galandiuk, S. (Eds.) Smoking and Inflammatory Bowel Disease: A Meta-Analysis. Mayo Clinic Proceedings; Elsevier: Rochester, MN, USA, 2006. [Google Scholar]
- Edinger, A.L.; Hoffman, T.L.; Sharron, M.; Lee, B.; O’Dowd, B.; Doms, R.W. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology 1998, 249, 367–378. [Google Scholar] [CrossRef][Green Version]
- Krumbiegel, M.; Kirchhoff, F. Coreceptor usage of BOB/GPR15 and Bonzo/STRL33 by primary isolates of human immunodeficiency virus type 1. J. Gen. Virol. 1999, 80, 1241–1251. [Google Scholar]
- Blaak, H.; Boers, P.; Gruters, R.; Schuitemaker, H.; Van Der Ende, M.; Osterhaus, A. CCR5, GPR15, and CXCR6 are major coreceptors of human immunodeficiency virus type 2 variants isolated from individuals with and without plasma viremia. J. Gen. Virol. 2005, 79, 1686–1700. [Google Scholar] [CrossRef]
- Cartwright, A.; Schmutz, C.; Askari, A.; Kuiper, J.H.; Middleton, J. Orphan receptor GPR15/BOB is up-reguLated in rheumatoid arthritis. Cytokine 2014, 67, 53–59. [Google Scholar] [CrossRef]
- Nguyen, L.P.; Pan, J.; Dinh, T.T.; Hadeiba, H.; O’Hara, E., 3rd; Ebtikar, A.; Hertweck, A.; Gökmen, M.R.; Lord, G.M.; Jenner, R.G.; et al. Role and species-specific expression of colon T cell homing receptor GPR15 in colitis. Nat. Immunol. 2015, 16, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Zundler, S.; Atreya, R.; Rath, T.; Voskens, C.; Hirschmann, S.; López-Posadas, R.; Watson, A.; Becker, C.; Schuler, G.; et al. Differential effects of α4β7 and GPR15 on homing of effector and reguLatory T cells from patients with UC to the inflamed gut in vivo. Gut 2015, 65, 1642–1664. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, A.; Gageik, D.; Frede, A.; Pastille, E.; Hansen, W.; Rueffer, A.; Buer, J.; Büning, J.; Langhorst, J.; Westendorf, A.M. Differential expression of GPR15 on T cells during uLcerative colitis. JCI Insight 2017, 2, 8. [Google Scholar] [CrossRef] [PubMed]
- Ammitzbøll, C.; Marina, R.; Börnsen, L.; Petersen, E.R.; McWilliam, O.; Ratzer, R.; Christensen, J.R.; Oturai, A.B.; Søndergaard, H.B.; Sellebjerg, F. GPR15+ T cells are Th17 like, increased in smokers and associated with multiple sclerosis. J. Autoimmun. 2018, 97, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, N.; Duan, Q.; Yang, H.; Wang, X.; Yang, P.; Zhang, M.; Liu, J.; Liu, Z.; Shao, Y.; et al. C10orf99 contributes to the development of psoriasis by promoting the proliferation of keratinocytes. Sci. Rep. 2018, 8, 8590. [Google Scholar] [CrossRef] [PubMed]
- Breitling, L.P.; Yang, R.; Korn, B.; Burwinkel, B.; Brenner, H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am. J Hum. Genet. 2011, 88, 450–457. [Google Scholar] [CrossRef]
- Wan, E.S.; Qiu, W.; Baccarelli, A.; Carey, V.J.; Bacherman, H.; Rennard, S.I.; Agusti, A.; Anderson, W.; Lomas, D.A.; DeMeo, D.L. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum. Mol. Genet. 2012, 21, 3073–3082. [Google Scholar] [CrossRef]
- Sun, Y.V.; Smith, A.K.; Conneely, K.N.; Chang, Q.; Li, W.; Lazarus, A.; Smith, J.A.; Almli, L.M.; Binder, E.B.; Klengel, T.; et al. Epigenomic association analysis identifies smoking-related DNA methylation sites in African Americans. Hum. Genet. 2013, 132, 1027–1037. [Google Scholar] [CrossRef]
- Zeilinger, S.; Kuhnel, B.; Klopp, N.; Baurecht, H.; Kleinschmidt, A.; Gieger, C.; Weidinger, S.; Lattka, E.; Adamski, J.; Peters, A.; et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE 2013, 8, e63812. [Google Scholar] [CrossRef]
- Gao, X.; Jia, M.; Zhang, Y.; Breitling, L.P.; Brenner, H. DNA methylation changes of whole blood cells in response to active smoking exposure in aduLts: A systematic review of DNA methylation studies. Clin. Epigenet. 2015, 7, 113. [Google Scholar] [CrossRef]
- Bauer, M.; Fink, B.; Thürmann, L.; Eszlinger, M.; Herberth, G.; Lehmann, I. Tobacco smoking differently influences cell types of the innate and adaptive immune system—indications from CpG site methylation. Clin. Epigenet. 2016, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.; Linsel, G.; Fink, B.; Offenberg, K.; Hahn, A.M.; Sack, U.; Knaack, H.; Eszlinger, M.; Herberth, G. A varying T cell subtype explains apparent tobacco smoking induced single CpG hypomethylation in whole blood. Clin. Epigenet. 2015, 7, 81. [Google Scholar] [CrossRef] [PubMed]
- Dogan, M.V.; Shields, B.; Cutrona, C.; Gao, L.; Gibbons, F.X.; Simons, R.; Monick, M.; Brody, G.H.; Tan, K.; Beach, S.R.; et al. The effect of smoking on DNA methylation of peripheral blood mononuclear cells from African American women. BMC Genom. 2014, 15, 151. [Google Scholar] [CrossRef] [PubMed]
- Dogan, M.V.; Xiang, J.H.; Beach, S.R.H.; Cutrona, C.; Gibbons, F.X.; Simons, R.L.; Brody, G.H.; Stapleton, J.T.; Philibert, R.A. Ethnicity and smoking-associated DNA methylation changes at HIV co-receptor GPR15. Front. Psychiatry 2015, 6, 132. [Google Scholar] [CrossRef] [PubMed]
- Joehanes, R.; Just, A.C.; Marioni, R.E.; Pilling, L.C.; Reynolds, L.M.; Mandaviya, P.R.; Guan, W.; Xu, T.; Elks, C.E.; Aslibekyan, S.; et al. Epigenetic Signatures of Cigarette Smoking. Circ. Cardiovasc. Genet. 2016, 9, 436–447. [Google Scholar] [CrossRef] [PubMed]
- Ladd-Acosta, C. Epigenetic Signatures as Biomarkers of Exposure. Curr. Environ. Health Rep. 2015, 2, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Andersen, A.; Dogan, M.; Beach, S.; Philibert, R. Current and Future Prospects for Epigenetic Biomarkers of Substance Use Disorders. Genes 2015, 6, 991–1022. [Google Scholar] [CrossRef] [PubMed]
- Philibert, R.; Beach, S.R.; Lei, M.-K.; Brody, G.H. Changes in DNA methylation at the aryl hydrocarbon receptor repressor may be a new biomarker for smoking. Clin. Epigenet. 2013, 5, 19. [Google Scholar] [CrossRef]
- Andersen, A.M.; Philibert, R.A.; Gibbons, F.X.; Simons, R.L.; Long, J. Accuracy and utility of an epigenetic biomarker for smoking in popuLations with varying rates of false self-report. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2017, 174, 641–650. [Google Scholar] [CrossRef]
- Philibert, R.; Dogan, M.V.; Noel, A.; Miller, S.; Krukow, B.; Papworth, E.; Cowley, J.; Long, J.D.; Beach, S.R.; Black, D.W. Dose response and prediction characteristics of a methylation sensitive digital PCR assay for cigarette consumption in aduLts. Front. Genet. Epigenet. 2018, 9, 137. [Google Scholar] [CrossRef]
- Philibert, R.; Hollenbeck, N.; Andersen, E.; Osborn, T.; Gerrard, M.; Gibbons, F.X.; Wang, K. A quantitative epigenetic approach for the assessment of cigarette consumption. Front. Psychol. 2015, 6, 656. [Google Scholar] [CrossRef] [PubMed]
- Marabita, F.; Almgren, M.; Sjöholm, L.K.; KuLar, L.; Liu, Y.; James, T.; Kiss, N.B.; Feinberg, A.P.; Olsson, T.; Kockum, I.; et al. Smoking induces DNA methylation changes in MuLtiple Sclerosis patients with exposure-response relationship. Sci. Rep. 2017, 7, 14589. [Google Scholar] [CrossRef] [PubMed]
- Guida, F.; Sandanger, T.M.; Castagne, R.; Campanella, G.; Polidoro, S.; Palli, D.; Krogh, V.; Tumino, R.; Sacerdote, C.; Panico, S.; et al. Dynamics of smoking-induced genome-wide methylation changes with time since smoking cessation. Hum. Mol. Genet. 2015, 24, 2349–2359. [Google Scholar] [CrossRef] [PubMed]
- Philibert, R.; Hollenbeck, N.; Andersen, E.; McElroy, S.; Wilson, S.; Vercande, K.; Beach, S.R.; Osborn, T.; Gerrard, M.; Gibbons, F.X.; et al. Reversion of AHRR demethylation is a quantitative biomarker of smoking cessation. Front. Psychiatry 2016, 7, 55. [Google Scholar] [CrossRef]
- Tsaprouni, L.G.; Yang, T.P.; Bell, J.; Dick, K.J.; Kanoni, S.; Nisbet, J.; Viñuela, A.; Grundberg, E.; Nelson, C.P.; Meduri, E.; et al. Cigarette smoking reduces DNA methylation levels at muLtiple genomic loci but the effect is partially reversible upon cessation. Epigenetics 2014, 9, 1382–1396. [Google Scholar] [CrossRef]
- Ambatipudi, S.; Cuenin, C.; Hernandez-Vargas, H.; Ghantous, A.; Le Calvez-Kelm, F.; Kaaks, R.; Barrdahl, M.; Boeing, H.; Aleksandrova, K.; Trichopoulou, A.; et al. Tobacco smoking-associated genome-wide DNA methylation changes in the EPIC study. Epigenomics 2016, 8, 599–618. [Google Scholar] [CrossRef]
- Moir, D.; Rickert, W.S.; Levasseur, G.; Larose, Y.; Maertens, R.; White, P.; Desjardins, S. A comparison of mainstream and sidestream marijuana and tobacco cigarette smoke produced under two machine smoking conditions. Chem. Res. Toxicol. 2008, 21, 494–502. [Google Scholar] [CrossRef]
- Vogel, C.F.; Haarmann-Stemmann, T. The aryl hydrocarbon receptor repressor–more than a simple feedback inhibitor of AhR signaling: Clues for its role in inflammation and cancer. Curr. Opin. Toxicol. 2017, 2, 109–119. [Google Scholar] [CrossRef]
- Ligthart, S.; Marzi, C.; Aslibekyan, S.; Mendelson, M.M.; Conneely, K.N.; Tanaka, T.; Colicino, E.; Waite, L.L.; Joehanes, R.; Guan, W.; et al. DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biol. 2016, 17, 255. [Google Scholar] [CrossRef]
- Zhang, Y.; Wilson, R.; Heiss, J.; Breitling, L.P.; Saum, K.-U.; Schöttker, B.; Holleczek, B.; Waldenberger, M.; Peters, A.; Brenner, H. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat. Commun. 2017, 8, 14617. [Google Scholar] [CrossRef]
- Bojesen, S.E.; Timpson, N.; Relton, C.; Smith, D.G.; Nordestgaard, B.G. AHRR (cg05575921) hypomethylation marks smoking behaviour, morbidity and mortality. Thorax 2017, 72, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.; Hackermüller, J.; Schor, J.; Schreiber, S.; Fink, B.; Pierzchalski, A.; Herberth, G. Specific induction of the unique GPR15 expression in heterogeneous blood lymphocytes by tobacco smoking. Biomarkers 2019, 24, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Wang, X.; Campbell, M.R.; Porter, D.K.; Pittman, G.S.; Bennett, B.D.; Wan, M.; Englert, N.A.; Crowl, C.L.; Gimple, R.N.; et al. Distinct epigenetic effects of tobacco smoking in whole blood and among leukocyte subtypes. PLoS ONE 2016, 11, e0166486. [Google Scholar] [CrossRef] [PubMed]
- Koks, G.; Uudelepp, M.L.; Limbach, M.; Peterson, P.; Reimann, E.; Koks, S. Smoking-induced expression of the GPR15 gene indicates its potential role in chronic inflammatory pathologies. Am. J. Pathol. 2015, 185, 2898–2906. [Google Scholar] [CrossRef]
- Kim, S.V.; Xiang, W.V.; Kwak, C.; Yang, Y.; Lin, X.W.; Ota, M.; Sarpel, U.; Rifkin, D.B.; Xu, R.; Littman, D.R. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science 2013, 340, 1456–1459. [Google Scholar] [CrossRef]
- Bauer, M.; Fink, B.; Seyfarth, H.-J.; Wirtz, H.; Frille, A. Tobacco-smoking induced GPR15-expressing T cells in blood do not indicate puLmonary damage. BMC Pulm. Med. 2017, 17, 159. [Google Scholar] [CrossRef]
- Russo, E.B.; McPartland, J.M. Cannabis is more than simply Δ9-tetrahydrocannabinol. Psychopharmacology 2003, 165, 431–432. [Google Scholar] [CrossRef]
- Padgett, D.A.; Glaser, R. How stress influences the immune response. Trends Immunol. 2003, 24, 444–448. [Google Scholar] [CrossRef]
- Steptoe, A.; Kunz-Ebrecht, S.; Brydon, L.; Wardle, J. Central adiposity and cortisol responses to waking in middle-aged men and women. Int. J. Obes. 2004, 28, 1168. [Google Scholar] [CrossRef][Green Version]
- Cho, J.Y. ImmunomoduLatory effect of nonsteroidal anti-inflammatory drugs (NSAIDs) at the clinically available doses. Arch. Pharmacal. Res. 2007, 30, 64. [Google Scholar] [CrossRef]
- Kayani, N.; Homan, S.G.; Yun, S. Racial disparities in smoking-attributable mortality and years of potential life lost-Missouri, 2003–2007. Morb. Mortal. Wkly. Rep. 2010, 59, 1518–1522. [Google Scholar]
- Haiman, C.A.; Stram, D.O.; Wilkens, L.R.; Pike, M.C.; Kolonel, L.N.; Henderson, B.E.; Le Marchand, L. Ethnic and racial differences in the smoking-related risk of lung cancer. N. Engl. J. Med. 2006, 354, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Paalani, M.; Lee, J.W.; Haddad, E.; Tonstad, S. Determinants of inflammatory markers in a bi-ethnic popuLation. Ethn. Dis. 2011, 21, 142. [Google Scholar] [PubMed]
- Kogan, S.M.; Lei, M.K.; Grange, C.R.; Simons, R.L.; Brody, G.H.; Gibbons, F.X.; Chen, Y.F. The contribution of community and family contexts to African American young aduLts’ romantic relationship health: A prospective analysis. J. Youth Adolesc. 2013, 42, 878–890. [Google Scholar] [CrossRef]
- Simons, R.L.; Simons, L.G.; Lei, M.K.; Landor, A.M. Relational schemas, hostile romantic relationships, and beliefs about marriage among young African American aduLts. J. Soc. Pers. Relatsh. 2012, 29, 77–101. [Google Scholar] [CrossRef]
- Simons, R.L.; Lei, M.-K.; Beach, S.R.; Barr, A.B.; Simons, L.G.; Gibbons, F.X.; Philibert, R.A. Discrimination, segregation, and chronic inflammation: Testing the weathering explanation for the poor health of Black Americans. Dev. Psychol. 2018, 54, 1993. [Google Scholar] [CrossRef]
- Kroenke, K.; Spitzer, R.L.; Williams, J.B. The PHQ-9: Validity of a brief depression severity measure. J. Gen. Intern. Med. 2001, 16, 606–613. [Google Scholar] [CrossRef]
- Cohen, S.; Kamarck, T.; Mermelstein, R. Perceived stress scale. Meas. Stress Guide Health Soc. Sci. 1994, 10, 235–283. [Google Scholar]
- Dandona, P.; Aljada, A.; Bandyopadhyay, A. Inflammation: The link between insulin resistance, obesity and diabetes. Trends Immun. 2004, 25, 4–7. [Google Scholar] [CrossRef]
- Philibert, R.A.; Beach, S.R.; Brody, G.H. Demethylation of the aryl hydrocarbon receptor repressor as a biomarker for nascent smokers. Epigenetics 2012, 7, 1331–1338. [Google Scholar] [CrossRef]
- RC Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2012. [Google Scholar]
- Chou, R.M.M.; Nakamoto, E.; Griffin, J. Analgesics for Osteoarthritis: An Update of the 2006 Comparative Effectiveness Review; Agency for Healthcare Research and Quality: Rockville, MD, USA, 2011. [Google Scholar]
- Groemping, U.; Matthias, L. Package ‘relaimpo’. 2018. [Google Scholar]
- Visser, M.; Bouter, L.M.; McQuillan, G.M.; Wener, M.H.; Harris, T.B. Elevated C-reactive protein levels in overweight and obese aduLts. Jama 1999, 282, 2131–2135. [Google Scholar] [CrossRef] [PubMed]
- Ford, E.S. Body mass index, diabetes, and C-reactive protein among US aduLts. Diabetes Care 1999, 22, 1971–1977. [Google Scholar] [CrossRef] [PubMed]
- Beach, S.R.H.; Lei, M.K.; Ong, M.L.; Brody, G.H.; Dogan, M.V.; Philibert, R.A. MTHFR methylation moderates the impact of smoking on DNA methylation at AHRR for African American young aduLts. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2017, 174, 608–618. [Google Scholar] [CrossRef] [PubMed]
- Andersen, A.M.; Lei, M.K.; Philibert, R.; Beach, S. Methylation of MTHFR moderates the effect of smoking on genomewide methylation among middle age African Americans. Front. Genet. 2018, 9, 622. [Google Scholar] [CrossRef] [PubMed]
- Houseman, E.A. DNA methylation and cell-type distribution. In Computational and Statistical Epigenomics; Teschendorff, A.E., Ed.; Translational Bioinformatics. 7; Springer: Dort, The Netherlands, 2015; pp. 35–50. [Google Scholar]

| Variable | M | SD |
|---|---|---|
| 1. Age (years) | 30.7 | 1.2 |
| 2. Gender (male) | 0.37 | - |
| 3. Race (African American) | 0.86 | - |
| 4. Body mass index (BMI) | 34.0 | 8.5 |
| 5. Perceived Stress Scale | 19.3 | 4.4 |
| 6. PHQ-9 total | 4.7 | 4.1 |
| 7. NSAID daily dose | 0.43 | 1.00 |
| 8. Any smoking (past year) | 0.55 | 0.50 |
| 9. Cannabis use (past month) | 0.32 | 0.47 |
| 10. Nicotine use (past week) | 0.31 | 0.47 |
| 11. Cotinine positivity | 0.31 | 0.47 |
| 12. THC positivity | 0.32 | 0.47 |
| 13. cg19859270 methylation | 93.4 | 2.4 |
| 14. cg05575921 methylation | 70.3 | 16.7 |
| 15. GPR15+ % (CD3+/CD4+) | 5.7 | 5.0 |
| Model | Model 1A | Model 1B | Model 1C | Model 1D | ||||
|---|---|---|---|---|---|---|---|---|
| Parameters | b | p-Value | b | p-Value | b | p-Value | b | p-Value |
| Cigarettes per day | 0.588 | 6.33 × 10−5 | 0.597 | 1.48 × 10−4 | 0.385 | 2.56 × 10−3 | 0.367 | 1.47 × 10−3 |
| Scaled cannabis use | 1.80 | 1.00 × 10−2 | 1.18 | 1.38 × 10−1 | 0.138 | 8.33 × 10−1 | 0.198 | 7.36 × 10−1 |
| Age (years) | 0.014 | 9.76 × 10−1 | 0.0365 | 9.20 × 10−1 | 0.0940 | 7.74 × 10−1 | ||
| Gender (male) | 2.47 | 4.51 × 10−2 | 1.58 | 1.08 × 10−1 | 1.38 | 1.18 × 10−1 | ||
| Race (African American) | 1.08 | 4.83 × 10−1 | 1.31 | 2.89 × 10−1 | 1.01 | 3.62 × 10−1 | ||
| BMI | 0.0766 | 2.40 × 10−1 | 0.105 | 4.57 × 10−2 | 0.0973 | 3.99 × 10−2 | ||
| Perceived Stress Scale | 0.0422 | 7.56 × 10−1 | 0.0152 | 8.89 × 10−1 | −0.0424 | 6.68 × 10−1 | ||
| PHQ-9 total | 0.145 | 3.21 × 10−1 | 0.0518 | 6.59 × 10−1 | 0.165 | 1.38 × 10−1 | ||
| NSAID daily dose | 0.0939 | 8.69 × 10−1 | 0.218 | 6.31 × 10−1 | 0.211 | 6.05 × 10−1 | ||
| cg19859270 methylation | −0.420 | 3.74 × 10−2 | −0.480 | 9.13 × 10−3 | ||||
| cg05575921 methylation | −0.139 | 4.70 × 10−5 | −0.115 | 2.10 × 10−4 | ||||
| cg19859270*cg05575921 | 1.44 | 6.67 × 10−4 | ||||||
| Constant | 3.65 | 4.18 × 10−7 | −2.42 | 8.72 × 10−1 | 46.9 | 3.23 × 10−2 | 49.6 | 1.23 × 10−2 |
| R-square | 0.357 | 0.348 | 0.590 | 0.670 | ||||
| p-value | 8.21 × 10−7 | 1.68 × 10−4 | 1.52 × 10−8 | 2.21 × 10−10 | ||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andersen, A.M.; Lei, M.-K.; Beach, S.R.H.; Philibert, R.A.; Sinha, S.; Colgan, J.D. Cigarette and Cannabis Smoking Effects on GPR15+ Helper T Cell Levels in Peripheral Blood: Relationships with Epigenetic Biomarkers. Genes 2020, 11, 149. https://doi.org/10.3390/genes11020149
Andersen AM, Lei M-K, Beach SRH, Philibert RA, Sinha S, Colgan JD. Cigarette and Cannabis Smoking Effects on GPR15+ Helper T Cell Levels in Peripheral Blood: Relationships with Epigenetic Biomarkers. Genes. 2020; 11(2):149. https://doi.org/10.3390/genes11020149
Chicago/Turabian StyleAndersen, Allan M., Man-Kit Lei, Steven R. H. Beach, Robert A. Philibert, Sushmita Sinha, and John D. Colgan. 2020. "Cigarette and Cannabis Smoking Effects on GPR15+ Helper T Cell Levels in Peripheral Blood: Relationships with Epigenetic Biomarkers" Genes 11, no. 2: 149. https://doi.org/10.3390/genes11020149
APA StyleAndersen, A. M., Lei, M.-K., Beach, S. R. H., Philibert, R. A., Sinha, S., & Colgan, J. D. (2020). Cigarette and Cannabis Smoking Effects on GPR15+ Helper T Cell Levels in Peripheral Blood: Relationships with Epigenetic Biomarkers. Genes, 11(2), 149. https://doi.org/10.3390/genes11020149






