Genome-Wide Association Study (GWAS) for Resistance to Sclerotinia sclerotiorum in Common Bean
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. White Mold Response Evaluation
2.3. Data Analysis
2.4. Genome-Wide Association Study
2.5. Quantitative Trait Intervals
2.6. Candidate Gene Identification
3. Results
3.1. White Mold Evaluation
3.2. Genome-Wide Association Study
3.3. Quantitative Trait Intervals
3.4. Candidate Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. FAOSTAT Statistics Database; FAO: Rome, Italy, 1998. [Google Scholar]
- Gepts, P.; Osborne, T.C.; Rashka, K.; Bliss, F.A. Phaseolin protein variability in wild forms and landraces of the common bean (Phaseolus vulgaris L.): Evidence for multiple centers of domestication. Econ. Bot. 1986, 40, 451–468. [Google Scholar] [CrossRef]
- Koenig, R.L.; Gepts, P. Allozyme diversity in wild Phaseolus vulgaris: Further evidence for two major centers of genetic diversity. Theor. Appl. Genet. 1989, 78, 809–817. [Google Scholar] [CrossRef]
- Singh, S.P.; Gepts, P.; Debouck, D.G. Races of common bean (Phaseolus vulgaris, Fabaceae). Econ. Bot. 1991, 45, 379–396. [Google Scholar] [CrossRef]
- Kwak, M.; Gepts, P. Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae). Theor. Appl. Genet. 2009, 118, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Bitocchi, E.; Bellucci, E.; Nanni, L.; Rau, D.; Attene, G.; Papa, R. Linkage disequilibrium and population structure in wild and domesticated populations of Phaseolus vulgaris L. Evol. Appl. 2009, 2, 504–522. [Google Scholar] [CrossRef] [PubMed]
- Campa, A.; Murube, E.; Ferreira, J.J. Genetic diversity, population structure, and linkage disequilibrium in a Spanish common bean diversity panel revealed through genotyping-by-sequencing. Genes 2018, 9, 518. [Google Scholar] [CrossRef] [PubMed]
- Bolland, G.J.; Hall, R. Index of plant host of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 1994, 16, 93–108. [Google Scholar] [CrossRef]
- Schwartz, H.F.; Steadman, J.R.; Hall, R.; Forster, R.L. Compendium of Bean Diseases, 2nd ed.; APS Press: Fort Collins, CO, USA, 2005; p. 109. ISBN 0890543283. [Google Scholar]
- Kabbage, M.; Yarden, O.; Dickman, M.B. Pathogenic attributes of Sclerotinia sclerotiorum: Switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 2015, 233, 53–60. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, L.-Y.; Cao, J.; Li, Y.-L.; Ding, L.-N.; Zhu, K.-M.; Yang, Y.-H.; Tan, X.-L. Recent advances in mechanism of plant defense to Sclerotinia sclerotiorum. Front. Plant Sci. 2019, 10, 1314. [Google Scholar] [CrossRef] [PubMed]
- Miklas, P.N.; Delorme, R.; Johnson, W.C.; Gepts, P. QTL conditioning physiological resistance and avoidance to white mold in dry bean. Crop Sci. 2001, 41, 309–315. [Google Scholar] [CrossRef]
- Park, O.S.; Coyne, D.P.; Steadman, J.R.; Skroch, P.W. Mapping of QTL for resistance to white mold disease in common bean. Crop Sci. 2001, 41, 1253–1262. [Google Scholar] [CrossRef]
- Kolkman, J.M.; Kelly, J.D. QTL conferring resistance and avoidance to white mold in common bean. Crop Sci. 2003, 43, 539–548. [Google Scholar] [CrossRef]
- Miklas, P.N.; Delorme, R. Identification of QTL conditioning resistance to white mold in snap bean. J. Am. Soc. Hortic. Sci. 2003, 128, 564–570. [Google Scholar] [CrossRef]
- Ender, M.; Kelly, J.D. Identification of QTL associated with white mold resistance in common bean. Crop Sci. 2005, 45, 2482–2490. [Google Scholar] [CrossRef]
- Maxwell, J.J.; Brick, M.A.; Byrne, P.F.; Schwartz, H.F.; Shan, X.; Ogg, J.B.; Hensen, R.A. Quantitative trait loci linked to white mold resistance in common bean. Crop Sci. 2007, 47, 2285–2294. [Google Scholar] [CrossRef]
- Miklas, P.N.; Larsen, K.M.; Terpstra, K.A.; Hauf, D.C.; Grafton, K.F.; Kelly, J.D. QTL analysis of ICA Bunsi-derived resistance to white mold in a Pinto 3 navy bean cross. Crop Sci. 2007, 47, 174–179. [Google Scholar] [CrossRef]
- Soule, M.; Porter, L.; Medina, J.; Santana, G.P.; Blair, M.W.; Miklas, P.N. Comparative QTL map for white mold resistance in common bean, and characterization of partial resistance in dry bean lines VA19 and I9365-31. Crop Sci. 2011, 51, 123–139. [Google Scholar] [CrossRef]
- Pérez-Vega, E.; Pascual, A.; Campa, A.; Giraldez, R.; Miklas, P.N.; Ferreira, J.J. Mapping quantitative trait loci conferring partial physiological resistance to white mold in the common bean RIL population Xana x Cornell49242. Mol. Breed. 2012, 29, 31–41. [Google Scholar] [CrossRef]
- Miklas, P.N.; Porter, L.D.; Kelly, J.D.; Myers, J.R. Characterization of white mold disease avoidance in common bean. Eur. J. Plant Pathol. 2013, 135, 525–543. [Google Scholar] [CrossRef]
- Hoyos-Villegas, V.; Mkwaila, W.; Cregan, P.B.; Kelly, J.D. Quantitative trait loci analysis of white mold avoidance in pinto bean. Crop Sci. 2015, 55, 2116–2129. [Google Scholar] [CrossRef]
- Vasconcellos, R.C.C.; Oraguzie, O.B.; Soler, A.; Arkwazee, H.; Myers, J.R.; Ferreira, J.J.; Song, Q.; McClean, P.; Miklas, P.N. Meta-QTL for resistance to white mold in common bean. PLoS ONE 2017, 12, e0171685. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Mao, Y.; Xie, C.; Smith, H.; Luo, L.; Xu, S. Mapping quantitative trait loci using naturally occurring genetic variance among commercial inbred lines of maize (Zea mays L.). Genetics 2005, 169, 2267–2275. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.B.; Feng, J.Y.; Ren, W.L.; Huang, B.; Zhou, L.; Wen, Y.J.; Zhang, J.; Dunwell, J.M.; Xu, S.; Zhang, Y.M. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep. 2016, 6, 19444. [Google Scholar] [CrossRef]
- Zhang, J.; Feng, J.Y.; Ni, Y.L.; Wen, Y.J.; Niu, Y.; Tamba, C.L.; Yue, C.; Song, Q.; Zhang, Y.M. pLARmEB: Integration of least angle regression with empirical bayes for multi-locus genome-wide association studies. Heredity 2017, 118, 517–524. [Google Scholar] [CrossRef]
- Tamba, C.L.; Ni, Y.L.; Zhang, Y.M. Iterative sure independence screening EM-Bayesian LASSO algorithm for multilocus genome-wide association studies. PLoS Comput. Biol. 2017, 13, e1005357. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Tamba, C.L. A fast mrMLM algorithm for multi-locus genome-wide association studies. bioRxiv 2018. [Google Scholar] [CrossRef]
- Wen, Y.J.; Zhang, H.; Ni, Y.L.; Huang, B.; Zhang, J.; Feng, J.Y.; Wang, S.B.; Dunwell, J.M.; Zhang, Y.M.; Wu, R. Methodological implementation of mixed linear models in multi-locus genome-wide association studies. Brief. Bioinform. 2018, 19, 700–712. [Google Scholar] [CrossRef]
- Ren, W.L.; Wen, Y.J.; Dunwell, J.M.; Zhang, Y.M. pKWmEB: Integration of Kruskal-Wallis test with empirical bayes under polygenic background control for multi-locus genome-wide association study. Heredity 2018, 120, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Arkwazee, H.; Hart, J.; Porch, T.; Griffiths, P.; Davis, J.; Myers, J.R. Genome wide association study (GWAS) for white mold resistance in snap bean. Annu. Rept. Bean Improv. Coop. 2018, 61, 85. [Google Scholar]
- Pérez-Vega, E.; Campa, A.; De la Rosa, L.; Giraldez, R.; Ferreira, J.J. Genetic diversity in a core collection established from the main genebank in Spain. Crop Sci. 2009, 49, 1377–1386. [Google Scholar] [CrossRef]
- Pascual, A.; Campa, A.; Pérez-Vega, E.; Giraldez, R.; Miklas, P.N.; Ferreira, J.J. Screening common bean for resistance to four Sclerotinia sclerotiorum isolates collected in northern Spain. Plant Dis. 2010, 94, 885–890. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Petzoldt, R.; Dickson, M.H. Straw test for resistance to white mold in beans. Annu. Rep. Bean Improv. Coop. Meet 1996, 39, 142–143. [Google Scholar]
- Miklas, P.N. Marker-assisted backcrossing QTL for partial resistance to Sclerotinia white mold in dry bean. Crop Sci. 2007, 47, 935–942. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; ISBN 3-900051-07-0. Available online: http://www.R-project.org (accessed on 1 April 2020).
- Zhang, Y.W.; Tamba, C.L.; Wen, Y.J.; Li, P.; Ren, W.L.; Ni, Y.L.; Gao, J.; Zhang, Y.M. mrMLM v4.0: An R platform for multi-locus genome-wide association studies. bioRxiv 2020. [Google Scholar] [CrossRef]
- Moghaddam, S.M.; Mamidi, S.; Osorno, J.M.; Lee, R.; Brick, M.; Kelly, J.; Miklas, P.; Urrea, C.; Song, Q.; Cregan, P.; et al. Genome-Wide association study identifies candidate loci underlying agronomic traits in a middle american diversity panel of common bean. Plant Genome 2016, 9. [Google Scholar] [CrossRef]
- Yu, Y.; Ouyang, Y.; Yao, W. ShinyCircos: An R/shiny application for interactive creation of circos plot. Bioinformatics 2018, 34, 1229–1231. [Google Scholar] [CrossRef] [PubMed]
- Fuller, P.A.; Coyne, D.P.; Steadman, J.R. Inheritance of resistance to white mold disease in a diallel cross of dry beans. Crop Sci. 1984, 24, 929–933. [Google Scholar] [CrossRef]
- Miklas, P.N.; Hauf, D.C.; Henson, R.A.; Grafton, K.F. Inheritance of ICA Bunsi derived resistance in a navy × pinto bean cross. Crop Sci. 2004, 44, 1584–1588. [Google Scholar] [CrossRef]
- Meziadi, C.; Richard, M.M.S.; Derquennes, A.; Thareau, V.; Blanchet, S.; Gratias, A.; Pflieger, S.; Geffroy, V. Development of molecular markers linked to disease resistance genes in common bean based on whole genome sequence. Plant Sci. 2016, 242, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Campa, A.; Ferreira, J.J. Gene coding for an elongation factor is involved in resistant against powdery mildew in common bean. Theor. Appl. Genet. 2017, 130, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Murube, E.; Campa, A.; Ferreira, J.J. Integrating genetic and physical positions of the anthracnose resistance genes described in bean chromosomes Pv01 and Pv04. PLoS ONE 2019, 14, e0212298. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.; Velasco, D.; Gepts, P. Mapping homologous sequences for determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris). J. Hered. 2008, 99, 283–291. [Google Scholar] [CrossRef]
- Repinski, S.L.; Kwak, M.; Gepts, P. The common bean growth habit gene PvTFL1y is a functional homolog of Arabidopsis TFL1. Theor. Appl. Genet. 2012, 124, 1539–1547. [Google Scholar] [CrossRef]
- Pandian, B.A.; Sathishraj, R.; Djanaguiraman, M.; Prasad, P.V.V.; Jugulam, M. Role of cytochrome P450 enzymes in plant stress response. Antioxidants 2020, 9, 454. [Google Scholar] [CrossRef]
- Yan, Q.; Cui, X.; Lin, S.; Gan, S.; Xing, H.; Dou, D. GmCYP82A3, a soybean cytochrome P450 family gene involved in the jasmonic acid and ethylene signaling pathway, enhances plant resistance to biotic and abiotic stresses. PLoS ONE 2016, 11, e0162253. [Google Scholar] [CrossRef]
- Sessa, G.; Martin, G.B. Protein kinases in the plant defence response. Adv. Bot. Res. 2000, 32, 379–404. [Google Scholar] [CrossRef]
- Marone, D.; Russo, M.A.; Laido, G.; Leonardis, A.M.; Mastrangelo, A.M. Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: Active guardians in host defense response. Int J. Mol. Sci. 2013, 14, 7302–7326. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Stotz, H.U. Defense against Sclerotinia sclerotiorum in arabidopsis is dependent on jasmonic acid, salicylic acid, and ethylene signaling xiaomei. Mol. Plant Microbe. Interact. 2007, 20, 1384–1395. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, J.; An, L.; Doerge, R.W.; Chen, Z.J.; Grau, C.R.; Meng, J.; Osborn, T.C. Analysis of gene expression profiles in response to Sclerotinia sclerotiorum in Brassica napus. Planta 2007, 227, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Kalunke, R.M.; Janni, M.; Sella, L.; David, P.; Geffroy, V.; Favaron, F.; D´Ovidio, R. Transcript analysis of the bean polygalacturonase-inhibiting protein gene family reveals that Pvpgip2 is expressed in the whole plant and is strongly induced by pathogen infection. J. Plant Pathol. 2011, 93, 141–148. [Google Scholar] [CrossRef]
- D´Ovidio, R.; Raiola, A.; Capodicasa, C.; Devoto, A.; Pontiggia, D.; Roberti, S.; Galletti, R.; Conti, E.; O’Sullivan, D.; De Lorenzo, G. Characterization of the complex locus of bean encoding Polygalacturonase-Inhibiting Proteins reveals subfunctionalization for defense against fungi and insects. Plant Physiol. 2004, 135, 2424–2435. [Google Scholar] [CrossRef]
- Vasconcellos, R.C.C.; Lima, T.F.C.; Fernandes-Brum, C.N.; Chalfun-Junior, A.; Santos, J.B. Expression and validation of PvPGIP genes for resistance to white mold (Sclerotinia sclerotiorum) in common beans (Phaseolus vulgaris L.). Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef]
- Porto, A.C.M.; Cardon, C.H.; Vasconcellos, R.C.C.; Novaes, E.; Leite, M.E.; Chalfun-Junior, A.; Pereira, W.A.; Santos, J.B. Expression of candidate genes related to white mold resistance in common beans. Trop. Plant Pathol. 2019, 44, 483–493. [Google Scholar] [CrossRef]
- Visel, A.; Rubin, E.M.; Pennacchio, L.A. Genomic views of distant-acting enhancers. Nature 2009, 461, 199–205. [Google Scholar] [CrossRef]
- Brodie, A.; Azaria, J.R.; Ofran, Y. How far from the SNP may the causative genes be? Nucleic Acids Res. 2016, 44, 6046–6054. [Google Scholar] [CrossRef]


| SDP | Accession/Variety | Type of Material | GP | GH | E1 | E2 | Em |
|---|---|---|---|---|---|---|---|
| SDP194 | BGE043359 | Spanish landrace | A | Ind | 4.0 | 4.0 | 4.0 |
| SDP217 | CN_227 | Spanish landrace | M | Ind | 4.0 | 4.0 | 4.0 |
| SDP289 | Sacha | Commercial variety | MA | Ind | 4.2 | 4.0 | 4.1 |
| SDP203 | BILMA | Commercial variety | MA | Ind | 4.3 | 4.0 | 4.1 |
| SDP216 | CN_226 | Spanish landrace | M | Ind | 4.3 | 4.0 | 4.1 |
| SDP246 | Helda | Commercial variety | MA | Ind | 4.1 | 4.1 | 4.1 |
| SDP274 | PLANETA | Commercial variety | MA | Ind | 4.1 | 4.3 | 4.2 |
| SDP226 | DONNA | Commercial variety | MA | Ind | 4.3 | 4.2 | 4.2 |
| SDP108 | BGE011762 | Spanish landrace | A | Ind | 4.3 | 4.3 | 4.3 |
| SDP039 | BGE003139 | Spanish landrace | MA | Ind | 4.2 | 4.4 | 4.3 |
| SDP133 | BGE022831 | Spanish landrace | MA | Ind | 4.7 | 4.0 | 4.3 |
| SDP305 | VITALIS | Commercial variety | MA | Ind | 4.5 | 4.1 | 4.3 |
| SDP234 | FLORENCIA | Commercial variety | MA | Ind | 4.4 | 4.3 | 4.4 |
| SDP304 | V381 | Spanish landrace | MA | Ind | 4.2 | 4.5 | 4.4 |
| SDP106 | BGE011736 | Spanish landrace | M | Det | 4.8 | 4.0 | 4.4 |
| SDP107 | BGE011758 | Spanish landrace | MA | Ind | 4.6 | 3.7 | 4.4 |
| SDP255 | MARCONI | Commercial variety | MA | Ind | 4.6 | 4.2 | 4.4 |
| SDP189 | BGE039982 | Spanish landrace | M | Ind | 4.6 | 4.1 | 4.4 |
| SDP025 | BGE002196 | Spanish landrace | M | Ind | 4.4 | 4.5 | 4.5 |
| SDP035 | BGE003121 | Spanish landrace | M | Ind | 4.5 | 4.5 | 4.5 |
| SDP053 | BGE003482 | Spanish landrace | A | Ind | 4.4 | 4.6 | 4.5 |
| SDP071 | BGE004000 | Spanish landrace | A | Ind | 5.0 | 4.0 | 4.5 |
| SDP088 | BGE005475 | Spanish landrace | MA | Ind | 5.2 | 3.8 | 4.5 |
| SDP221 | CN_241 | Spanish landrace | A | Det | 4.6 | 4.4 | 4.5 |
| SDP295 | Tendergreen | Commercial variety | A | Det | 5.0 | 4.1 | 4.5 |
| SDP110 | BGE013962 | Spanish landrace | M | Ind | 4.9 | 4.0 | 4.5 |
| SDP150 | BGE025745 | Spanish landrace | A | Ind | 5.0 | 4.1 | 4.5 |
| SDP024 | BGE002189 | Spanish landrace | MA | Ind | 5.0 | 4.0 | 4.5 |
| AB136 | Resistant check | MA | Ind | 4.0 | 4.0 | 4.0 | |
| Cornell49242 | Susceptible check | MA | Ind | 8.9 | 9.0 | 9.0 |
| E1 | E2 | Em | |||
|---|---|---|---|---|---|
| QTN | SNP | GWAS Method | −log10(p) | −log10(p) | −log10(p) |
| WM1_42.55 | s1_42558186 | MLM | 3 | - | 3 |
| WM1_42.85 | s1_42857105 | MLM | 4 | - | 3 |
| pLARmEB | 7 | - | 6 | ||
| WM1_42.91 | s1_42916300 | FASTmrEMMA | 7 | - | 5 |
| WM1_45.45 | s1_45457672 | MLM | - | 4 | 3 |
| WM1_45.59 | s1_45594842 | MLM | - | 6 | 4 |
| mrMLM | - | 7 | 5 | ||
| FASTmrMLM | - | 9 | 10 | ||
| ISIS EM-BLASSO | - | 11 | 11 | ||
| FASTmrEMMA | - | 7 | 5 | ||
| pLARmEB | - | 6 | 8 | ||
| pLARmEB | 5 | - | 7 | ||
| WM1_51.06 | s1_51067156 | ISIS EM-BLASSO | 3 | - | 3 |
| WM2_4.62 | s2_4625990 | MLM | 9 | - | 5 |
| WM2_5.57 | s2_5576016 | pLARmEB | 3 | - | 3 |
| WM2_34.64 | s2_34644216 | MLM | - | 5 | 4 |
| WM2_34.73 | s2_34738605 | mrMLM | 3 | - | 3 |
| WM3_0.9 | s3_947347 | FASTmrMLM | - | 4 | 5 |
| WM3_39.95 | s3_39951398 | MLM | 7 | - | 7 |
| pLARmEB | 3 | - | 3 | ||
| WM3_40.53 | s3_40535470 | MLM | - | 6 | 5 |
| WM4_2.11 | s4_2115855 | ISIS EM-BLASSO | - | 4 | 3 |
| WM4_2.37 | s4_2371248 | MLM | - | 7 | 6 |
| pKWmEB | - | 4 | 8 | ||
| pLARmEB | 4 | 3 | 5 | ||
| WM8_13.31 | s8_13317583 | MLM | - | 3 | 3 |
| WM8_37.13 | s8_37139214 | MLM | 4 | - | 4 |
| WM8_52.39 | s8_52395252 | MLM | 7 | - | 7 |
| FASTmrMLM | 6 | 6 | 7 | ||
| ISIS EM-BLASSO | 10 | - | 10 | ||
| FASTmrEMMA | - | 5 | 5 | ||
| pKWmEB | 5 | - | 4 | ||
| pLARmEB | - | 7 | 6 | ||
| WM8_55.56 | s8_55569419 | pKWmEB | 3 | - | 3 |
| WM9_11.70 | s9_11707260 | MLM | - | 5 | 4 |
| WM9_16.78 | s9_16783907 | mrMLM | - | 8 | 8 |
| WM9_36.67 | s9_36677215 | pLARmEB | 3 | - | 3 |
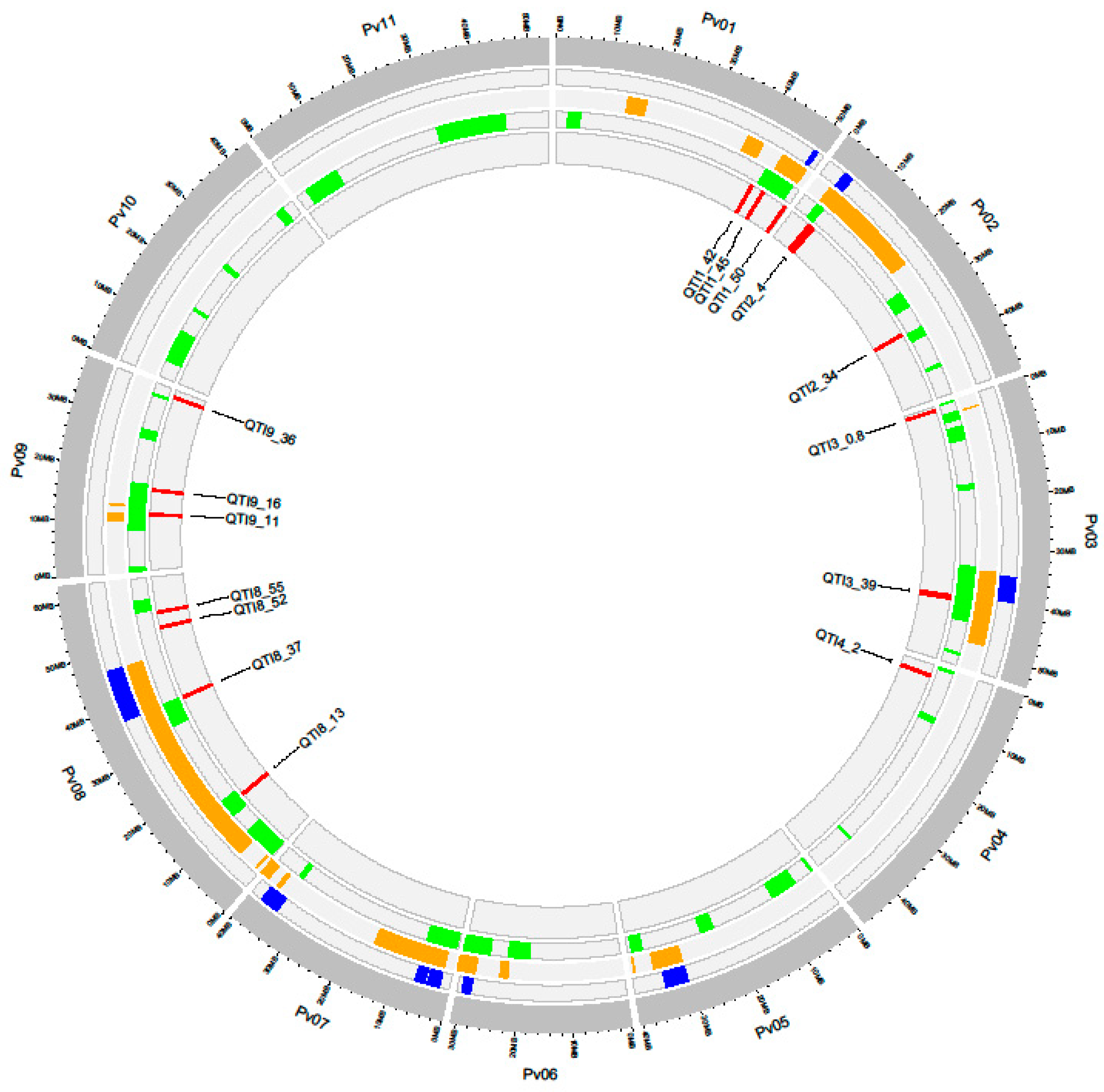
| QTI | Chr | Start-End | QTNs | Nº Annotated Genes | Nº Candidate Genes | Stage of Resistance Response (Number of Genes) |
|---|---|---|---|---|---|---|
| QTI1_42 | Pv01 | 42.46−43.02 | WM1_42.55, WM11_42.85, WM1_42.91 | 46 | 8 | Recognition (6), Signaling (2) |
| QTI1_45 | Pv01 | 45.36−45.69 | WM1_45.45, WM1_45.59 | 36 | 4 | Recognition (1), Signaling (1), Defense (2) |
| QTI1_50 | Pv01 | 50.97–51.17 | WM1_51.06 | 28 | 2 | Recognition (1), Defense (1) |
| QTI2_4 | Pv02 | 4.53–5.68 | WM2_4.62, WM2_5.57 | 74 | 7 | Recognition (3), Signaling (2), Defense (2) |
| QTI2_34 | Pv02 | 34.54–34.84 | WM2_34.64, WM2_34.73 | 23 | - | - |
| QTI3_0.9 | Pv03 | 0.80–1.05 | WM3_0.9 | 29 | 6 | Recognition (1), Defense (5) |
| QTI3_39 | Pv03 | 39.85–40.64 | WM3_39.95, WM3_40.53 | 77 | 7 | Recognition (2), Signaling (1), Defense (4) |
| QTI4_2 | Pv04 | 2.02–2.47 | WM4_2.11, WM4_2.37 | 42 | 12 | Recognition (1), Defense (11) |
| QTI8_13 | Pv08 | 13.22–13.42 | WM8_13.31 | 6 | - | - |
| QTI8_37 | Pv08 | 37.04–37.24 | WM8_37.13 | 2 | - | - |
| QTI8_52 | Pv08 | 52.30–52.50 | WM8_52.39 | 24 | 5 | Recognition (4), Defense (1) |
| QTI8_55 | Pv08 | 55.47–55.67 | WM8_55.56 | 22 | 1 | Signaling (1) |
| QTI9_11 | Pv09 | 11.61–11.88 | WM9_11.70 | 24 | 2 | Recognition (2) |
| QTI9_16 | Pv09 | 16.68–16.88 | WM9_16.78 | 15 | 3 | Recognition (1), Defense (2) |
| QTI9_36 | Pv09 | 36.58–36.78 | WM9_36.67 | 20 | 4 | Signaling (1), Defense (3) |
| 468 | 61 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campa, A.; García-Fernández, C.; Ferreira, J.J. Genome-Wide Association Study (GWAS) for Resistance to Sclerotinia sclerotiorum in Common Bean. Genes 2020, 11, 1496. https://doi.org/10.3390/genes11121496
Campa A, García-Fernández C, Ferreira JJ. Genome-Wide Association Study (GWAS) for Resistance to Sclerotinia sclerotiorum in Common Bean. Genes. 2020; 11(12):1496. https://doi.org/10.3390/genes11121496
Chicago/Turabian StyleCampa, Ana, Carmen García-Fernández, and Juan José Ferreira. 2020. "Genome-Wide Association Study (GWAS) for Resistance to Sclerotinia sclerotiorum in Common Bean" Genes 11, no. 12: 1496. https://doi.org/10.3390/genes11121496
APA StyleCampa, A., García-Fernández, C., & Ferreira, J. J. (2020). Genome-Wide Association Study (GWAS) for Resistance to Sclerotinia sclerotiorum in Common Bean. Genes, 11(12), 1496. https://doi.org/10.3390/genes11121496





