Molecular Mechanism of Slow Vegetative Growth in Populus Tetraploid
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Measurement of Plant Growth Rate, Leaf Greenness, Leaf Area, and Photosynthesis
2.3. Transcriptome Analysis
2.4. RT-PCR Validation of Differentially Expressed Genes (DEGs)
2.5. Gene Ontology (GO) Analysis
2.6. Phytohormone Analysis
2.7. Measurements of Chlorophyll, Starch, and Sugar Contents
2.8. Enzyme Assay
2.9. Analysis of Mesophyll Cell Suspensions to Count Chloroplasts
2.10. Transmission Electron Microscopy (TEM) Observations of Chloroplasts
2.11. Statistical Analysis
3. Results
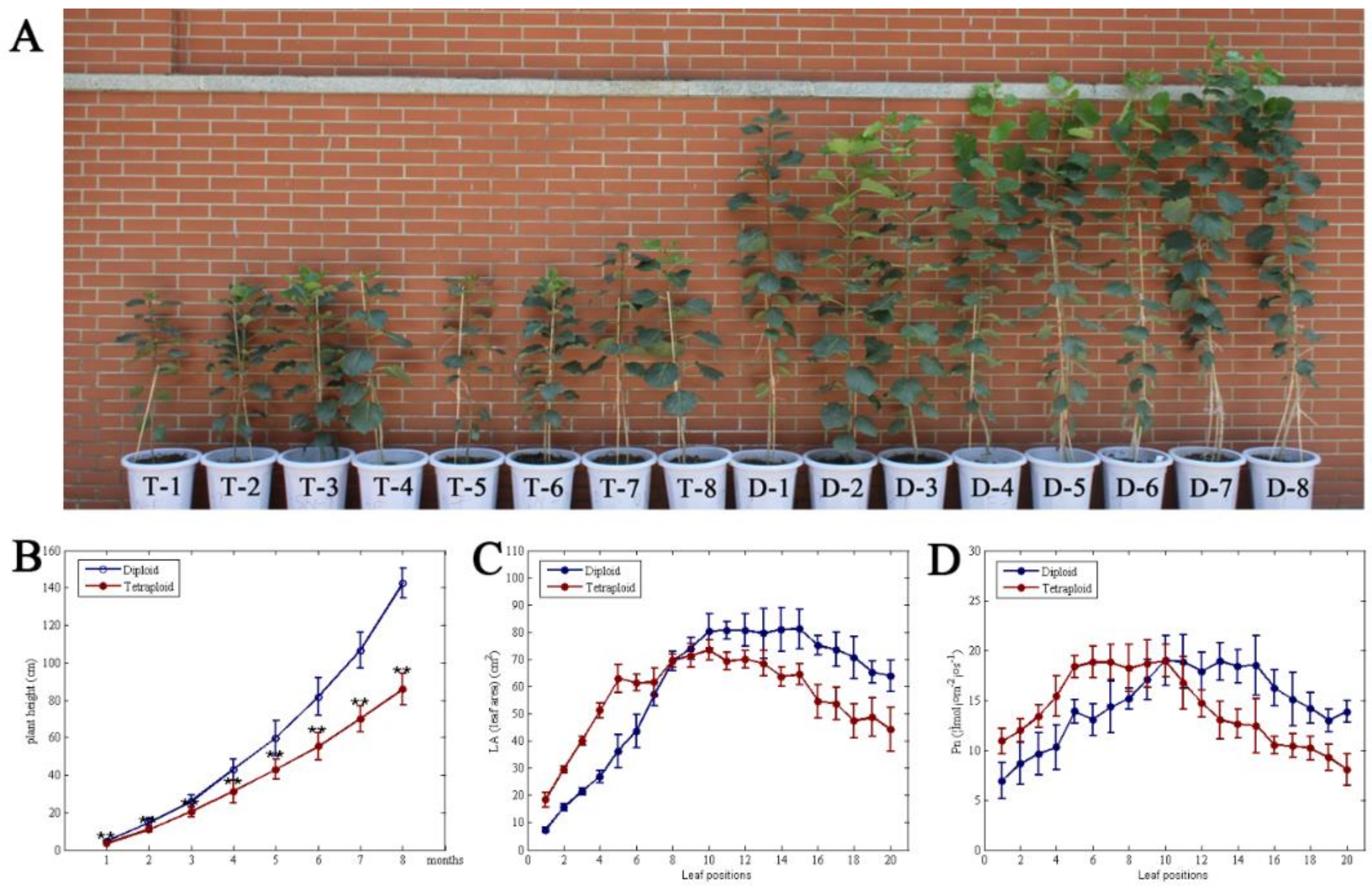
3.1. Comparative Analysis of Plant Height, Leaf Area, and Photosynthetic Rate between Tetraploid and Diploid Plants
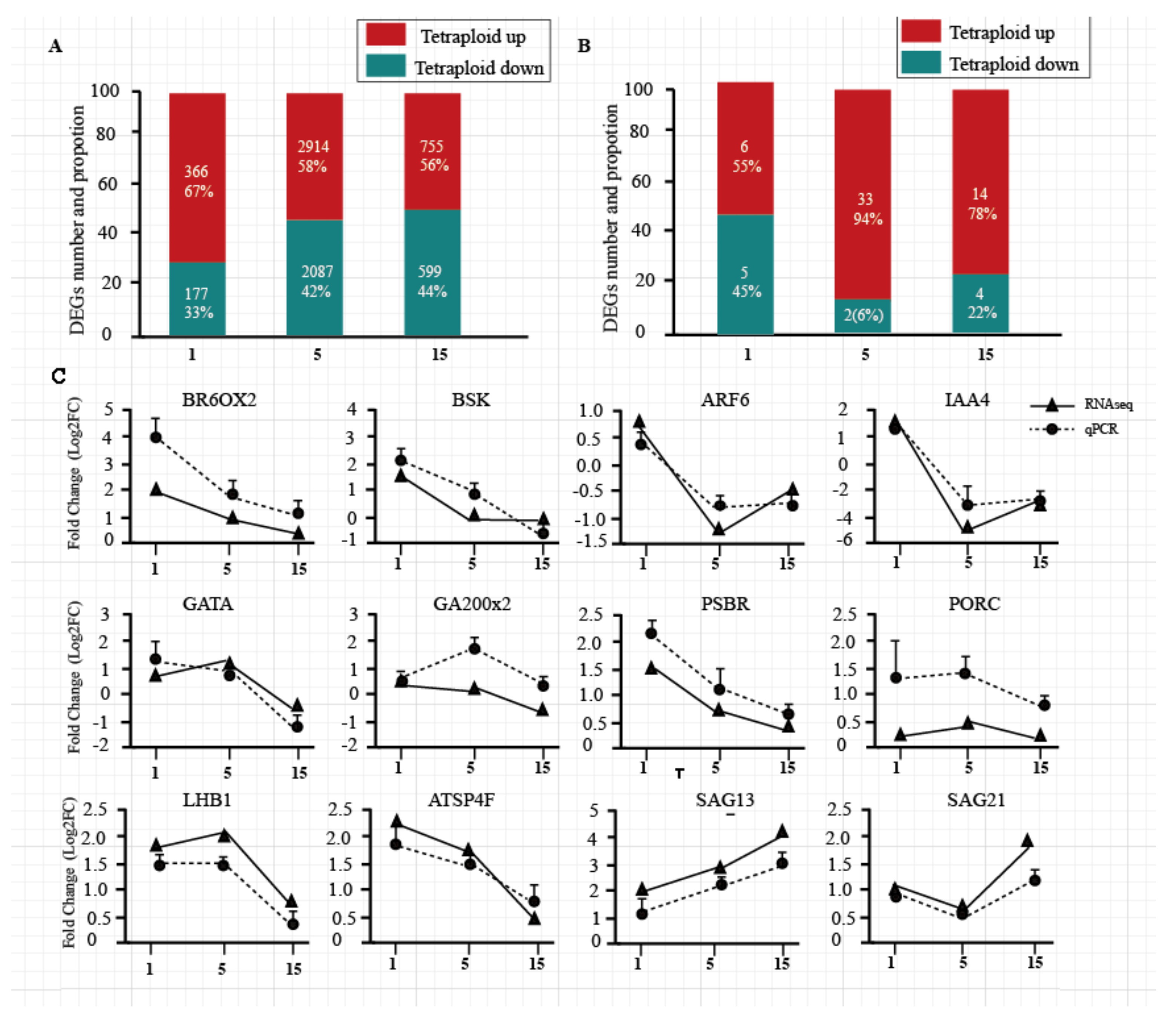
3.2. Verification of DEG Analysis Results by RT-PCR
3.3. DEGs Associated with Hormone Synthesis and Signal Transduction in Tetraploids and Diploids
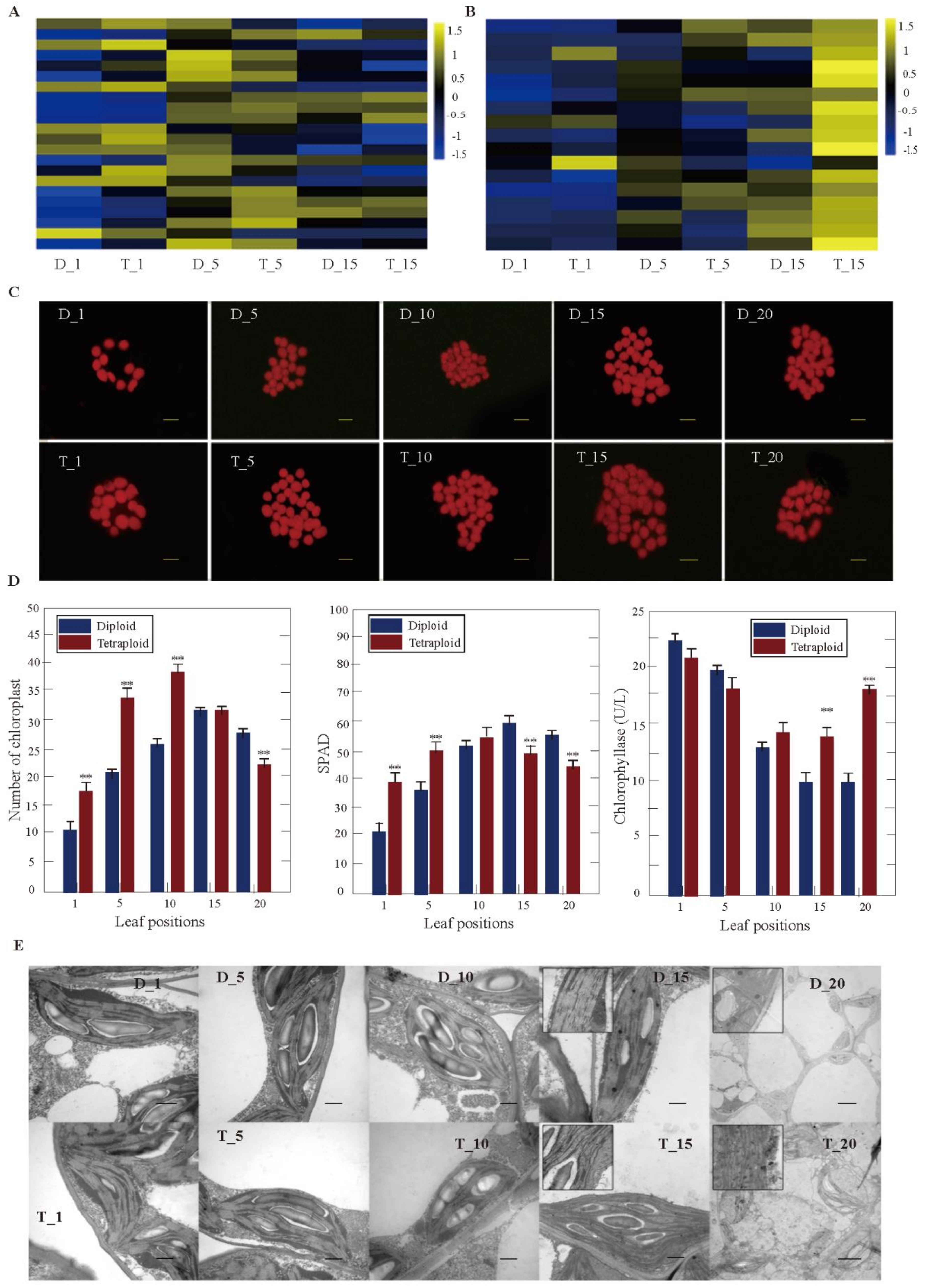
3.4. DEGs Related to Chlorophyll Synthesis and Decomposition between Tetraploids and Diploids
3.5. DEGs Related to Photosynthesis between Tetraploids and Diploids
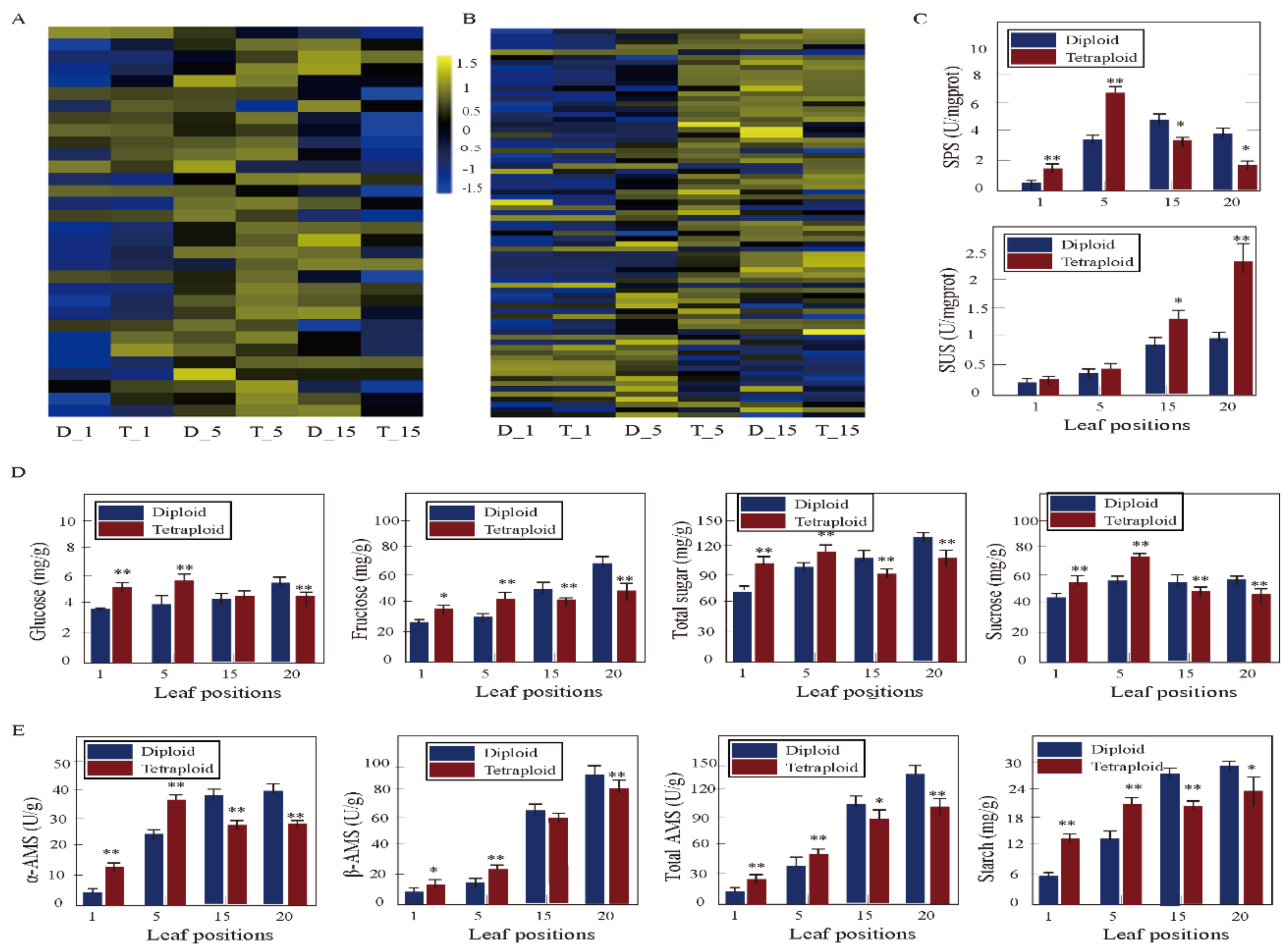
3.6. DEGs Associated with Sucrose and Starch Metabolism between Tetraploids and Diploids
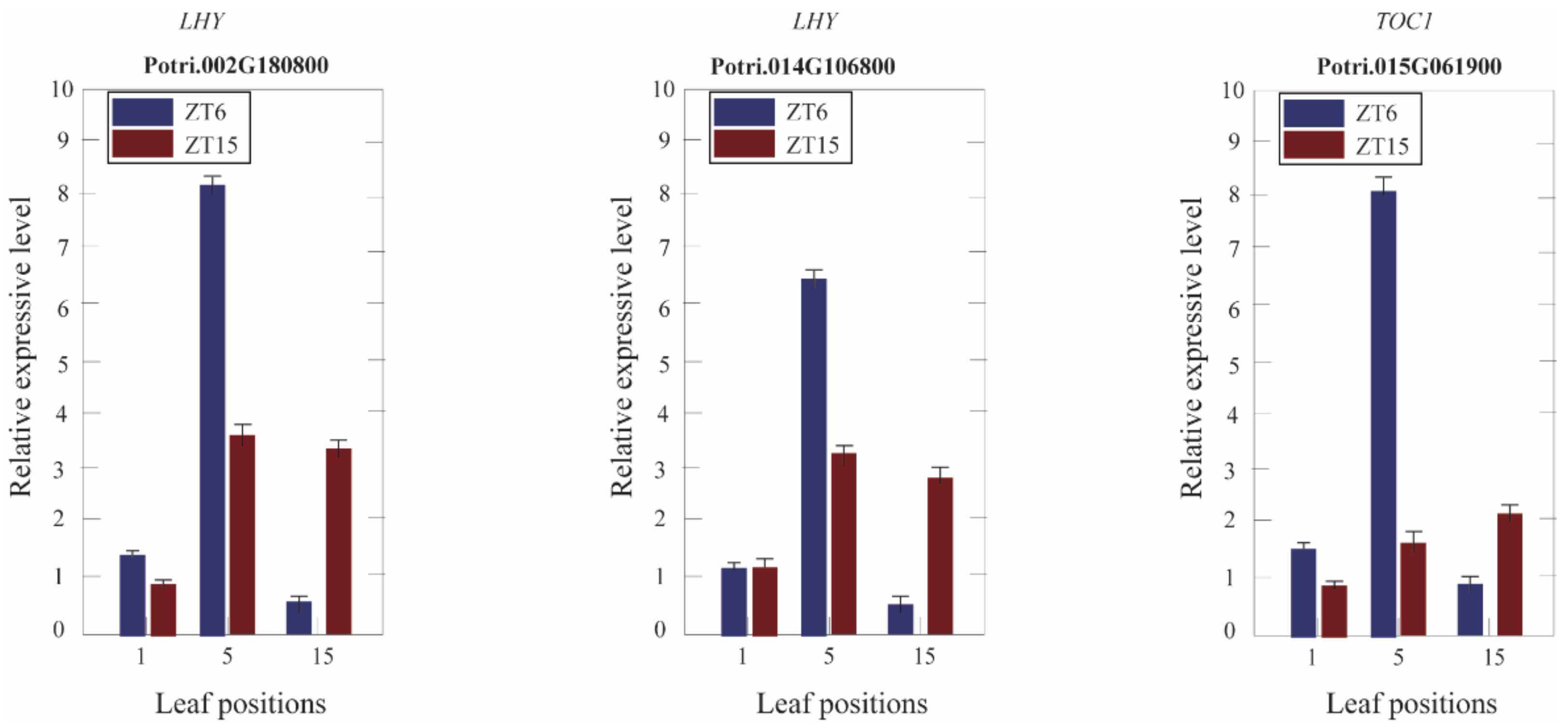
3.7. Expression of Circadian Clock Genes in Different Leaf Canopy Positions
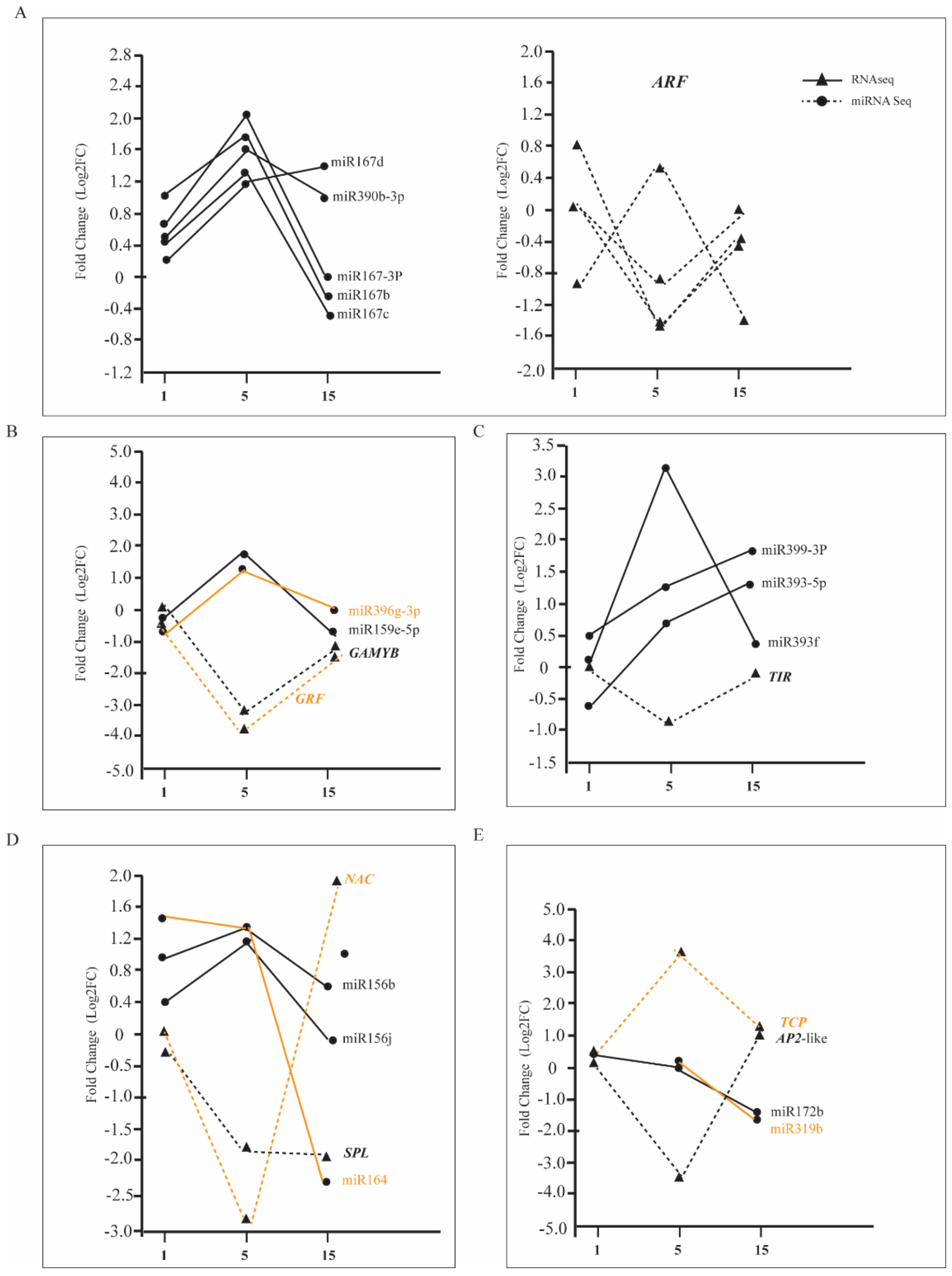
3.8. Differential Expression of miRNA between Tetraploids and Diploids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Masterson, J. Stomatal size in fossil plants: Evidence for polyploidy in majority of angiosperms. Science 1994, 264, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Comai, L. The advantages and disadvantages of being polyploid. Nature Rev. Genet. 2005, 6, 836. [Google Scholar] [CrossRef] [PubMed]
- Mayrose, I.; Zhan, S.H.; Rothfels, C.J.; Magnuson-Ford, K.; Barker, M.S.; Rieseberg, L.H.; Otto, S.P. Recently formed polyploid plants diversify at lower rates. Science 2011, 333, 1257. [Google Scholar] [CrossRef]
- Adams, K.L.; Wendel, J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005, 8, 135–141. [Google Scholar] [CrossRef]
- Chen, Z.J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 2007, 58, 377–406. [Google Scholar] [CrossRef]
- Ni, Z.; Kim, E.D.; Ha, M.; Lackey, E.; Liu, J.; Zhang, Y.; Chen, Z.J. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 2009, 457, 327. [Google Scholar] [CrossRef]
- Allario, T.; Brumos, J.; Colmenero-Flores, J.M.; Tadeo, F.; Froelicher, Y.; Talon, M.; Morillon, R. Large changes in anatomy and physiology between diploid Rangpur lime (Citrus limonia) and its autotetraploid are not associated with large changes in leaf gene expression. J. Exp. Bot. 2011, 62, 2507–2519. [Google Scholar] [CrossRef]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Dai, F.; Wang, Z.; Luo, G.; Tang, C. Phenotypic and transcriptomic analyses of autotetraploid and diploid mulberry (Morus alba L.). Int. J. Mol. Sci. 2015, 16, 22938–22956. [Google Scholar] [CrossRef]
- Kondorosi, E.; Roudier, F.; Gendreau, E. Plant cell-size control: Growing by ploidy? Curr. Opin. Plant Biol. 2000, 3, 488–492. [Google Scholar] [CrossRef]
- Vyas, P.; Bisht, M.S.; Miyazawa, S.I.; Yano, S.; Noguchi, K.; Terashima, I.; Funayama-Noguchi, S. Effects of polyploidy on photosynthetic properties and anatomy in leaves of Phlox drummondii. Funct. Plant Biol. 2007, 34, 673–682. [Google Scholar] [CrossRef]
- Miller, M.; Zhang, C.; Chen, Z.J. Ploidy and hybridity effects on growth vigor and gene expression in Arabidopsis thaliana hybrids and their parents. G3 Genes Genomes Genet. 2012, 2, 505–513. [Google Scholar] [CrossRef]
- Austin, R.B.; Morgan, C.L.; Ford, M.A.; Bhagwat, S.G. Flag leaf photosynthesis of Triticum aestivum and related diploid and tetraploid species. Annu. Bot. 1982, 49, 177–189. [Google Scholar] [CrossRef]
- Stupar, R.M.; Bhaskar, P.B.; Yandell, B.S.; Rensink, W.A.; Hart, A.L.; Ouyang, S.; Jiang, J. Phenotypic and transcriptomic changes associated with potato autopolyploidization. Genetics 2007, 176, 2055–2067. [Google Scholar] [CrossRef]
- Mu, H.Z.; Liu, Z.J.; Lin, L.; Li, H.Y.; Jiang, J.; Liu, G.F. Transcriptomic analysis of phenotypic changes in birch (Betula platyphylla) autotetraploids. Int. J. Mol. Sci. 2012, 13, 13012–13029. [Google Scholar] [CrossRef]
- Lange, T.; Kappler, J.; Fischer, A.; Frisse, A.; Padeffke, T.; Schmidtke, S.; Lange, M.J. Gibberellin biosynthesis in developing pumpkin seedlings. Plant Physiol. 2005, 139, 213–223. [Google Scholar] [CrossRef]
- Moon, J.; Zhu, L.; Shen, H.; Huq, E. PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 9433–9438. [Google Scholar] [CrossRef]
- Chandler, P.M.; Harding, C.A.; Ashton, A.R.; Mulcair, M.D.; Dixon, N.E.; Mander, L.N. Characterization of gibberellin receptor mutants of barley (Hordeum vulgare L.). Mol. Plant 2008, 1, 285–294. [Google Scholar] [CrossRef]
- Shi, H.; Reiter, R.J.; Tan, D.X.; Chan, Z. INDOLE-3-ACETIC ACID INDUCIBLE 17 positively modulates natural leaf senescence through melatonin-mediated pathway in Arabidopsis. J. Pineal Res. 2015, 58, 26–33. [Google Scholar] [CrossRef]
- Du, H.; Wu, N.; Fu, J.; Wang, S.; Li, X.; Xiao, J.; Xiong, L. A GH3 family member; OsGH3-2.; modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 2012, 63, 6467–6480. [Google Scholar] [CrossRef]
- Tabata, R.; Ikezaki, M.; Fujibe, T.; Aida, M.; Tian, C.E.; Ueno, Y.; Yamamoto, K.T.; Machida, Y.; Nakamura, K.; Ishiguro, S. Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 2010, 51, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef] [PubMed]
- Ru, P.; Xu, L.; Ma, H.; Huang, H. Plant fertility defects induced by the enhanced expression of microRNA167. Cell Res. 2006, 16, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.G.; Gao, Z.; Zhao, Z.; Liu, X.; Li, Y.; Zhang, Y.; Liu, X.; Sun, Y.; Tang, W. BZR1 Family Transcription Factors Function Redundantly and Indispensably in BR Signaling but Exhibit BRI1-Independent Function in Regulating Anther Development in Arabidopsis. Mol Plant. 2019, 12, 1408–1415. [Google Scholar] [CrossRef]
- Liu, J.; Yang, R.; Jian, N.; Wei, L.; Ye, L.; Wang, R.; Gao, H.; Zheng, Q. Putrescine metabolism modulates the biphasic effects of brassinosteroids on canola and Arabidopsis salt tolerance. Plant Cell Environ. 2020, 43, 1348–1359. [Google Scholar] [CrossRef]
- Bollivar, D.W. Recent advances in chlorophyll biosynthesis. Photosynth. Res. 2007, 90, 173–194. [Google Scholar] [CrossRef]
- Liu, J.; Yang, H.; Lu, Q.; Wen, X.; Chen, F.; Peng, L.; Zhang, L.; Lu, C. PSBP-DOMAIN PROTEIN1, a Nuclear-Encoded Thylakoid Lumenal Protein, Is Essential for Photosystem I Assembly in Arabidopsis. Plant Cell 2012, 24, 4992–5006. [Google Scholar] [CrossRef]
- Allahverdiyeva, Y.; Suorsa, M.; Rossi, F.; Pavesi, A.; Kater, M.M.; Antonacci, A.; Tadini, L.; Pribil, M.; Schneider, A.; Wanner, G.; et al. Arabidopsis plants lacking PsbQ and PsbR subunits of the oxygen-evolving complex show altered PSII super-complex organization and short-term adaptive mechanisms. Plant J. 2013, 75, 671–684. [Google Scholar] [CrossRef]
- Szydlowski, N.; Ragel, P.; Raynaud, S.; Lucas, M.M.; Roldán, I.; Montero, M.; Muñoz, F.J.; Ovecka, M.; Bahaji, A.; Planchot, V. Starch Granule Initiation in Arabidopsis Requires the Presence of Either Class IV or Class III Starch Synthases. Plant Cell 2009, 21, 2443–2457. [Google Scholar] [CrossRef]
- Maloney, V.; Park, J.-Y.; Unda, F.; Mansfield, S.D. Sucrose phosphate synthase and sucrose phosphate phosphatase interact in planta and promote plant growth and biomass accumulation. J. Exp. Bot. 2015, 66, 4383–4394. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, H.; Zhang, L.; Zhang, F.; Ou, C.; Wang, F.; Zhang, Z. Involvement of auxin and brassinosteroid in dwarfism of autotetraploid apple (Malus× domestica). Sci. Rep. 2016, 6, 26719. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Huang, Z.; Liao, T.; Li, Y.; Kang, X. In vitro tetraploid plants regeneration from leaf explants of multiple genotypes in Populus. Plant Cell Tissue Organ Cult. (PCTOC) 2016, 125, 1–9. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562. [Google Scholar] [CrossRef]
- Pan, X.; Welti, R.; Wang, X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nat. Protoc. 2010, 5, 986. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, D.K.; Singh, V.P.; Prasad, S.M.; Chauhan, D.K.; Dubey, N.K. Silicon nanoparticles (SiNp) alleviate chromium (VI) phytotoxicity in Pisum sativum (L.) seedlings. Plant Physiol. Biochem. 2015, 96, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Kang, X.Y. In vitro tetraploid induction from leaf explants of Populus pseudo-simonii Kitag. Plant Cell Rep. 2011, 30, 1771. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Kato, A.; Mooney, B.; Birchler, J.A. Phenotypic and gene expression analyses of a ploidy series of maize inbred Oh43. Plant Mol. Biol. 2011, 75, 237–251. [Google Scholar] [CrossRef]
- Dudits, D.; Török, K.; Cseri, A.; Paul, K.; Nagy, A.V.; Nagy, B.; Vass, I. Response of organ structure and physiology to autotetraploidization in early development of energy willow Salix viminalis. Plant Physiol. 2016, 170, 1504–1523. [Google Scholar] [CrossRef]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M. Ethylene role in plant growth, development and senescence: Interaction with other phytohormones. Front. Plant Sci. 2017, 8, 475. [Google Scholar] [CrossRef]
- Wang, L.; Wu, L.M.; Greaves, I.K.; Zhu, A.; Dennis, E.S.; Peacock, W.J. PIF4-controlled auxin pathway contributes to hybrid vigor in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, E3555–E3562. [Google Scholar] [CrossRef]
- Sun, T.P. The molecular mechanism and evolution of the GA–GID1–DELLA signaling module in plants. Curr. Biol. 2011, 21, R338–R345. [Google Scholar] [CrossRef] [PubMed]
- Pandey, N.; Iqbal, Z.; Pandey, B.K.; Sawant, S.V. Phytohormones and drought stress: Plant responses to transcriptional regulation. Mech. Plant Horm. Signal. Stress 2017, 2, 477–504. [Google Scholar]
- Lim, P.O.; Woo, H.R.; Nam, H.G. Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci. 2003, 8, 272–278. [Google Scholar] [CrossRef]
- Curaba, J.; Singh, M.B.; Bhalla, P.L. miRNAs in the crosstalk between phytohormone signalling pathways. J. Exp. Bot. 2014, 65, 1425–1438. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef]
- Lor, V.S.; Olszewski, N.E. GA signalling and cross-talk with other signalling pathways. Essays Biochem. 2015, 58, 49–60. [Google Scholar]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Chu, C. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef]
- Halliday, K.J. Plant hormones: The interplay of brassinosteroids and auxin. Curr. Biol. 2004, 14, R1008–R1010. [Google Scholar] [CrossRef]
- Park, S.Y.; Yu, J.W.; Park, J.S.; Li, J.; Yoo, S.C.; Lee, N.Y.; Lee, S.K.; Jeong, S.W.; Seo, H.S.; Koh, H.J.; et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef]
- Matile, P.; Hörtensteiner, S.; Thomas, H. Chlorophyll degradation. Annu. Rev. Plant Biol. 1999, 50, 67–95. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef] [PubMed]
- Carrington, J.C.; Ambros, V. Role of microRNAs in plant and animal development. Science 2003, 301, 336–338. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.F.; Tian, Q.; Reed, J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 2006, 133, 4211–4218. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Wang, H.; Song, A.; Liu, T.; Chen, Y.; Fang, W.; Jiang, J. miRNAs are involved in determining the improved vigor of autotetrapoid Chrysanthemum nankingense. Front. Plant Sci. 2016, 7, 1412. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, H.; Guo, S.; Wang, B.; Li, Z.; Chong, K.; Xu, Y. OsmiR396d affects gibberellin and brassinosteroid signaling to regulate plant architecture in rice. Plant Physiol. 2018, 176, 946–959. [Google Scholar] [CrossRef] [PubMed]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522. [Google Scholar] [CrossRef]
- Chen, X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. 2009, 25, 21–44. [Google Scholar] [CrossRef]
- D’Ario, M.; Griffiths-Jones, S.; Kim, M. Small RNAs: Big impact on plant development. Trends Plant Sci. 2017, 22, 1056–1068. [Google Scholar] [CrossRef]
- Chuck, G.; Candela, H.; Hake, S. Big impacts by small RNAs in plant development. Curr. Opin. Plant Biol. 2009, 12, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Mlotshwa, S.; Yang, Z.; Kim, Y.; Chen, X. Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana. Plant Mol. Biol. 2006, 61, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Usami, T.; Horiguchi, G.; Yano, S.; Tsukaya, H. The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for squamosa promoter binding protein-like genes in the control of heteroblasty. Development 2009, 136, 955–964. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, C.; Zhang, Y.; Han, Q.; Kang, X. Molecular Mechanism of Slow Vegetative Growth in Populus Tetraploid. Genes 2020, 11, 1417. https://doi.org/10.3390/genes11121417
Xu C, Zhang Y, Han Q, Kang X. Molecular Mechanism of Slow Vegetative Growth in Populus Tetraploid. Genes. 2020; 11(12):1417. https://doi.org/10.3390/genes11121417
Chicago/Turabian StyleXu, Congping, Ying Zhang, Qiang Han, and Xiangyang Kang. 2020. "Molecular Mechanism of Slow Vegetative Growth in Populus Tetraploid" Genes 11, no. 12: 1417. https://doi.org/10.3390/genes11121417
APA StyleXu, C., Zhang, Y., Han, Q., & Kang, X. (2020). Molecular Mechanism of Slow Vegetative Growth in Populus Tetraploid. Genes, 11(12), 1417. https://doi.org/10.3390/genes11121417



