Improving the Management of Patients with Hearing Loss by the Implementation of an NGS Panel in Clinical Practice
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Panel Design
2.3. Library Preparation and Sequencing
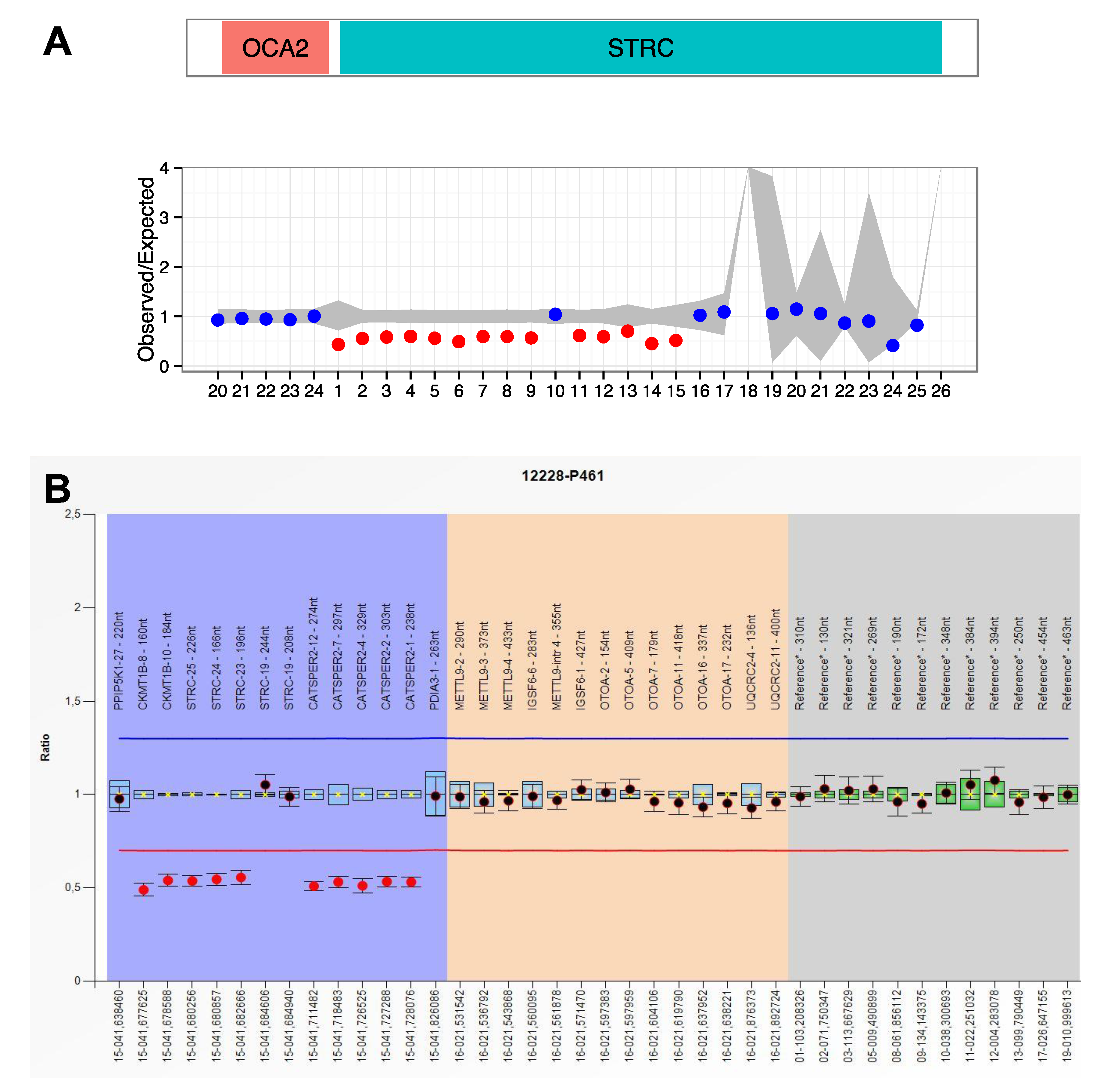
2.4. Data Analysis
3. Results
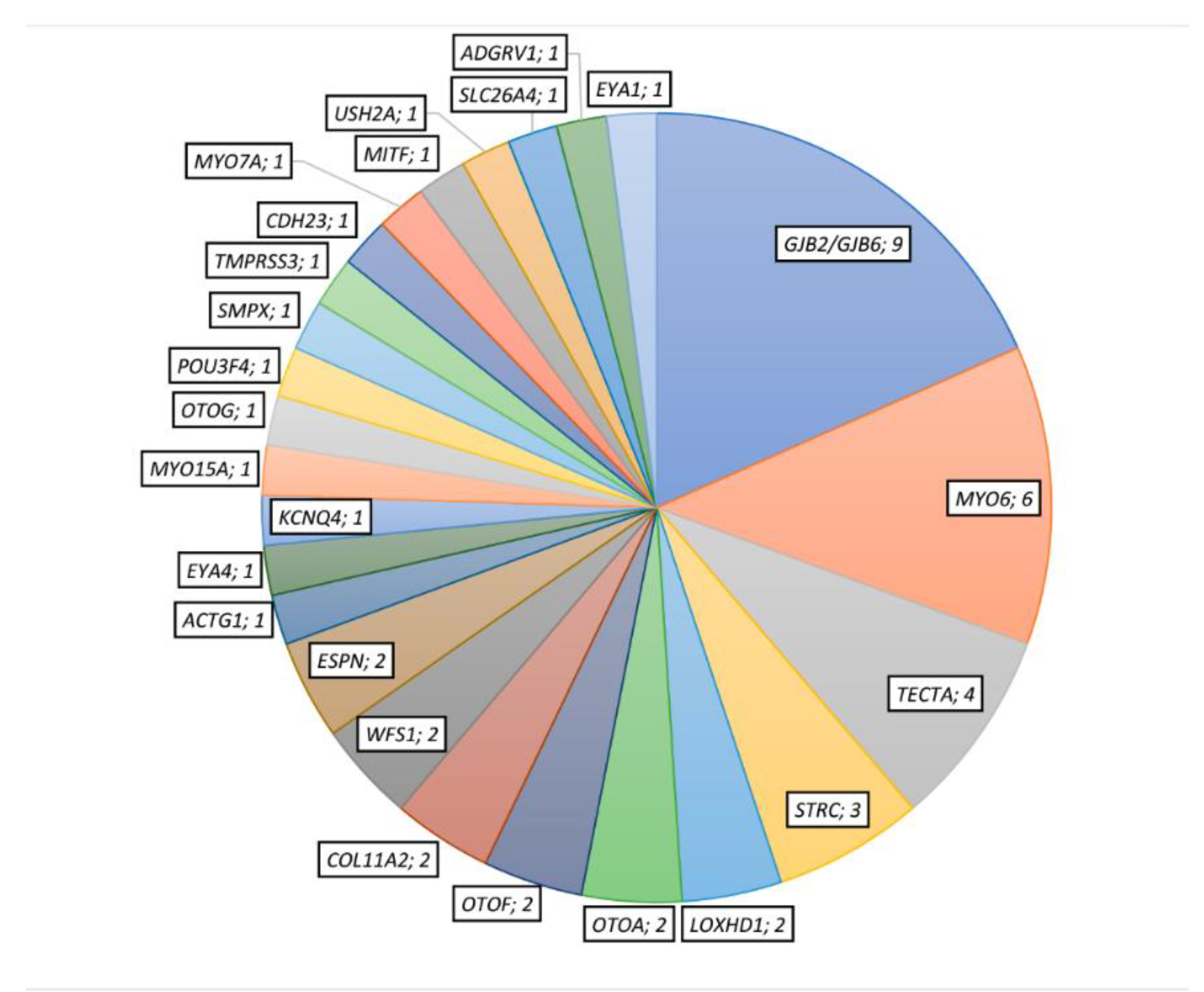
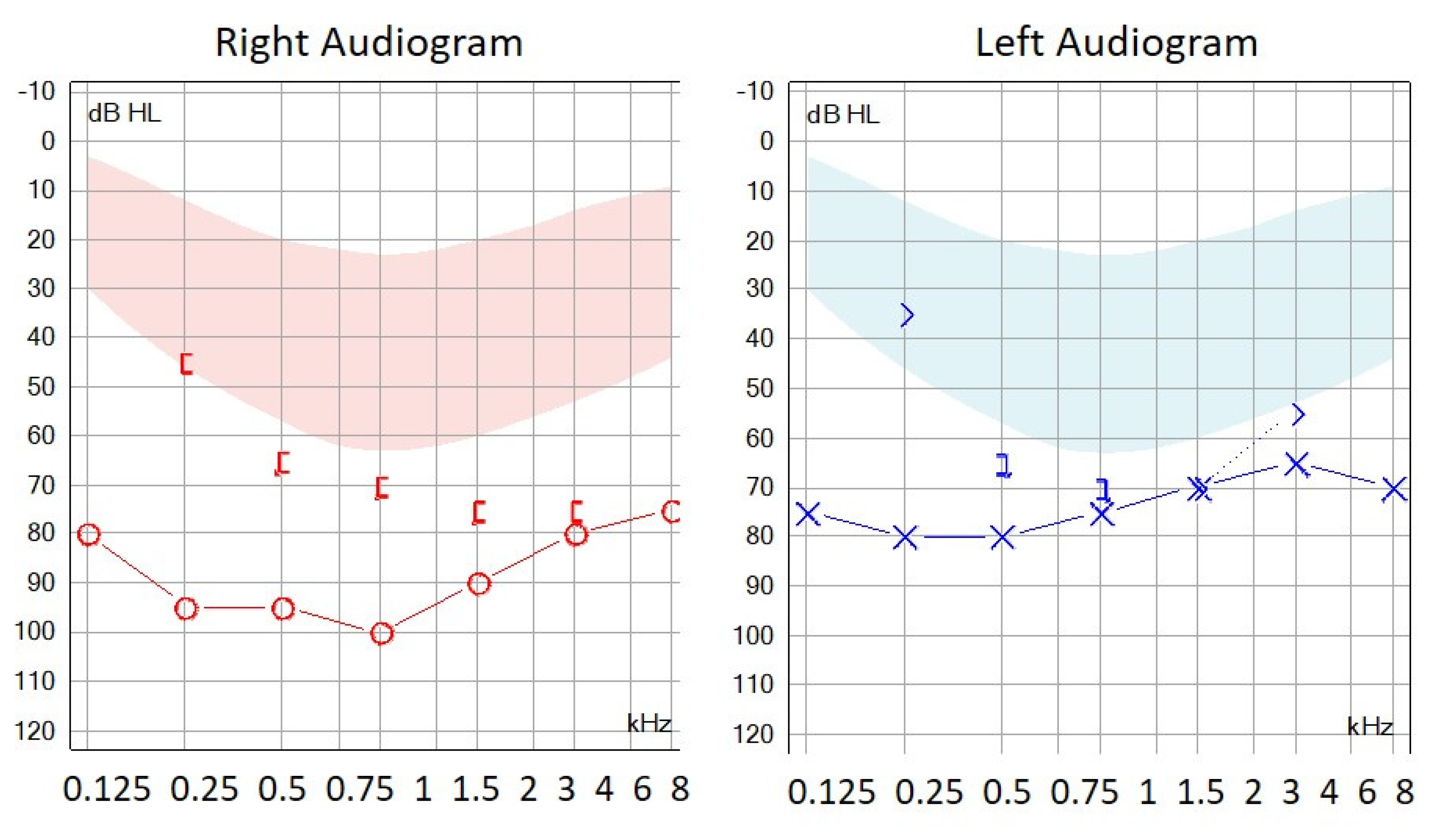
3.1. Autosomal Recessive HL
3.2. Autosomal Dominant HL
3.3. X-Linked HL
3.4. Partially Diagnosed Patients
4. Discussion
4.1. Novel VUS/Likely Pathogenic Variants
4.2. Patients with Pathogenic Variants in Two Different Genes
4.3. Syndromic Cases
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Shearer, A.E.; Smith, R.J.H. Massively Parallel Sequencing for Genetic Diagnosis of Hearing Loss: The New Standard of Care. Otolaryngol. Head Neck Surg. 2015, 153, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Hoefsloot, L.H.; Feenstra, I.; Kunst, H.P.M.; Kremer, H. Genotype phenotype correlations for hearing impairment: Approaches to management. Clin. Genet. 2014, 85, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Morton, C.C.; Nance, W.E. Newborn hearing screening—A silent revolution. N. Engl. J. Med. 2006, 354, 2151–2164. [Google Scholar] [CrossRef]
- Smith, R.J.H.; Bale, J.F.; White, K.R. Sensorineural hearing loss in children. Lancet 2005, 365, 879–890. [Google Scholar] [CrossRef]
- Kochhar, A.; Hildebrand, M.S.; Smith, R.J.H. Clinical aspects of hereditary hearing loss. Genet. Med. 2007, 9, 393–408. [Google Scholar] [CrossRef] [PubMed]
- Alford, R.L.; Arnos, K.S.; Fox, M.; Lin, J.W.; Palmer, C.G.; Pandya, A.; Rehm, H.L.; Robin, N.H.; Scott, D.A.; Yoshinaga-Itano, C.; et al. American College of Medical Genetics and Genomics guideline for the clinical evaluation and etiologic diagnosis of hearing loss. Genet. Med. Off. J. Am. Coll. Med. Genet. 2014, 16, 347–355. [Google Scholar] [CrossRef]
- Delmaghani, S.; El-Amraoui, A. Inner Ear Gene Therapies Take Off: Current Promises and Future Challenges. J. Clin. Med. 2020, 9, 2309. [Google Scholar] [CrossRef]
- Sloan-Heggen, C.M.; Bierer, A.O.; Shearer, A.E.; Kolbe, D.L.; Nishimura, C.J.; Frees, K.L.; Ephraim, S.S.; Shibata, S.B.; Booth, K.T.; Campbell, C.A.; et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum. Genet. 2016, 135, 441–450. [Google Scholar] [CrossRef]
- Yan, D.; Xiang, G.; Chai, X.; Qing, J.; Shang, H.; Zou, B.; Mittal, R.; Shen, J.; Smith, R.J.H.; Fan, Y.-S.; et al. Screening of deafness-causing DNA variants that are common in patients of European ancestry using a microarray-based approach. PLoS ONE 2017, 12, e0169219. [Google Scholar] [CrossRef]
- Domínguez-Ruíz, M. Estudio Molecular de Genes Implicados en Hipoacusia No Sindrómica Autosómica Recesiva Mediante Secuenciación Sanger y de Nueva Generación; UAM: Madrid, Spain, 2015. [Google Scholar]
- Baux, D.; Vaché, C.; Blanchet, C.; Willems, M.; Baudoin, C.; Moclyn, M.; Faugère, V.; Touraine, R.; Isidor, B.; Dupin-Deguine, D.; et al. Combined genetic approaches yield a 48% diagnostic rate in a large cohort of French hearing-impaired patients. Sci. Rep. 2017, 7, 16783. [Google Scholar] [CrossRef]
- Liquori, A.; Vaché, C.; Baux, D.; Blanchet, C.; Hamel, C.; Malcolm, S.; Koenig, M.; Claustres, M.; Roux, A.-F. Whole USH2A Gene Sequencing Identifies Several New Deep Intronic Mutations. Hum. Mutat. 2016, 37, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Vaché, C.; Besnard, T.; Le Berre, P.; García-García, G.; Baux, D.; Larrieu, L.; Abadie, C.; Blanchet, C.; Bolz, H.J.; Millan, J.; et al. Usher syndrome type 2 caused by activation of an USH2A pseudoexon: Implications for diagnosis and therapy. Hum. Mutat. 2012, 33, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Fowler, A.; Mahamdallie, S.; Ruark, E.; Seal, S.; Ramsay, E.; Clarke, M.; Uddin, I.; Wylie, H.; Strydom, A.; Lunter, G.; et al. Accurate clinical detection of exon copy number variants in a targeted NGS panel using DECoN. Wellcome Open Res. 2016, 1, 20. [Google Scholar] [CrossRef] [PubMed]
- del Castillo, F.J.; Rodríguez-Ballesteros, M.; Alvarez, A.; Hutchin, T.; Leonardi, E.; de Oliveira, C.A.; Azaiez, H.; Brownstein, Z.; Avenarius, M.R.; Marlin, S.; et al. A novel deletion involving the connexin-30 gene, del(GJB6-d13s1854), found in trans with mutations in the GJB2 gene (connexin-26) in subjects with DFNB1 non-syndromic hearing impairment. J. Med. Genet. 2005, 42, 588–594. [Google Scholar] [CrossRef]
- Zelante, L.; Gasparini, P.; Estivill, X.; Melchionda, S.; D’Agruma, L.; Govea, N.; Milá, M.; Monica, M.D.; Lutfi, J.; Shohat, M.; et al. Connexin26 mutations associated with the most common form of non-syndromic neurosensory autosomal recessive deafness (DFNB1) in Mediterraneans. Hum. Mol. Genet. 1997, 6, 1605–1609. [Google Scholar] [CrossRef]
- Green, G.E.; Scott, D.A.; McDonald, J.M.; Woodworth, G.G.; Sheffield, V.C.; Smith, R.J. Carrier rates in the midwestern United States for GJB2 mutations causing inherited deafness. JAMA 1999, 281, 2211–2216. [Google Scholar] [CrossRef]
- Marlin, S.; Garabédian, E.N.; Roger, G.; Moatti, L.; Matha, N.; Lewin, P.; Petit, C.; Denoyelle, F. Connexin 26 gene mutations in congenitally deaf children: Pitfalls for genetic counseling. Arch. Otolaryngol. Head Neck Surg. 2001, 127, 927–933. [Google Scholar] [CrossRef][Green Version]
- Denoyelle, F.; Marlin, S.; Weil, D.; Moatti, L.; Chauvin, P.; Garabédian, E.N.; Petit, C. Clinical features of the prevalent form of childhood deafness, DFNB1, due to a connexin-26 gene defect: Implications for genetic counselling. Lancet 1999, 353, 1298–1303. [Google Scholar] [CrossRef]
- Kelsell, D.P.; Dunlop, J.; Stevens, H.P.; Lench, N.J.; Liang, J.N.; Parry, G.; Mueller, R.F.; Leigh, I.M. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature 1997, 387, 80–83. [Google Scholar] [CrossRef]
- Brobby, G.W.; Müller-Myhsok, B.; Horstmann, R.D. Connexin 26 R143W mutation associated with recessive nonsyndromic sensorineural deafness in Africa. N. Engl. J. Med. 1998, 338, 548–550. [Google Scholar] [CrossRef]
- del Castillo, I.; Villamar, M.; Moreno-Pelayo, M.A.; del Castillo, F.J.; Alvarez, A.; Tellería, D.; Menéndez, I.; Moreno, F. A deletion involving the connexin 30 gene in nonsyndromic hearing impairment. N. Engl. J. Med. 2002, 346, 243–249. [Google Scholar] [CrossRef]
- Zhang, Y.; Malekpour, M.; Al-Madani, N.; Kahrizi, K.; Zanganeh, M.; Lohr, N.J.; Mohseni, M.; Mojahedi, F.; Daneshi, A.; Najmabadi, H.; et al. Sensorineural deafness and male infertility: A contiguous gene deletion syndrome. J. Med. Genet. 2007, 44, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ballesteros, M.; del Castillo, F.J.; Martín, Y.; Moreno-Pelayo, M.A.; Morera, C.; Prieto, F.; Marco, J.; Morant, A.; Gallo-Terán, J.; Morales-Angulo, C.; et al. Auditory neuropathy in patients carrying mutations in the otoferlin gene (OTOF). Hum. Mutat. 2003, 22, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Migliosi, V.; Modamio-Høybjør, S.; Moreno-Pelayo, M.A.; Rodríguez-Ballesteros, M.; Villamar, M.; Tellería, D.; Menéndez, I.; Moreno, F.; Del Castillo, I. Q829X, a novel mutation in the gene encoding otoferlin (OTOF), is frequently found in Spanish patients with prelingual non-syndromic hearing loss. J. Med. Genet. 2002, 39, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Eppsteiner, R.W.; Shearer, A.E.; Hildebrand, M.S.; Deluca, A.P.; Ji, H.; Dunn, C.C.; Black-Ziegelbein, E.A.; Casavant, T.L.; Braun, T.A.; Scheetz, T.E.; et al. Prediction of cochlear implant performance by genetic mutation: The spiral ganglion hypothesis. Hear. Res. 2012, 292, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Shahin, H.; Walsh, T.; Rayyan, A.A.; Lee, M.K.; Higgins, J.; Dickel, D.; Lewis, K.; Thompson, J.; Baker, C.; Nord, A.S.; et al. Five novel loci for inherited hearing loss mapped by SNP-based homozygosity profiles in Palestinian families. Eur. J. Hum. Genet. EJHG 2010, 18, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Wattenhofer, M.; Di Iorio, M.V.; Rabionet, R.; Dougherty, L.; Pampanos, A.; Schwede, T.; Montserrat-Sentis, B.; Arbones, M.L.; Iliades, T.; Pasquadibisceglie, A.; et al. Mutations in the TMPRSS3 gene are a rare cause of childhood nonsyndromic deafness in Caucasian patients. J. Mol. Med. Berl. Ger. 2002, 80, 124–131. [Google Scholar] [CrossRef]
- Miyagawa, M.; Nishio, S.; Ikeda, T.; Fukushima, K.; Usami, S. Massively parallel DNA sequencing successfully identifies new causative mutations in deafness genes in patients with cochlear implantation and EAS. PLoS ONE 2013, 8, e75793. [Google Scholar] [CrossRef]
- Kalay, E.; Uzumcu, A.; Krieger, E.; Caylan, R.; Uyguner, O.; Ulubil-Emiroglu, M.; Erdol, H.; Kayserili, H.; Hafiz, G.; Başerer, N.; et al. MYO15A (DFNB3) mutations in Turkish hearing loss families and functional modeling of a novel motor domain mutation. Am. J. Med. Genet. Part A 2007, 143, 2382–2389. [Google Scholar] [CrossRef]
- Pera, A.; Villamar, M.; Viñuela, A.; Gandía, M.; Medà, C.; Moreno, F.; Hernández-Chico, C. A mutational analysis of the SLC26A4 gene in Spanish hearing-impaired families provides new insights into the genetic causes of Pendred syndrome and DFNB4 hearing loss. Eur. J. Hum. Genet. EJHG 2008, 16, 888–896. [Google Scholar] [CrossRef]
- Hutchin, T.; Coy, N.N.; Conlon, H.; Telford, E.; Bromelow, K.; Blaydon, D.; Taylor, G.; Coghill, E.; Brown, S.; Trembath, R.; et al. Assessment of the genetic causes of recessive childhood non-syndromic deafness in the UK—Implications for genetic testing. Clin. Genet. 2005, 68, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, X.M.; Yan, D.; Du, L.L.; Hejtmancik, J.F.; Jacobson, S.G.; Nance, W.E.; Li, A.R.; Angeli, S.; Kaiser, M.; Newton, V.; et al. Characterization of Usher syndrome type I gene mutations in an Usher syndrome patient population. Hum. Genet. 2005, 116, 292–299. [Google Scholar] [CrossRef]
- Kothiyal, P.; Cox, S.; Ebert, J.; Husami, A.; Kenna, M.A.; Greinwald, J.H.; Aronow, B.J.; Rehm, H.L. High-throughput detection of mutations responsible for childhood hearing loss using resequencing microarrays. BMC Biotechnol. 2010, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Nájera, C.; Beneyto, M.; Blanca, J.; Aller, E.; Fontcuberta, A.; Millán, J.M.; Ayuso, C. Mutations in myosin VIIA (MYO7A) and usherin (USH2A) in Spanish patients with Usher syndrome types I and II, respectively. Hum. Mutat. 2002, 20, 76–77. [Google Scholar] [CrossRef]
- García-García, G.; Besnard, T.; Baux, D.; Vaché, C.; Aller, E.; Malcolm, S.; Claustres, M.; Millan, J.M.; Roux, A.-F. The contribution of GPR98 and DFNB31 genes to a Spanish Usher syndrome type 2 cohort. Mol. Vis. 2013, 19, 367–373. [Google Scholar] [PubMed]
- Besnard, T.; Vaché, C.; Baux, D.; Larrieu, L.; Abadie, C.; Blanchet, C.; Odent, S.; Blanchet, P.; Calvas, P.; Hamel, C.; et al. Non-USH2A mutations in USH2 patients. Hum. Mutat. 2012, 33, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Astuto, L.M.; Bork, J.M.; Weston, M.D.; Askew, J.W.; Fields, R.R.; Orten, D.J.; Ohliger, S.J.; Riazuddin, S.; Morell, R.J.; Khan, S.; et al. CDH23 mutation and phenotype heterogeneity: A profile of 107 diverse families with Usher syndrome and nonsyndromic deafness. Am. J. Hum. Genet. 2002, 71, 262–275. [Google Scholar] [CrossRef]
- Aller, E.; Jaijo, T.; Beneyto, M.; Nájera, C.; Oltra, S.; Ayuso, C.; Baiget, M.; Carballo, M.; Antiñolo, G.; Valverde, D.; et al. Identification of 14 novel mutations in the long isoform of USH2A in Spanish patients with Usher syndrome type II. J. Med. Genet. 2006, 43, e55. [Google Scholar] [CrossRef][Green Version]
- Neuhaus, C.; Eisenberger, T.; Decker, C.; Nagl, S.; Blank, C.; Pfister, M.; Kennerknecht, I.; Müller-Hofstede, C.; Charbel Issa, P.; Heller, R.; et al. Next-generation sequencing reveals the mutational landscape of clinically diagnosed Usher syndrome: Copy number variations, phenocopies, a predominant target for translational read-through, and PEX26 mutated in Heimler syndrome. Mol. Genet. Genomic Med. 2017, 5, 531–552. [Google Scholar] [CrossRef]
- Sanggaard, K.M.; Kjaer, K.W.; Eiberg, H.; Nürnberg, G.; Nürnberg, P.; Hoffman, K.; Jensen, H.; Sørum, C.; Rendtorff, N.D.; Tranebjaerg, L. A novel nonsense mutation in MYO6 is associated with progressive nonsyndromic hearing loss in a Danish DFNA22 family. Am. J. Med. Genet. Part A 2008, 146, 1017–1025. [Google Scholar] [CrossRef]
- Kwon, T.-J.; Oh, S.-K.; Park, H.-J.; Sato, O.; Venselaar, H.; Choi, S.Y.; Kim, S.; Lee, K.-Y.; Bok, J.; Lee, S.-H.; et al. The effect of novel mutations on the structure and enzymatic activity of unconventional myosins associated with autosomal dominant non-syndromic hearing loss. Open Biol. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Donaudy, F.; Zheng, L.; Ficarella, R.; Ballana, E.; Carella, M.; Melchionda, S.; Estivill, X.; Bartles, J.R.; Gasparini, P. Espin gene (ESPN) mutations associated with autosomal dominant hearing loss cause defects in microvillar elongation or organisation. J. Med. Genet. 2006, 43, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, C.; Riahi, Z.; Chantot-Bastaraud, S.; Smagghe, L.; Letexier, M.; Marcaillou, C.; Lefèvre, G.M.; Hardelin, J.-P.; El-Amraoui, A.; Singh-Estivalet, A.; et al. An innovative strategy for the molecular diagnosis of Usher syndrome identifies causal biallelic mutations in 93% of European patients. Eur. J. Hum. Genet. EJHG 2016, 24, 1730–1738. [Google Scholar] [CrossRef]
- Plantinga, R.F.; de Brouwer, A.P.M.; Huygen, P.L.M.; Kunst, H.P.M.; Kremer, H.; Cremers, C.W.R.J. A novel TECTA mutation in a Dutch DFNA8/12 family confirms genotype-phenotype correlation. J. Assoc. Res. Otolaryngol. JARO 2006, 7, 173–181. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hildebrand, M.S.; Morín, M.; Meyer, N.C.; Mayo, F.; Modamio-Hoybjor, S.; Mencía, A.; Olavarrieta, L.; Morales-Angulo, C.; Nishimura, C.J.; Workman, H.; et al. DFNA8/12 caused by TECTA mutations is the most identified subtype of nonsyndromic autosomal dominant hearing loss. Hum. Mutat. 2011, 32, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Pelayo, M.A.; del Castillo, I.; Villamar, M.; Romero, L.; Hernández-Calvín, F.J.; Herraiz, C.; Barberá, R.; Navas, C.; Moreno, F. A cysteine substitution in the zona pellucida domain of alpha-tectorin results in autosomal dominant, postlingual, progressive, mid frequency hearing loss in a Spanish family. J. Med. Genet. 2001, 38, E13. [Google Scholar] [CrossRef] [PubMed]
- Brunner, H.G.; van Beersum, S.E.; Warman, M.L.; Olsen, B.R.; Ropers, H.H.; Mariman, E.C. A Stickler syndrome gene is linked to chromosome 6 near the COL11A2 gene. Hum. Mol. Genet. 1994, 3, 1561–1564. [Google Scholar] [CrossRef]
- Sun, Y.; Cheng, J.; Lu, Y.; Li, J.; Lu, Y.; Jin, Z.; Dai, P.; Wang, R.; Yuan, H. Identification of two novel missense WFS1 mutations, H696Y and R703H, in patients with non-syndromic low-frequency sensorineural hearing loss. J. Genet. Genom. Yi Chuan Xue Bao 2011, 38, 71–76. [Google Scholar] [CrossRef]
- Escribano, L.B. Epidemiología Genética en Pacientes Españoles con Hipoacusia de Herencia Autosómica Dominante, Sindrómica y No Sindrómica, Utilizando Herramientas de Nueva Generación: Array-CGH y Secuenciación Masiva; UAM: Madrid, Spain, 2016. [Google Scholar]
- Shinagawa, J.; Moteki, H.; Nishio, S.-Y.; Ohyama, K.; Otsuki, K.; Iwasaki, S.; Masuda, S.; Oshikawa, C.; Ohta, Y.; Arai, Y.; et al. Prevalence and clinical features of hearing loss caused by EYA4 variants. Sci. Rep. 2020, 10, 3662. [Google Scholar] [CrossRef] [PubMed]
- Nobukuni, Y.; Watanabe, A.; Takeda, K.; Skarka, H.; Tachibana, M. Analyses of loss-of-function mutations of the MITF gene suggest that haploinsufficiency is a cause of Waardenburg syndrome type 2A. Am. J. Hum. Genet. 1996, 59, 76–83. [Google Scholar]
- Wu, C.-C.; Tsai, C.-Y.; Lin, Y.-H.; Chen, P.-Y.; Lin, P.-H.; Cheng, Y.-F.; Wu, C.-M.; Lin, Y.-H.; Lee, C.-Y.; Erdenechuluun, J.; et al. Genetic Epidemiology and Clinical Features of Hereditary Hearing Impairment in the Taiwanese Population. Genes 2019, 10, 772. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Vidal, C.; González-Del Pozo, M.; Vela-Boza, A.; Santoyo-López, J.; López-Domingo, F.J.; Vázquez-Marouschek, C.; Dopazo, J.; Borrego, S.; Antiñolo, G. Whole-exome sequencing identifies novel compound heterozygous mutations in USH2A in Spanish patients with autosomal recessive retinitis pigmentosa. Mol. Vis. 2013, 19, 2187–2195. [Google Scholar] [PubMed]
- Bernal, S.; Medà, C.; Solans, T.; Ayuso, C.; Garcia-Sandoval, B.; Valverde, D.; Del Rio, E.; Baiget, M. Clinical and genetic studies in Spanish patients with Usher syndrome type II: Description of new mutations and evidence for a lack of genotype—Phenotype correlation. Clin. Genet. 2005, 68, 204–214. [Google Scholar] [CrossRef]
- Adato, A.; Weil, D.; Kalinski, H.; Pel-Or, Y.; Ayadi, H.; Petit, C.; Korostishevsky, M.; Bonne-Tamir, B. Mutation profile of all 49 exons of the human myosin VIIA gene, and haplotype analysis, in Usher 1B families from diverse origins. Am. J. Hum. Genet. 1997, 61, 813–821. [Google Scholar] [CrossRef]
- Xu, Y.; Guan, L.; Shen, T.; Zhang, J.; Xiao, X.; Jiang, H.; Li, S.; Yang, J.; Jia, X.; Yin, Y.; et al. Mutations of 60 known causative genes in 157 families with retinitis pigmentosa based on exome sequencing. Hum. Genet. 2014, 133, 1255–1271. [Google Scholar] [CrossRef]
- Abe, S.; Usami, S.; Shinkawa, H.; Kelley, P.M.; Kimberling, W.J. Prevalent connexin 26 gene (GJB2) mutations in Japanese. J. Med. Genet. 2000, 37, 41–43. [Google Scholar] [CrossRef]
- Rabionet, R.; Zelante, L.; López-Bigas, N.; D’Agruma, L.; Melchionda, S.; Restagno, G.; Arbonés, M.L.; Gasparini, P.; Estivill, X. Molecular basis of childhood deafness resulting from mutations in the GJB2 (connexin 26) gene. Hum. Genet. 2000, 106, 40–44. [Google Scholar] [CrossRef]
- Wagatsuma, M.; Kitoh, R.; Suzuki, H.; Fukuoka, H.; Takumi, Y.; Usami, S. Distribution and frequencies of CDH23 mutations in Japanese patients with non-syndromic hearing loss. Clin. Genet. 2007, 72, 339–344. [Google Scholar] [CrossRef]
- de Heer, A.-M.R.; Collin, R.W.J.; Huygen, P.L.M.; Schraders, M.; Oostrik, J.; Rouwette, M.; Kunst, H.P.M.; Kremer, H.; Cremers, C.W.R.J. Progressive sensorineural hearing loss and normal vestibular function in a Dutch DFNB7/11 family with a novel mutation in TMC1. Audiol. Neurootol. 2011, 16, 93–105. [Google Scholar] [CrossRef]
- Duman, D.; Tekin, M. Autosomal recessive nonsyndromic deafness genes: A review. Front. Biosci. 2012, 17, 2213–2236. [Google Scholar] [CrossRef] [PubMed]
- Shearer, A.E.; Hildebrand, M.S.; Smith, R.J. Hereditary Hearing Loss and Deafness Overview. In GeneReviews; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J., Stephens, K., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Mandelker, D.; Amr, S.S.; Pugh, T.; Gowrisankar, S.; Shakhbatyan, R.; Duffy, E.; Bowser, M.; Harrison, B.; Lafferty, K.; Mahanta, L.; et al. Comprehensive diagnostic testing for stereocilin: An approach for analyzing medically important genes with high homology. J. Mol. Diagn. 2014, 16, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Zwaenepoel, I.; Mustapha, M.; Leibovici, M.; Verpy, E.; Goodyear, R.; Liu, X.Z.; Nouaille, S.; Nance, W.E.; Kanaan, M.; Avraham, K.B.; et al. Otoancorin, an inner ear protein restricted to the interface between the apical surface of sensory epithelia and their overlying acellular gels, is defective in autosomal recessive deafness DFNB22. Proc. Natl. Acad. Sci. USA 2002, 99, 6240–6245. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, G.M.D.; Ramos, P.Z.; Castilho, A.M.; Guimarães, A.C.; Sartorato, E.L. Molecular study of patients with auditory neuropathy. Mol. Med. Rep. 2016, 14, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Yokota, Y.; Moteki, H.; Nishio, S.; Yamaguchi, T.; Wakui, K.; Kobayashi, Y.; Ohyama, K.; Miyazaki, H.; Matsuoka, R.; Abe, S.; et al. Frequency and clinical features of hearing loss caused by STRC deletions. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Yazdanpanahi, N.; Tabatabaiefar, M.A.; Bagheri, N.; Azadegan Dehkordi, F.; Farrokhi, E.; Hashemzadeh Chaleshtori, M. The role and spectrum of SLC26A4 mutations in Iranian patients with autosomal recessive hereditary deafness. Int. J. Audiol. 2015, 54, 124–130. [Google Scholar] [CrossRef]
- Jaijo, T.; Aller, E.; García-García, G.; Aparisi, M.J.; Bernal, S.; Avila-Fernández, A.; Barragán, I.; Baiget, M.; Ayuso, C.; Antiñolo, G.; et al. Microarray-based mutation analysis of 183 Spanish families with Usher syndrome. Invest. Ophthalmol. Vis. Sci. 2010, 51, 1311–1317. [Google Scholar] [CrossRef]
- Blanco-Kelly, F.; Jaijo, T.; Aller, E.; Avila-Fernandez, A.; López-Molina, M.I.; Giménez, A.; García-Sandoval, B.; Millán, J.M.; Ayuso, C. Clinical aspects of Usher syndrome and the USH2A gene in a cohort of 433 patients. JAMA Ophthalmol. 2015, 133, 157–164. [Google Scholar] [CrossRef]
- Butz, M.; McDonald, A.; Lundquist, P.A.; Meyer, M.; Harrington, S.; Kester, S.; Stein, M.I.; Mistry, N.A.; Zimmerman Zuckerman, E.; Niu, Z.; et al. Development and Validation of a Next-Generation Sequencing Panel for Syndromic and Nonsyndromic Hearing Loss. J. Appl. Lab. Med. 2020, 5, 467–479. [Google Scholar] [CrossRef]
- Cabanillas, R.; Diñeiro, M.; Cifuentes, G.A.; Castillo, D.; Pruneda, P.C.; Álvarez, R.; Sánchez-Durán, N.; Capín, R.; Plasencia, A.; Viejo-Díaz, M.; et al. Comprehensive genomic diagnosis of non-syndromic and syndromic hereditary hearing loss in Spanish patients. BMC Med. Genom. 2018, 11, 58. [Google Scholar] [CrossRef]
- Diaz-Horta, O.; Duman, D.; Foster, J.; Sırmacı, A.; Gonzalez, M.; Mahdieh, N.; Fotouhi, N.; Bonyadi, M.; Cengiz, F.B.; Menendez, I.; et al. Whole-exome sequencing efficiently detects rare mutations in autosomal recessive nonsyndromic hearing loss. PLoS ONE 2012, 7, e50628. [Google Scholar] [CrossRef] [PubMed]
- Tekin, D.; Yan, D.; Bademci, G.; Feng, Y.; Guo, S.; Foster, J.; Blanton, S.; Tekin, M.; Liu, X. A next-generation sequencing gene panel (MiamiOtoGenes) for comprehensive analysis of deafness genes. Hear. Res. 2016, 333, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Mei, X.; Yang, W.; Zhu, R.; Yang, T.; Hu, H. Whole-exome sequencing identifies rare pathogenic and candidate variants in sporadic Chinese Han deaf patients. Clin. Genet. 2020, 97, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Vears, D.F.; Sénécal, K.; Borry, P. Reporting practices for variants of uncertain significance from next generation sequencing technologies. Eur. J. Med. Genet. 2017, 60, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Hoppman, N.; Aypar, U.; Brodersen, P.; Brown, N.; Wilson, J.; Babovic-Vuksanovic, D. Genetic testing for hearing loss in the United States should include deletion/duplication analysis for the deafness/infertility locus at 15q15.3. Mol. Cytogenet. 2013, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Kremer, H. Hereditary hearing loss; about the known and the unknown. Hear. Res. 2019, 376, 58–68. [Google Scholar] [CrossRef] [PubMed]
| Gene | Phenotype | Gene | Phenotype |
|---|---|---|---|
| ACTG1 | NSHL | TRIOBP | NSHL |
| CEP250 | NSHL | CDH23 | USH/NSHL |
| CHD7 | CHARGE | CIB2 | USH/NSHL |
| CISD2 | NSHL | DFNB31 | USH/NSHL |
| CLDN14 | NSHL | MYO7A | USH/NSHL |
| COCH | NSHL | PCDH15 | USH/NSHL |
| DFNA5 | NSHL | USH1C | USH/NSHL |
| DFNB59 | NSHL | USH1G | USH/NSHL |
| ESPN | NSHL | EDN3 | WS |
| EYA4 | NSHL | EDNRB | WS |
| GJB2 | NSHL | MITF | WS |
| GJB6 | NSHL | PAX3 | WS |
| KCNQ4 | NSHL | SNAI2 | WS |
| LHFPL5 | NSHL | SOX10 | WS |
| LOXHD1 | NSHL | EYA1 | BOR |
| LRTOMT | NSHL | SIX1 | BOR |
| MYH9 | NSHL | SIX5 | BOR |
| MYH14 | NSHL | ADGRV1 | USH |
| MYO6 | NSHL | CLRN1 | USH |
| MYO15A | NSHL | USH2A | USH |
| OTOA | NSHL | KCNE1 | JLNS |
| OTOF | NSHL | KCNQ1 | JLNS |
| OTOG | NSHL | COL11A2 | Stickler/NSHL |
| OTOGL | NSHL | SEMA3E | CHARGE |
| POU3F4 | NSHL | SLC26A4 | Pendred/NSHL |
| PTPRQ | NSHL | WFS1 | WF/NSHL |
| SMPX | NSHL | chr1:215827262-215827362 | USH |
| STRC | NSHL | chr1:215967733-215967833 | USH |
| TECTA | NSHL | chr1:216039671-216039771 | USH |
| TIMM8A | NSHL | chr1:216064520-216064560 | USH |
| TMC1 | NSHL | chr1:216247426-216247526 | USH |
| TMPRSS3 | NSHL | ||
| TPRN | NSHL |
| (A) Patients Diagnosed with Autosomal Recessive Deafness | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family | Patient | Sex | Age | Diagnosis | Gene | Allele 1 | Allele 2 | Phenotype | ||||
| 1 | 33311 | M | 1 | NSHL | GJB2 NM_004004.5 | c.35del/p.(Gly12Valfs *2) [16] | c.35del/p.(Gly12Valfs *2) [16] | SNHL, bilateral, symmetrical, prelingual, severe, stable | ||||
| 2 | 35961 | F | 2 | NSHL | GJB2 NM_004004.5 | c.35del/p.(Gly12Valfs *2) [16] | c.35del/p.(Gly12Valfs *2) [16] | SNHL, bilateral, symmetrical, prelingual, moderate, stable | ||||
| 3 | 39026 | F | 0 | NSHL | GJB2 NM_004004.5 | c.35del/p.(Gly12Valfs *2) [16] | c.35del/p.(Gly12Valfs *2) [16] | SNHL, bilateral, symmetrical, prelingual, severe-profound, stable | ||||
| 4 | 39611 | F | 5 | NSHL | GJB2 NM_004004.5 | c.596C > T/p.(Ser199Phe) [17] | c.35del/p.(Gly12Valfs *2) [16] | SNHL, bilateral, symmetrical, postlingual, severe, stable | ||||
| 5 | 40372 | M | 5 | NSHL | GJB2 NM_004004.5 | c.617A > G/p.(Asn206Ser) [18] | c.269dup/p.(Val91Serfs *11) [19] | SNHL, bilateral, symmetrical, postlingual, mild–moderate, stable | ||||
| 6 | 42105 | M | 0 | NSHL | GJB2 NM_004004.5 | c.101T > C/p.(Met34Thr) [20] | c.427C > T/p.(Arg143Trp) [21] | SNHL, bilateral, symmetrical, prelingual, moderate, stable | ||||
| 7 | 28981 | M | 0 | NSHL | GJB2 NM_004004.5 | c.35del/p.(Gly12Valfs *2) [16] | SNHL, bilateral, symmetrical, prelingual, profound, stable | |||||
| GJB6 NM_001110219.2 | del(GJB6-D13S1830) [22] | |||||||||||
| 8 | 34307 | M | 0 | NSHL | GJB2 NM_004004.5 | c.269dup/p.(Val91Serfs *11) [19] | SNHL, bilateral, symmetrical, prelingual, profound, stable | |||||
| GJB6 NM_001110219.2 | del(GJB6-D13S1830) [22] | |||||||||||
| 9 | 37468 | M | 1 | NSHL | GJB2 NM_004004.5 | c.617A > G/p.(Asn206Ser) [18] | SNHL, bilateral, symmetrical, prelingual, profound, stable | |||||
| GJB6 NM_001110219.2 | del(GJB6-D13S1830) [22] | |||||||||||
| 10 | 37439 | F | 5 | NSHL | STRC NM_153700.2 | Whole gene deletion (15q15) [23] | Whole gene deletion (15q15) [23] | SNHL, bilateral, symmetrical, postlingual, moderate, stable | ||||
| GJB2 NM_004004.5 | c.101T > C/p.(Met34Thr) [20] | |||||||||||
| 11 | 33416 | F | 7 | NSHL | STRC NM_153700.2 | Whole gene deletion (15q15) [23] | Whole gene deletion(15q15) [23] | SNHL, bilateral, symmetrical, postlingual, moderate, stable | ||||
| 12 | 37112 | M | 4 | NSHL | STRC NM_153700.2 | Whole gene deletion (15q15) [23] | Whole gene deletion (15q15) [23] | SNHL, bilateral, symmetrical, postlingual, moderate, stable | ||||
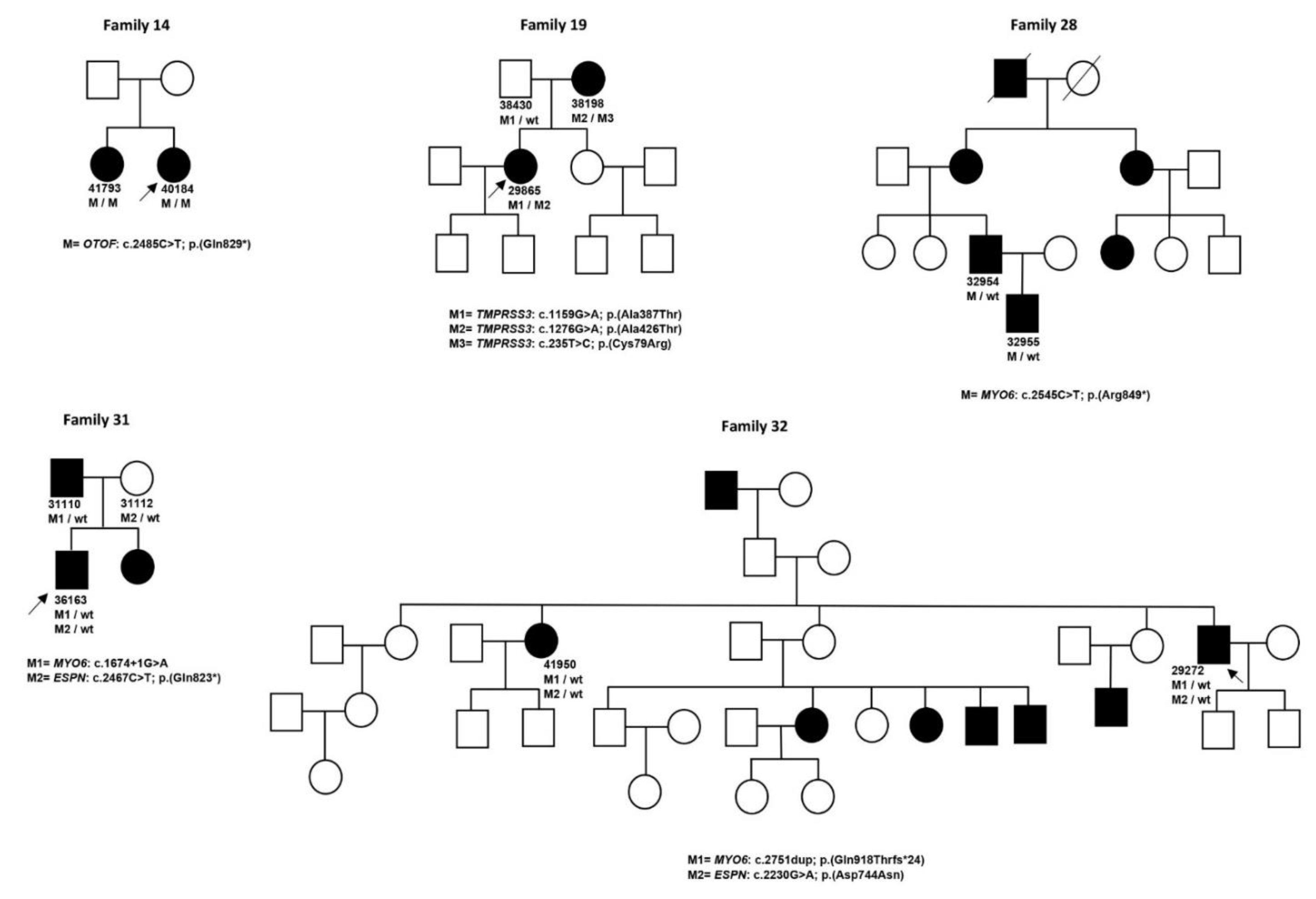
| 13 | 31410 | M | 5 | NSHL | OTOF NM_194248.2 | c.4275G > A/p.(Trp1425 *) [24] | c.2485C > T/p.(Gln829 *) [25] | SNHL, bilateral, symmetrical, prelingual, profound, stable | ||||
| 14 | 40184 | F | 0 | NSHL | OTOF NM_194248.2 | c.2485C > T/p.(Gln829 *) [25] | c.2485C > T/p.(Gln829 *) [25] | SNHL, bilateral, symmetrical, prelingual, profound, stable | ||||
| 14 | 41793 | F | 0 | NSHL | OTOF NM_194248.2 | c.2485C > T/p.(Gln829 *) [25] | c.2485C > T/p.(Gln829 *) [25] | SNHL, bilateral, symmetrical, prelingual, profound, stable | ||||
| 15 | 34197 | F | 54 | NSHL | LOXHD1 NM_144612.6 | c.3419dup/p.(Leu1140Phefs *5) | c.3419dup/p.(Leu1140Phefs *5) | SNHL, bilateral, symmetrical, postlingual, moderate–severe, stable | ||||
| 16 | 34865 | M | 7 | NSHL | LOXHD1 NM_144612.6 | c.4480C > T/p.(Arg1494 *) [26] | c.4480C > T/p.(Arg1494 *) [26] | SNHL, bilateral, symmetrical, postlingual, moderate, stable | ||||
| 17 | 29440 | M | 33 | NSHL | OTOA NM_144672.3 | c.877C > T/p.(Gln293 *) | Whole gene deletion (16q12.2 region) [27] | SNHL, bilateral, postlingual, moderate, stable | ||||
| 18 | 37140 | M | 4 | NSHL | OTOA NM_144672.3 | Whole gene deletion (16q12.2 region) [27] | Whole gene deletion (16q12.2 region) [27] | SNHL, bilateral, symmetrical, postlingual, moderate, stable | ||||
| 19 | 29865 | F | 46 | NSHL | TMPRSS3 NM_024022.2 | c.1276G > A/p.(Ala426Thr) [28] | c.1159G > A/p.(Ala387Thr) [29] | SNHL, bilateral, symmetrical, postlingual, mild–moderate, progressive | ||||
| 19 | 38198 | F | 40 | NSHL | TMPRSS3 NM_024022.2 | c.1276G > A/p.(Ala426Thr) [28] | c.235T > C/p.(Cys79Arg) | SNHL, bilateral, symmetrical, postlingual, profound, progressive | ||||
| 20 | 42108 | F | 1 | NSHL | MYO15A NM_016239.3 | c.8968-1G > T [30] | c.8968-1G > T [30] | SNHL, bilateral, symmetrical, prelingual, severe, stable | ||||
| 21 | 37513 | M | 4 | NSHL/EVA | SLC26A4 NM_000441.1 | c.1540C > A/p.(Gln514Lys) [31] | c.1540C > A/p.(Gln514Lys) [31] | SNHL, bilateral, asymmetrical, postlingual, Right: profound Left: moderate, stable, EVA | ||||
| 22 | 36777 | F | 1 | NSHL | OTOG NM_001277269.1 | c.2140dup/p.(Ser714Lysfs *22) | c.2140dup/p.(Ser714Lysfs *22) | SNHL, bilateral, symmetrical, prelingual, moderate, stable | ||||
| 23 | 39949 | F | 18 | NSHL | TECTA NM_005422.2 | c.4055G > A/p.(Cys1352Tyr) [32] | c.4055G > A/p.(Cys1352Tyr) [32] | SNHL, bilateral, moderate | ||||
| MYO7A NM_000260.3 | c.5648G > A/p.(Arg1883Gln) [33] | |||||||||||
| 24 | 40453 | F | 40 | NSHL | MYO7A NM_000260.3 | c.1232T > C/p.(Val411Ala) [34] | c.6025del/p.(Ala2009Profs *32) [35] | SNHL, bilateral, symmetrical, postlingual, mild, stable | ||||
| 25 | 27862 | M | 30 | USH | ADGRV1 NM_032119.3 | c.12528-1G > T [36] | c.17933A > G/p.(His5978Arg) [37] | SNHL, congenital, moderate, retinitis pigmentosa | ||||
| 26 | 30816 | F | 1 | USH | CDH23 NM_022124.5 | c.310G > T/p.(Glu104*) | c.2289 + 1G > A [38] | SNHL, bilateral, symmetrical, prelingual, profound, stable, bilateral, vestibular areflexia, retinitis pigmentosa | ||||
| 27 | 27734 | F | 49 | USH | USH2ANM_206933.2 | c.9799T > C/p.(Cys3267Arg) [39] | c.9676C > T/p.(Arg3226 *) [40] | SNHL, bilateral, symmetrical, postlingual, moderate, stable, retinitis pigmentosa | ||||
| (B) Patients Diagnosed with Autosomal Dominant Deafness | ||||||||||||
| Family | Patient | Sex | Age | Diagnosis | Gene | Allele 1 | Allele 2 | Phenotype | ||||
| 28 | 32954 | M | 46 | NSHL | MYO6 NM_004999.3 | c.2545C > T/p.(Arg849 *) [41] | SNHL, bilateral, symmetrical, postlingual, moderate, stable | |||||
| 28 | 32955 | F | 15 | NSHL | MYO6 NM_004999.3 | c.2545C > T/p.(Arg849 *) [41] | SNHL, bilateral, symmetrical, moderate, stable | |||||
| 29 | 35197 | F | 37 | NSHL | MYO6 NM_004999.3 | c.1666C > T/p.(Arg556 *) | SNHL, bilateral, symmetrical, postlingual, moderate, stable | |||||
| 30 | 40488 | F | 30 | NSHL | MYO6 NM_004999.3 | c.1224-9del | SNHL, bilateral, asymmetrical, postlingual, severe–profound, progressive | |||||
| 31 | 31110 | M | 42 | NSHL | MYO6 NM_004999.3 | c.1674 + 1G > A | SNHL, bilateral, symmetrical, postlingual, moderate, stable | |||||
| 31 | 36163 | M | 32 | NSHL | MYO6 NM_004999.3 | c.1674 + 1G > A | SNHL, bilateral, symmetrical, postlingual, moderate, stable | |||||
| ESPN NM_031475.2 | c.2467C > T/p.(Gln823 *) | |||||||||||
| 32 | 29272 | M | 46 | NSHL | MYO6 NM_004999.3 | c.2751dup/p.(Gln918Thrfs *24)[42] | SNHL, postlingual | |||||
| ESPN NM_031475.2 | c.2230G > A/p.(Asp744Asn) [43] | |||||||||||
| 32 | 41950 | F | 61 | NSHL | MYO6 NM_004999.3 | c.2751dup/p.(Gln918Thrfs *24) [42] | SNHL, postlingual | |||||
| ESPN NM_031475.2 | c.2230G > A/p.(Asp744Asn) [43] | |||||||||||
| 33 | 41268 | M | 18 | NSHL | MYO6 NM_004999.3 | c.494T > G/p.(Leu165Arg) | SNHL, bilateral, symmetrical, postlingual, profound, progressive, tinnitus | |||||
| MYO7A NM_000260.3 | c.1997G > A/p.(Arg666Gln) [44] | c.3527G > A/p.(Ser1176Asn) [8] | ||||||||||
| 34 | 33945 | M | 3 | NSHL | TECTA NM_005422.2 | c.5668C > T/p.(Arg1890Cys) [45] | SNHL, bilateral, asymmetrical prelingual, stable | |||||
| 35 | 35453 | M | 2 | NSHL | TECTA NM_005422.2 | c.5383 + 5_5383 + 8del [46] | SNHL, bilateral, symmetrical, prelingual, moderate, stable | |||||
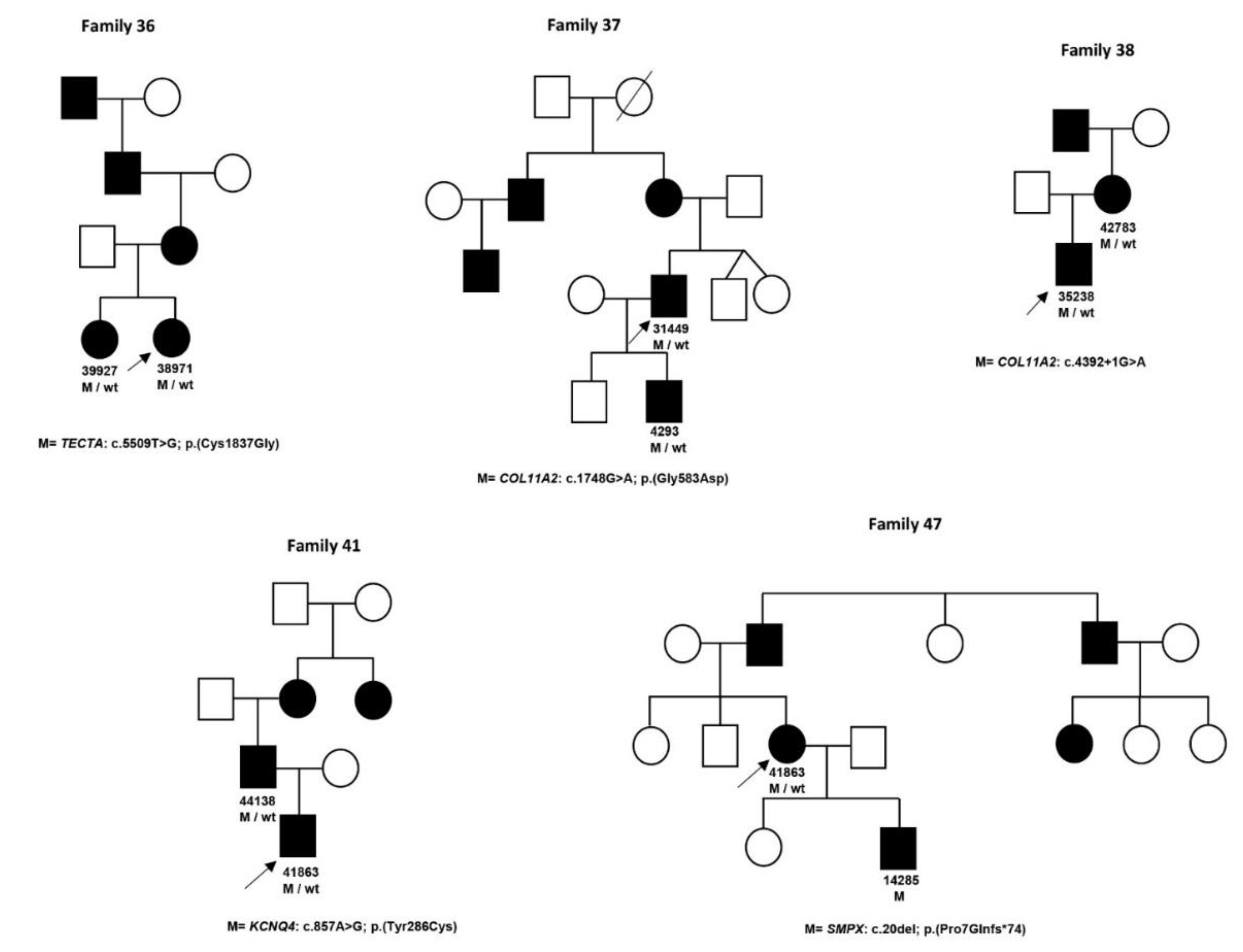
| 36 | 38971 | F | 0 | NSHL | TECTA NM_005422.2 | c.5509T > G/p.(Cys1837Gly) [47] | SNHL, bilateral, symmetrical, prelingual, moderate, stable | |||||
| 36 | 39927 | F | 0 | NSHL | TECTA NM_005422.2 | c.5509T > G/p.(Cys1837Gly)[47] | SNHL, unilateral, asymmetrical, prelingual, moderate–severe, progressive | |||||
| 37 | 4293 | M | 6 | NSHL | COL11A2 NM_080680.2 | c.1748G > A/p.(Gly583Asp) | SNHL, bilateral, symmetrical, postlingual, stable | |||||
| 37 | 31449 | M | 35 | NSHL | COL11A2 NM_080680.2 | c.1748G > A/p.(Gly583Asp) | SNHL, bilateral, asymmetrical, postlingual, Right: mild–moderate; Left: moderate–severe, stable | |||||
| 38 | 35238 | M | 6 | NSHL/Stickler | COL11A2 NM_080680.2 | c.4392 + 1G > A [48] | SNHL, bilateral, symmetrical, postlingual, moderate, stable, flattened facial profile, sunken nasal root, short nose with anteverted nostrils, osteorticular problems | |||||
| 38 | 42783 | F | 37 | NSHL/Stickler | COL11A2 NM_080680.2 | c.4392 + 1G > A [48] | SNHL, flattened facial profile, osteorticular problems and maxillofacial alterations | |||||
| 39 | 40431 | M | 5 | NSHL | WFS1 NM_006005.3 | c.1463_1474dup/p.(Val491_Pro492insLeuIleThrVal) | SNHL, bilateral, asymmetrical, postlingual, Right: profound Left: severe, progressive | |||||
| 40 | 42125 | F | 5 | NSHL | WFS1 NM_006005.3 | c.2108G > A/p.(Arg703His) [49] | SNHL, bilateral, symmetrical, postlingual, severe–profound, progressive | |||||
| 41 | 36655 | M | 7 | NSHL | KCNQ4 NM_004700.3 | c.857A > G/p.(Tyr286Cys) [50] | SNHL, bilateral, symmetrical, postlingual, moderate, stable | |||||
| 41 | 44138 | M | 46 | NSHL | KCNQ4 NM_004700.3 | c.857A > G/p.(Tyr286Cys) [50] | ||||||
| 42 | 39490 | F | 45 | NSHL | ACTG1 NM_001199954.1 | c.895C > G/p.(Leu299Val) [29] | SNHL, bilateral, asymmetrical, postlingual, right: moderate left: severe, progressive | |||||
| 43 | 40519 | M | 40 | NSHL | EYA4 NM_004100.4 | c.988C > T/p.(Gln330 *) [51] | SNHL, bilateral, asymmetrical, postlingual, right: profound left: severe, progressive, tinnitus, decrease in size of both cochlear nerves | |||||
| 44 | 12227 | M | 34 | WS | MITF NM_198159.2 | c.943C > T/p.(Arg315 *) [52] | HL, prelingual, White forelock, Heterochromia iridis | |||||
| GJB6 NM_001110219.2 | del(GJB6-D13S1830) [22] | |||||||||||
| 45 | 37350 | M | 2 | BOR | EYA1 NM_000503.5 | c.1540_1542del/p.(Leu514del) [53] | Mixed HL, bilateral, symmetrical, prelingual, severe, stable, 2nd branchial arch fistula, facial dysmorphia | |||||
| (C) Patients Diagnosed with X-Linked Deafness | ||||||||||||
| Family | Patient | Sex | Age | Diagnosis | Gene | Allele 1 | Allele 2 | Phenotype | ||||
| 46 | 34796 | M | 1 | NSHL | POU3F4 (XLR) NM_000307.4 | c.977T > C/p.(Phe326Ser) | Mixed HL, bilateral symmetrical, prelingual, moderate, stable, bilateral corkscrew cochlea, incomplete splitting of turns, absence of meatus and stapes fixation | |||||
| 47 | 14285 | M | 3 | NSHL | SMPX (XLD) NM_014332.2 | c.20del/p.(Pro7Glnfs *74) | SNHL, bilateral, symmetrical, postlingual, moderate, stable | |||||
| 47 | 41863 | F | 30 | NSHL | SMPX (XLD) NM_014332.2 | c.20del/p.(Pro7Glnfs *74) | SNHL, bilateral, symmetrical, postlingual, moderate, stable | |||||
| Variant | Frequency | Pathogenicity Scores | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Nucleotide | Protein | Classification | GnomAD Exomes | GnomAD Genomes | Deafness Variation Database | Missense Pathogenicity Scores | Conservation Score (GERP) | MaxEnt |
| LOXHD1 NM_144612.6 | c.3419dup | p.(Leu1140Phefs *5) | Pathogenic | 0.0000267 | NF | NF | NA | 5.05 | - |
| OTOA NM_144672.3 | c.877C > T | p.(Gln293 *) | Pathogenic | NF | NF | NF | NA | 5.41 | - |
| TMPRSS3 NM_024022.2 | c.235T > C | p.(Cys79Arg) | Likely Pathogenic | NF | NF | NF | 11/13 | 5.23 | - |
| OTOG NM_001277269.1 | c.2140dup | p.(Ser714Lysfs *22) | Pathogenic | NF | NF | NF | NA | 4.9 | - |
| CDH23 NM_022124.5 | c.310G > T | p.(Glu104 *) | Pathogenic | NF | NF | NF | NA | 5.43 | - |
| MYO6 NM_004999.3 | c.1666C > T | p.(Arg556 *) | Pathogenic | 0.0000119 | NF | Unknown significance–Impact High | NA | 5.77 | - |
| MYO6 NM_004999.3 | c.1224-9del | - | VUS | NF | NF | NF | NA | 5.23 | AS broken (from 7.08 to −4.37) |
| MYO6 NM_004999.3 | c.1674 + 1G > A | - | Pathogenic | 0.00000736 | NF | Unknown significance-Impact High | NA | 5.77 | DS broken (from 7.94 to −0.24) |
| ESPN NM_031475.2 | c.2467C > T | p.(Gln823 *) | Pathogenic | NF | NF | NF | NA | 4.28 | - |
| MYO6 NM_004999.3 | c.494T > G | p.(Leu165Arg) | VUS | NF | NF | NF | 13/13 | 5.45 | - |
| COL11A2 NM_080680.2 | c.1748G > A | p.(Gly583Asp) | Likely Pathogenic | NF | NF | NF | 11/11 | 3.89 | - |
| WFS1 NM_006005.3 | c.1463_1474dup | p.(Val491_Pro492insLeuIleThrVal) | VUS | NF | NF | NF | NA | 4.25 | - |
| POU3F4 NM_000307.4 | c.977T > C | p.(Phe326Ser) | Likely Pathogenic | NF | NF | NF | 10/10 | 5.07 | - |
| SMPX NM_014332.2 | c.20del | p.(Pro7Glnfs *74) | Pathogenic | NF | NF | NF | NA | 5.78 | - |
| Patient | Diagnosis | Gene | Allele 1 |
|---|---|---|---|
| 40056 | NSHL | USH2A NM_206933.2 | c.4325T > C/p.(Phe1442Ser) [55] |
| 31443 | USH | USH2A NM_206933.2 | c.2431_2432del/p.(Lys811Aspfs*11) [35] |
| 28523 | NSHL | USH2A NM_206933.2 | c.2135del/p.(Ser712*) [56] |
| MYO7A NM_000260.3 | c.5581C > T/p.(Arg1861*) [57] | ||
| 37248 | NSHL | USH2A NM_206933.2 | c.9244A > G/p.(Ile3082Val) [58] |
| GJB2 NM_004004.5 | c.109G > A/p.(Val37Ile) [59] | ||
| 37986 | NSHL | GJB2 NM_004004.5 | c.269T > C/(p.Leu90Pro) [19] |
| 39353 | NSHL | GJB2 NM_004004.5 | c.445G > A/p.(Ala149Thr) [60] |
| 12228 | NSHL | STRC NM_153700.2 | Complex rearrangement |
| 28358 | NSHL | OTOF NM_194248.2 | c.2485C > T/p.(Gln829*) [25] |
| 35862 | NSHL | OTOF NM_194248.2 | c.2485C > T/p.(Gln829*) [25] |
| CDH23 NM_022124.5 | c.4762C > T/p.(Arg1588Trp) [61] | ||
| 33335 | USH | CDH23 NM_022124.5 | c.2289 + 1G > A [38] |
| 34978 | NSHL | TMC1 NM_138691.2 | c.1763 + 3A > G [62] |
| TMPRSS3 NM_024022.2 | c.280G > A/p.(Gly94Arg) [29] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-García, G.; Berzal-Serrano, A.; García-Díaz, P.; Villanova-Aparisi, R.; Juárez-Rodríguez, S.; de Paula-Vernetta, C.; Cavallé-Garrido, L.; Jaijo, T.; Armengot-Carceller, M.; Millán, J.M.; et al. Improving the Management of Patients with Hearing Loss by the Implementation of an NGS Panel in Clinical Practice. Genes 2020, 11, 1467. https://doi.org/10.3390/genes11121467
García-García G, Berzal-Serrano A, García-Díaz P, Villanova-Aparisi R, Juárez-Rodríguez S, de Paula-Vernetta C, Cavallé-Garrido L, Jaijo T, Armengot-Carceller M, Millán JM, et al. Improving the Management of Patients with Hearing Loss by the Implementation of an NGS Panel in Clinical Practice. Genes. 2020; 11(12):1467. https://doi.org/10.3390/genes11121467
Chicago/Turabian StyleGarcía-García, Gema, Alba Berzal-Serrano, Piedad García-Díaz, Rebeca Villanova-Aparisi, Sara Juárez-Rodríguez, Carlos de Paula-Vernetta, Laura Cavallé-Garrido, Teresa Jaijo, Miguel Armengot-Carceller, José M Millán, and et al. 2020. "Improving the Management of Patients with Hearing Loss by the Implementation of an NGS Panel in Clinical Practice" Genes 11, no. 12: 1467. https://doi.org/10.3390/genes11121467
APA StyleGarcía-García, G., Berzal-Serrano, A., García-Díaz, P., Villanova-Aparisi, R., Juárez-Rodríguez, S., de Paula-Vernetta, C., Cavallé-Garrido, L., Jaijo, T., Armengot-Carceller, M., Millán, J. M., & Aller, E. (2020). Improving the Management of Patients with Hearing Loss by the Implementation of an NGS Panel in Clinical Practice. Genes, 11(12), 1467. https://doi.org/10.3390/genes11121467






