The Genomic Impact of Selection for Virulence against Resistance in the Potato Cyst Nematode, Globodera pallida
Abstract
1. Introduction
2. Materials and Methods
2.1. G. pallida Populations
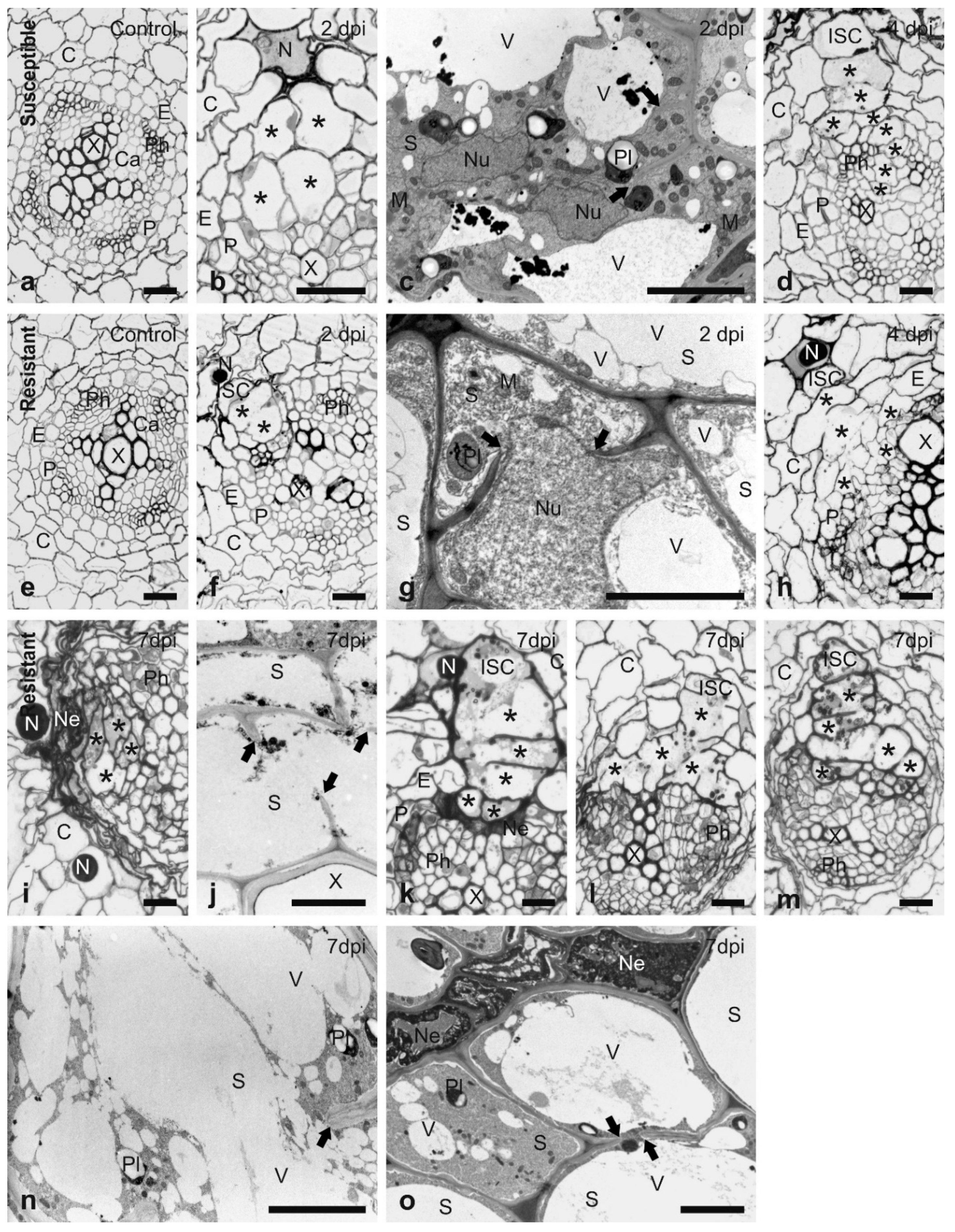
2.2. Histological Analysis of the H3 Resistant Response
2.3. Comparison of G. pallida Reproduction on 3 Potato Genotypes and Collection of Females
2.4. DNA Extraction and Quantification
2.5. Statistical Analysis
2.6. Targeted Sequencing, Library Preparation and Enrichment (PenSeq)
Bait Design
2.7. PenSeq Library Preparation and Sequencing
2.8. Variant Calling of the PenSeq Samples
2.9. Real-Time PCR (qPCR) Analysis of Enriched Samples
2.10. Whole-Genome Scanning (ReSeq)
Library Preparation for ReSeq and Sequencing
2.11. Variant Calling of the ReSeq Samples
3. Results
3.1. Histological Analysis of the Resistant Response to H3
3.2. The Reproductive Ability of Selected Nematode Populations Is Dependent on the Genetic Background of the Potato Genotype
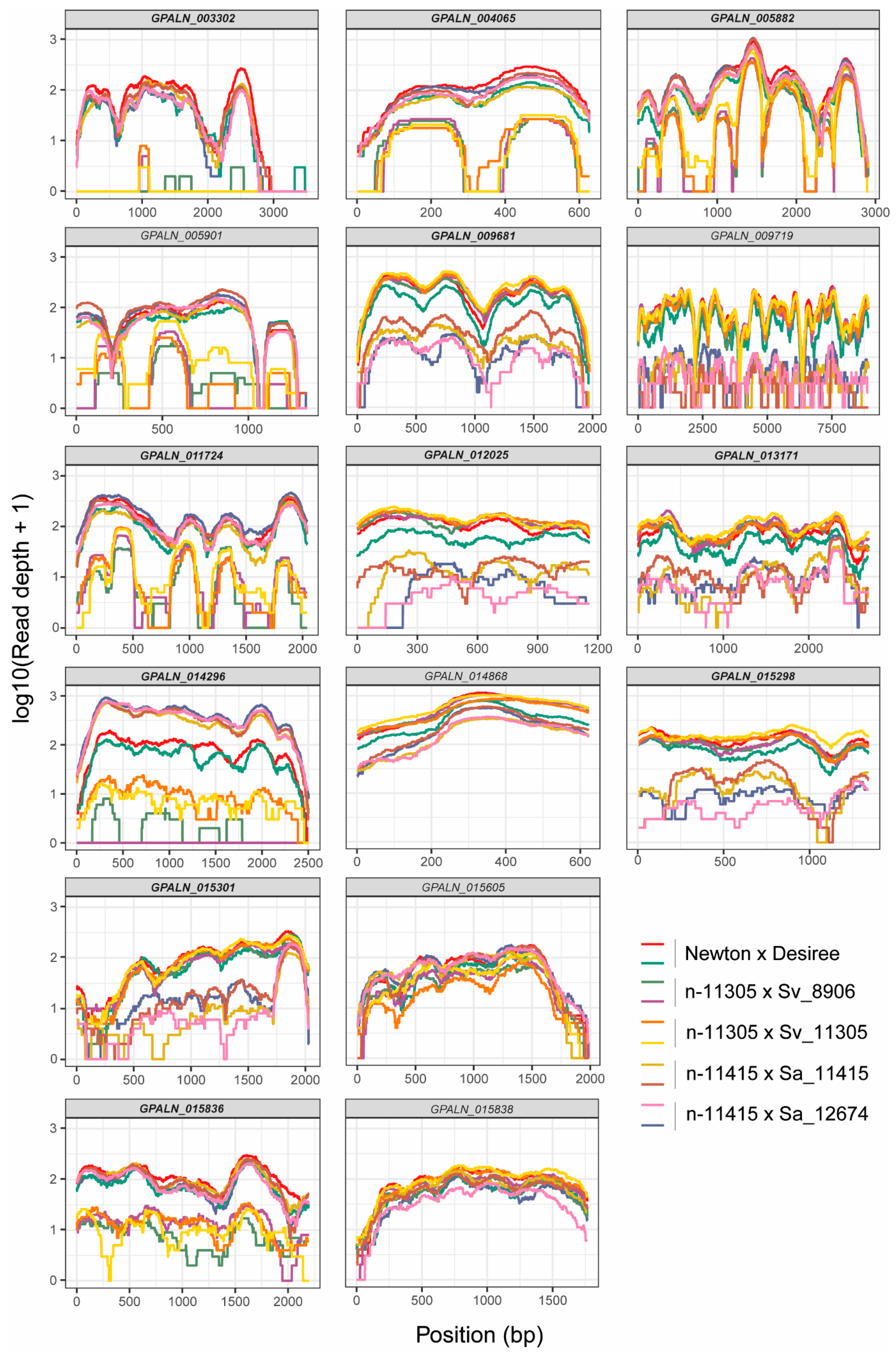
3.3. Post-Enriched Captured Libraries (PenSeq) Are Highly Enriched for Known Effectors
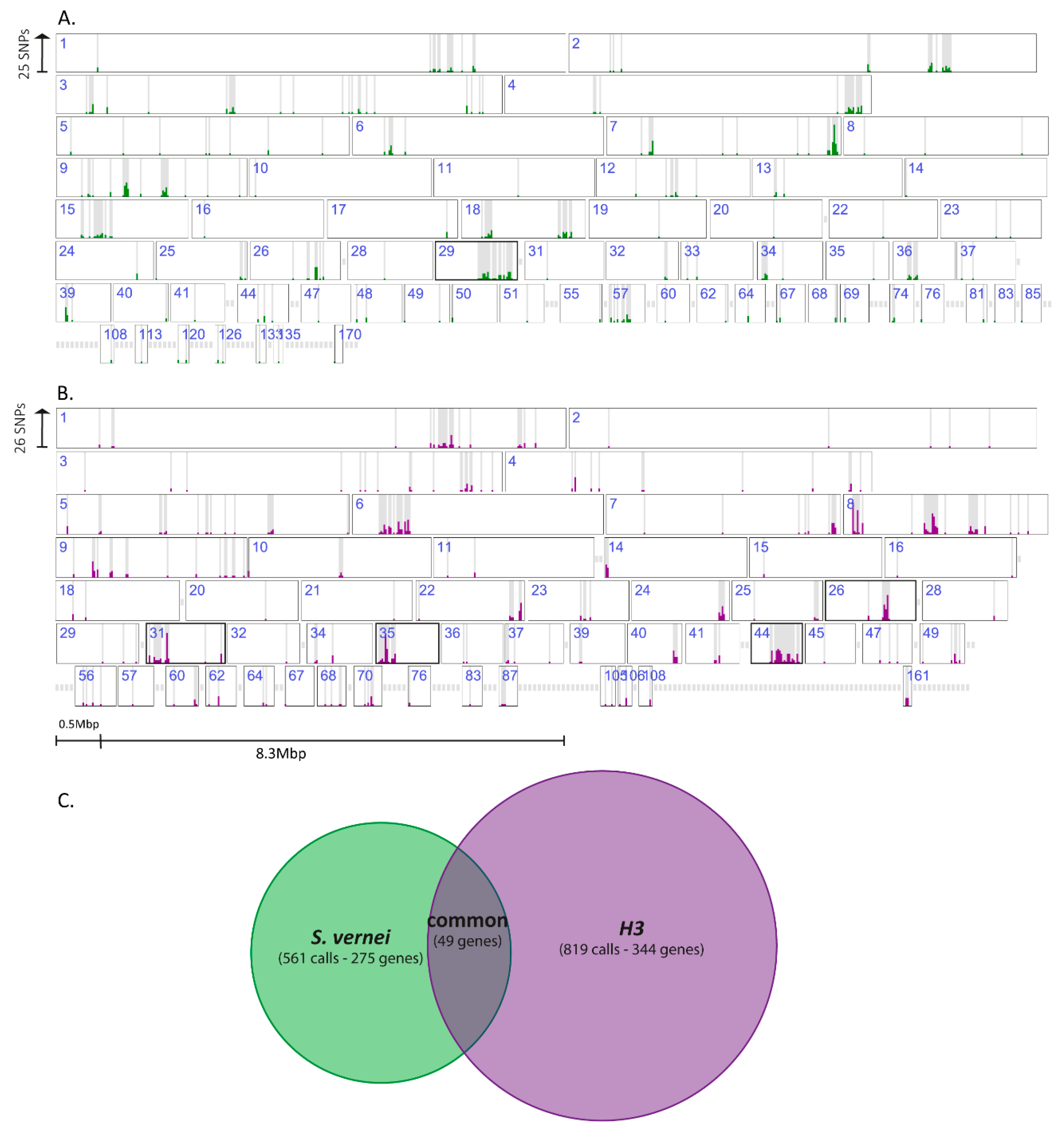
3.4. Genomic Analyses Allow Identification of High Confidence Variant Genes
3.5. Proteins Containing a SPRY Domain Were the Most Abundant Identified as Variants in the Selected Populations
3.6. Most of the Identified Variants in Both Genomic Approaches Were Associated with H3-Selected Populations
3.7. Variant Genes with Complete Drop of the Reference Allele Represent Potential Avr Genes
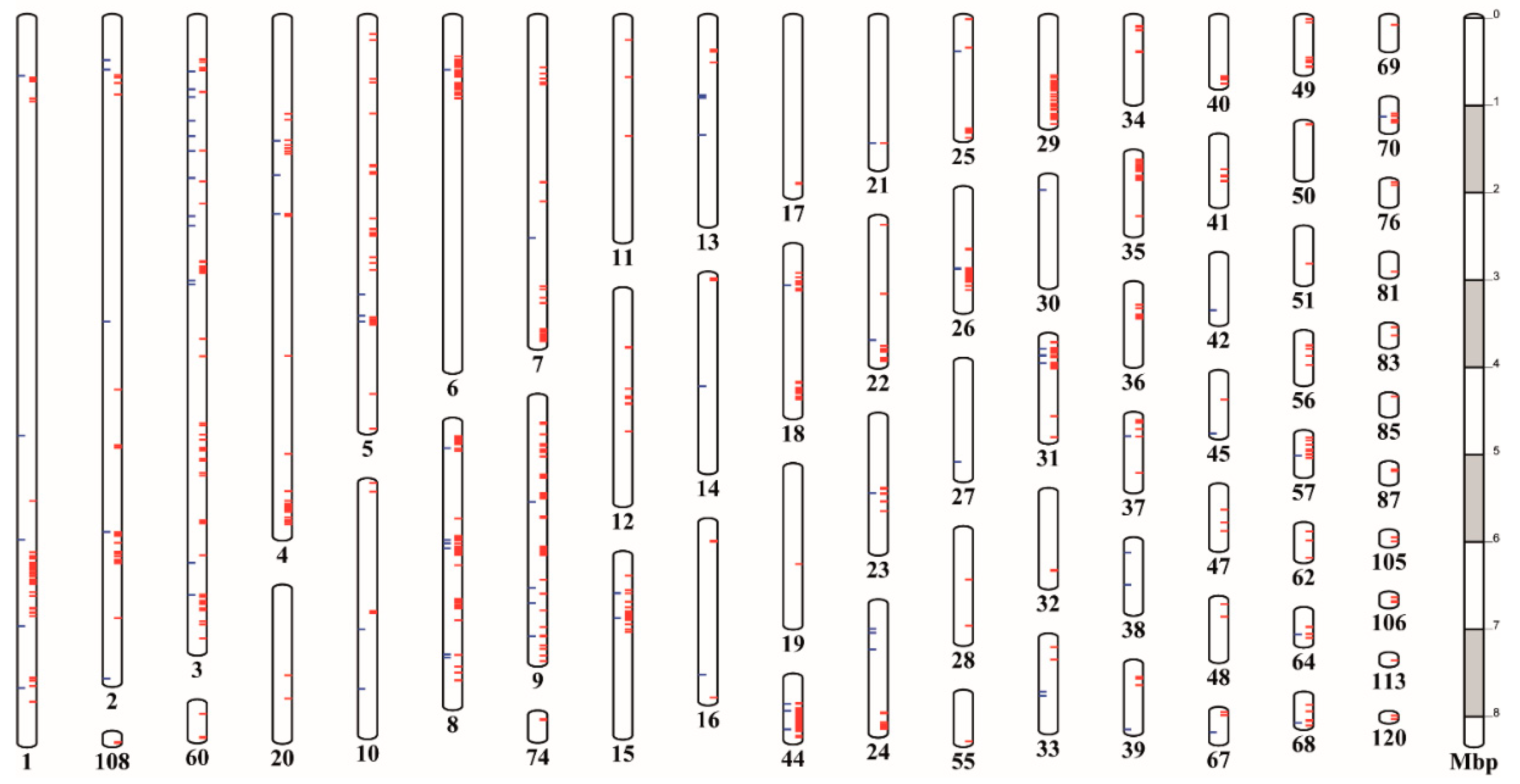
3.8. Variants Are Not Located Randomly in the G. pallida Genome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the Plant Immune System from Dissection to Deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nat. Cell Biol. 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Bent, A.F.; Mackey, D. Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 2007, 45, 399–436. [Google Scholar] [CrossRef]
- Sacco, M.A.; Koropacka, K.; Grenier, E.; Jaubert, M.J.; Blanchard, A.; Goverse, A.; Smant, G.; Moffett, P. The Cyst Nematode SPRYSEC Protein RBP-1 Elicits Gpa2- and RanGAP2-Dependent Plant Cell Death. PLoS Pathog. 2009, 5, e1000564. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.J.; Subbotin, S.A. Cyst nematodes. In Plant Nematology; Perry, R.N., Moens, M., Eds.; CAB International: London, UK, 2013. [Google Scholar]
- De Scurrah, M.M.; Evans, K.; Franco, J. Distribution of Species of Potato Cyst-Nematodes in South America. Nematology 1975, 21, 365–369. [Google Scholar] [CrossRef]
- Bakker, E.; Borm, T.; Prins, P.; Van Der Vossen, E.; Uenk, G.; Arens, M.; De Boer, J.; Van Eck, H.J.; Muskens, M.; Vossen, J.; et al. A genome-wide genetic map of NB-LRR disease resistance loci in potato. Theor. Appl. Genet. 2011, 123, 493–508. [Google Scholar] [CrossRef] [PubMed]
- Lilley, C.J.; Atkinson, H.J.; Urwin, P.E. Molecular aspects of cyst nematodes. Mol. Plant Pathol. 2005, 6, 577–588. [Google Scholar] [CrossRef]
- Dale, M.F.B.; Phillips, M.S. An investigation of resistance to the white potato cyst-nematode. J. Agric. Sci. 1982, 99, 325–328. [Google Scholar] [CrossRef]
- Phillips, M.S.; Blok, V.C. Selection for reproductive ability in Globodera pallida populations in relation to quantitative resistance from Solanum vernei and S. tuberosum ssp. andigena CPC2802. Plant Pathol. 2008, 57, 573–580. [Google Scholar] [CrossRef]
- Strachan, S.M.; Armstrong, M.R.; Kaur, A.; Wright, K.M.; Lim, T.Y.; Baker, K.; Jones, J.; Bryan, G.; Blok, V.; Hein, I. Mapping the H2 resistance effective against Globodera pallida pathotype Pa1 in tetraploid potato. Theor. Appl. Genet. 2019, 132, 1283–1294. [Google Scholar] [CrossRef]
- Paal, J.; Henselewski, H.; Muth, J.; Meksem, K.; Menéndez, C.M.; Salamini, F.; Ballvora, A.; Gebhardt, C. Molecular cloning of the potato Gro1-4 gene conferring resistance to pathotype Ro1 of the root cyst nematode Globodera rostochiensis, based on a candidate gene approach. Plant J. 2004, 38, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Adillah Tan, M.Y.; Park, T.-H.; Alles, R.; Hutten, R.C.B.; Visser, R.G.F.; van Eck, H.J. GpaXItarl originating from Solanum tarijense is a major resistance locus to Globodera pallida and is localised on chromosome 11 of potato. Theor. Appl. Genet. 2009, 119, 1477–1487. [Google Scholar] [CrossRef] [PubMed]
- Caromel, B.; Mugniery, D.; Kerlan, M.C.; Andrzejewski, S.; Palloix, A.; Ellisseche, D.; Rousselle-Bourgeois, F.; Lefebvre, V. Resistance quantitative trait loci originating from Solanum sparsipilum act independently on the sex ration of Globodera pallida and together for developing a necrotic reaction. MPMI 2005, 18, 1186–1194. [Google Scholar] [CrossRef] [PubMed]
- Plantard, O.; Picard, D.; Valette, S.; Scurrah, M.; Grenier, E.; Mugniéry, D. Origin and genetic diversity of Western European populations of the potato cyst nematode (Globodera pallida) inferred from mitochondrial sequences and microsatellite loci. Mol. Ecol. 2008, 17, 2208–2218. [Google Scholar] [CrossRef]
- Picard, D.; Sempere, T.; Plantard, O. A northward colonisation of the Andes by the potato cyst nematode during geological times suggests multiple host-shifts from wild to cultivated potatoes. Mol. Phylogenetics Evol. 2007, 42, 308–316. [Google Scholar] [CrossRef]
- Akker, S.E.-V.D.; Lilley, C.J.; Jones, J.T.; Urwin, P.E. Plant-parasitic nematode feeding tubes and plugs: New perspectives on function. Nematology 2015, 17, 1–9. [Google Scholar] [CrossRef]
- Sobczak, M.; Golinowski, W. Cyst Nematodes and Syncytia. In Genomics and Molecular Genetics of Plant-Nematode Interactions; Jones, J.T., Gheysen, G., Fenoll, C., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 61–82. [Google Scholar]
- Bleve-Zacheo, T.; Melillo, M.T.; Zacheo, G. Ultrastructural response of potato roots resistant to cyst nematode Globodera rostochiensis pathotype Ro1. Rev. Nematol. 1990, 13, 29–36. [Google Scholar]
- Rice, S.L.; Leadbeater, B.; Stone, A. Changes in cell structure in roots of resistant potatoes parasitized by potato cyst-nematodes. I. Potatoes with resistance gene H1 derived from Solanum tuberosum ssp. andigena. Physiol. Plant Pathol. 1985, 27, 219–234. [Google Scholar] [CrossRef]
- Turner, S.J.; Fleming, C.C. Multiple selection of potato cyst nematode Globodera pallida virulence on a range of potato species. I.Serial selection on Solanum-hybrids. Eur. J. Plant Pathol. 2002, 108, 461–467. [Google Scholar] [CrossRef]
- Eoche-Bosy, D.; Gauthier, J.; Juhel, A.S.; Esquibet, M.; Fournet, S.; Grenier, E.; Montarry, J. Experimentally evolved populations of the potato cyst nematode Globodera pallida allow the targeting of genomic footprints of selection due to host adaptation. Plant Pathol. 2016, 66, 1022–1030. [Google Scholar] [CrossRef]
- Eoche-Bosy, D.; Gautier, M.; Esquibet, M.; Legeai, F.; Bretaudeau, A.; Bouchez, O.; Fournet, S.; Grenier, E.; Montarry, J. Genome scans on experimentally evolved populations reveal candidate regions for adaptation to plant resistance in the potato cyst nematode Globodera pallida. Mol. Ecol. 2017, 26, 4700–4711. [Google Scholar] [CrossRef] [PubMed]
- Luikart, G.; England, P.R.; Tallmon, D.A.; Jordan, S.; Taberlet, P. The power and promise of population genomics: From genotyping to genome typing. Nat. Rev. Genet. 2003, 4, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Beniers, A.; Mulder, A.; Schouten, H. Selection for virulence of Globodera pallida by potato cultivars. Fundam. Appl. Nematol. 1995, 18, 497–500. [Google Scholar]
- Fournet, S.; Eoche-Bosy, D.; Renault, L.; Hamelin, F.M.; Montarry, J. Adaptation to resistant hosts increases fitness on susceptible hosts in the plant parasitic nematode Globodera pallida. Ecol. Evol. 2016, 6, 2559–2568. [Google Scholar] [CrossRef]
- Varypatakis, K.; Jones, J.T.; Blok, V.C. Susceptibility of potato varieties to populations of Globodera pallida selected for increased virulence. Nematology 2019, 21, 995–998. [Google Scholar] [CrossRef]
- Gao, B.; Allen, R.; Maier, T.; Davis, E.L.; Baum, T.; Hussey, S. The parasitome of the phytonematde Heterodera glycines. MPMI 2003, 16, 720–726. [Google Scholar] [CrossRef]
- Jones, J.T.; Kumar, A.; Pylypenko, L.A.; Thirugnanasambandam, A.; Castelli, L.; Chapman, S.; Cock, P.J.A.; Grenier, E.; Lilley, C.J.; Phillips, M.S.; et al. Identification and functional characterization of effectors in expressed sequence tags from various life cycle stages of the potato cyst nematode Globodera pallida. Mol. Plant Pathol. 2009, 10, 815–828. [Google Scholar] [CrossRef]
- Cotton, J.A.; Lilley, C.J.; Jones, L.M.; Kikuchi, T.; Reid, A.J.; Thorpe, P.; Tsai, I.J.; Beasley, H.; Blok, V.; Cock, P.; et al. The genome and life-stage specific transcriptomes of Globodera pallida elucidate key aspects of plant parasitism by a cyst nematode. Genome Biol. 2014, 15, R43. [Google Scholar] [CrossRef]
- Akker, S.E.-V.D.; Laetsch, D.R.; Thorpe, P.; Lilley, C.J.; Danchin, E.G.J.; Da Rocha, M.; Rancurel, C.; Holroyd, N.E.; Cotton, J.A.; Szitenberg, A.; et al. The genome of the yellow potato cyst nematode, Globodera rostochiensis, reveals insights into the basis of parasitism and virulence. Genome Biol. 2016, 17, 1–23. [Google Scholar] [CrossRef]
- Thorpe, P.; Mantelin, S.; Cock, P.J.; Blok, V.; Coke, M.C.; Eves-van den Akker, S.; Guzeeva, E.; Lilley, C.J.; Smant, G.; Reid, A.J.; et al. Genomic characterization of the effector complement of the potato cyst nematode Globodera pallida. BMC Genom. 2014, 15, 923. [Google Scholar] [CrossRef]
- Kort, J.; Ross, H.; Stone, A.; Rumpenhorst, H. An international scheme for identifying and classifying pathotypes of potato cyst-nematodes Globodera rostochiensis and G. pallida. Nematology 1977, 23, 333–339. [Google Scholar] [CrossRef]
- Thilliez, G.; Armstrong, M.; Lim, T.; Baker, K.; Jouet, A.; Ward, B.; Van Oosterhout, C.; Jones, J.D.G.; Huitema, E.; Birch, P.R.J.; et al. Pathogen enrichment sequencing (PenSeq) enables population genomic studies in oomycetes. New Phytol. 2019, 221, 1634–1648. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, J.E.; Ramsay, G. Utilisation of the Commonwealth Potato Collection in potato breeding. Euphytica 2005, 146, 9–19. [Google Scholar] [CrossRef]
- Sobczak, M.; Avrova, A.; Jupowicz, J.; Phillips, M.S.; Ernst, K.; Kumar, A. Characterization of susceptibility and resistance responses to potato cyst nematode (Globodera spp.) infection of tomato lines in the absence and presence of the broad-spectrum nematode resistance Hero gene. Mol. Plant-Microbe Int. 2005, 18, 158–168. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinform 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Thorpe, P.; Varypatakis, K.; Cock, P.J.; Blok, V.C.; Jones, J.T.; Eves-van den Akker, S. Long read assembly of highly heterozygous Globodera pallida population. Unpublished, Paper in preparation.
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Sembdner, G. Anatomie Der Heterodera-Rostochiensis-Gallen an Tomatenwurzeln 1). Nematology 1963, 9, 55–64. [Google Scholar] [CrossRef]
- Mei, Y.; Thorpe, P.; Guzha, A.; Haegeman, A.; Blok, V.C.; MacKenzie, K.; Gheysen, G.; Jones, J.T.; Mantelin, S. Only a small subset of the SPRY domain gene family in Globodera pallida is likely to encode effectors, two of which suppress host defences induced by the potato resistance gene Gpa2. Nematology 2015, 17, 409–424. [Google Scholar] [CrossRef]
- Haegeman, A.; Mantelin, S.; Jones, J.T.; Gheysen, G. Functional roles of effectors of plant-parasitic nematodes. Gene 2012, 492, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Rice, S.L.; Stone, A.; Leadbeater, B. Changes in cell structure in roots of resistant potatoes parasitized by potato cyst nematodes. 2. Potatoes with resistance derived from Solanum vernei. Physiol. Mol. Plant Pathol. 1987, 31, 1–14. [Google Scholar] [CrossRef]
- Jones, M.G.; Northcote, D.H. Nematode-induced syncytium—A multinucleate transfer cell. J. Cell Sci. 1972, 10, 789–809. [Google Scholar] [PubMed]
- Melillo, M.T.; Bleve-Zacheo, T.; Zacheo, G. Ultrastructural response of potato roots susceptible to cyst nematode Globodera pallida pathotype Pa3. Revue Nematol. 1990, 13, 17–28. [Google Scholar]
- Castelli, L.; Bryan, G.; Blok, V.G.; Ramsay, G.; Sobczak, M.; Gillespie, T.; Phillips, M.S. nvestigation of Globodera pallida invasion and syncytia formation within roots of the susceptible potato cultivar Desiree and resistant species Solanum canasense. Nematology 2006, 8, 103–110. [Google Scholar] [CrossRef]
- Fournet, S.; Kerlan, M.-C.; Renault, L.; Dantec, J.-P.; Rouaux, C.; Montarry, J. Selection of nematodes by resistant plants has implications for local adaptation and cross-virulence. Plant Pathol. 2012, 62, 184–193. [Google Scholar] [CrossRef]
- Bryan, G.; McLean, K.; Bradshaw, J.; De Jong, W.; Phillips, M.; Castelli, L.; Waugh, R. Mapping QTLs for resistance to the cyst nematode Globodera pallida derived from the wild potato species Solanum vernei. Theor. Appl. Genet. 2002, 105, 68–77. [Google Scholar] [CrossRef]
- Bryan, G.J.; McLean, K.; Pande, B.; Purvis, A.; Hackett, C.A.; Bradshaw, J.E.; Waugh, R. Genetical dissection of H3-mediated polygenic PCN resistance in a heterozygous autotetraploid potato population. Mol. Breed. 2004, 14, 105–116. [Google Scholar] [CrossRef]
- Van Der Voort, J.R.; Van Der Vossen, E.; Bakker, E.; Overmars, H.; Van Zandvoort, P.; Hutten, R.; Lankhorst, R.K.; Bakker, J. Two additive QTLs conferring broad-spectrum resistance in potato to Globodera pallida are localized on resistance gene clusters. Theor. Appl. Genet. 2000, 101, 1122–1130. [Google Scholar] [CrossRef]
- Davis, E.L.; Hussey, R.S.; Mitchum, M.G.; Baum, T.J. Parasitism proteins in nematode–plant interactions. Curr. Opin. Plant Biol. 2008, 11, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Gheysen, G.; Mitchum, M.G. How nematodes manipulate plant development pathways for infection. Curr. Opin. Plant Biol. 2011, 14, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Lilley, C.J.; Maqbool, A.; Wu, D.; Yusup, H.B.; Jones, L.M.; Birch, P.R.J.; Banfield, M.J.; Urwin, P.E.; Akker, S.E.-V.D. Effector gene birth in plant parasitic nematodes: Neofunctionalization of a housekeeping glutathione synthetase gene. PLoS Genet. 2018, 14, e1007310. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Granados, A.; Petrescu, A.-J.; Goverse, A.; Smant, G. SPRYSEC Effectors: A Versatile Protein-Binding Platform to Disrupt Plant Innate Immunity. Front. Plant Sci. 2016, 7, 1575. [Google Scholar] [CrossRef] [PubMed]
- Perfetto, L.; Gherardini, P.F.; Davey, N.E.; Diella, F.; Helmer-Citterich, M.; Cesareni, G. Exploring the diversity of SPRY/B30.2-mediated interactions. Trends Biochem. Sci. 2013, 38, 38–46. [Google Scholar] [CrossRef]
- Rehman, S.; Butterbach, P.; Popeijus, H.; Overmars, H.; Davis, E.L.; Jones, J.T.; Goverse, A.; Bakker, J.; Smant, G. Identification and Characterization of the Most Abundant Cellulases in Stylet Secretions from Globodera rostochiensis. Phytopathology 2009, 99, 194–202. [Google Scholar] [CrossRef]
- Carpentier, J.; Esquibet, M.; Fouville, D.; Manzanares-Dauleux, M.J.; Kerlan, M.; Grenier, E. he evolution of the Gp-Rbp-1 gene in Globodera pallida includes multiple selective replacements. Mol. Plant Pathol. 2011, 13, 546–555. [Google Scholar] [CrossRef]
- Zhou, C.; Luo, J.; He, X.; Zhou, Q.; He, Y.; Wang, X.; Ma, L. The NALCN channel regulator UNC-80 functions in a subset of interneurons to regulate Caenorhabditis elegans reversal behavior. Genes Genomes Genet. 2019, 10, 199–210. [Google Scholar] [CrossRef]
- Palomares-Rius, J.E.; Hedley, P.E.; Cock, P.J.A.; Morris, J.A.; Jones, J.T.; Blok, V.C. Gene expression changes in diapause or quiescent potato cyst nematode, Globodera pallida, eggs after hydration or exposure to tomato root diffusate. PeerJ 2016, 4, e1654. [Google Scholar] [CrossRef]
- Palomares-Rius, J.E.; Hedley, P.E.; Cock, P.J.; Morris, J.A.; Jones, J.T.; Vovlas, N.; Blok, V. Comparison of transcript profiles in different life stages of the nematode Globodera pallida under different host potato genotypes. Mol. Plant Pathol. 2012, 13, 1120–1134. [Google Scholar] [CrossRef] [PubMed]
| “Désirée” | Sv_8906 | Sv_11305 | Sa_11415 | Sa_12674 | |
|---|---|---|---|---|---|
| Newton | 141.0 ± 30.3 a | 23.3 ± 0.7 b | 23.5 ± 3.8 b | 28.4 ± 6.2 b | 13.1 ± 3.3 b |
| n-11305 | 150.4 ± 18.9 a | 74.5 ± 5.0 c | 107.0 ± 10.5 b | 43.5 ± 2.3 c | 4.9 ± 0.3 d |
| n-11415 | 84.4 ± 13.2 a | 5.1 ± 3.4 c | 9.9 ± 2.5 c | 109.0 ± 6.2 a | 34.6 ± 10.0 b |
| Common Genes | Description | Similarity to Gene Annotated in [27] |
|---|---|---|
| GPALN_004071 | Unknown function | GPLIN_001153200 |
| GPALN_004129 | TWiK family of potassium channels | GPLIN_001292400 |
| GPALN_015295 | SPRY domain protein | GPLIN_000507800 |
| GPALN_007439 | SPRY domain protein | GPLIN_000800200 |
| GPALN_009681 | SPRY domain protein | GPLIN_000909700 |
| GPALN_009697 | RNA polymerase II subunit A C-terminal domain phosphatase SSU72 | GPLIN_001349800 |
| GPALN_012007 | SPRY domain protein | GPLIN_000437400 |
| GPALN_012018 | SPRY domain protein | GPLIN_000413600 |
| GPALN_013052 | Protein unc-80 | GPLIN_000425400 |
| GPALN_013436 | SPRY domain protein | GPLIN_000433800 |
| GPALN_015286 | Unknown function | GPLIN_000807100 |
| GPALN_010968 * | BTB/POZ domain-containing protein 3 | GPLIN_000507800 |
| GPALN_006664 * | SPRY domain protein | GPLIN_000716900 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varypatakis, K.; Véronneau, P.-Y.; Thorpe, P.; Cock, P.J.A.; Lim, J.T.-Y.; Armstrong, M.R.; Janakowski, S.; Sobczak, M.; Hein, I.; Mimee, B.; et al. The Genomic Impact of Selection for Virulence against Resistance in the Potato Cyst Nematode, Globodera pallida. Genes 2020, 11, 1429. https://doi.org/10.3390/genes11121429
Varypatakis K, Véronneau P-Y, Thorpe P, Cock PJA, Lim JT-Y, Armstrong MR, Janakowski S, Sobczak M, Hein I, Mimee B, et al. The Genomic Impact of Selection for Virulence against Resistance in the Potato Cyst Nematode, Globodera pallida. Genes. 2020; 11(12):1429. https://doi.org/10.3390/genes11121429
Chicago/Turabian StyleVarypatakis, Kyriakos, Pierre-Yves Véronneau, Peter Thorpe, Peter J. A. Cock, Joanne Tze-Yin Lim, Miles R. Armstrong, Sławomir Janakowski, Mirosław Sobczak, Ingo Hein, Benjamin Mimee, and et al. 2020. "The Genomic Impact of Selection for Virulence against Resistance in the Potato Cyst Nematode, Globodera pallida" Genes 11, no. 12: 1429. https://doi.org/10.3390/genes11121429
APA StyleVarypatakis, K., Véronneau, P.-Y., Thorpe, P., Cock, P. J. A., Lim, J. T.-Y., Armstrong, M. R., Janakowski, S., Sobczak, M., Hein, I., Mimee, B., Jones, J. T., & Blok, V. C. (2020). The Genomic Impact of Selection for Virulence against Resistance in the Potato Cyst Nematode, Globodera pallida. Genes, 11(12), 1429. https://doi.org/10.3390/genes11121429





