MicroRNA-214 Inhibits Chicken Myoblasts Proliferation, Promotes Their Differentiation, and Targets the TRMT61A Gene
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Animals and RNA Extraction
2.3. cDNA Synthesis and Design of the Primers for Quantitative Real-Time PCR (qPCR)
2.4. Isolation and Culture of CPMs
2.5. CCK-8 Assay
2.6. Cell Cycle Analysis
2.7. EdU Assay
2.8. Construction and Transfection of Plasmid
2.9. Immunofluorescence
2.10. Western Blotting Assay
2.11. Statistical Analysis
3. Results
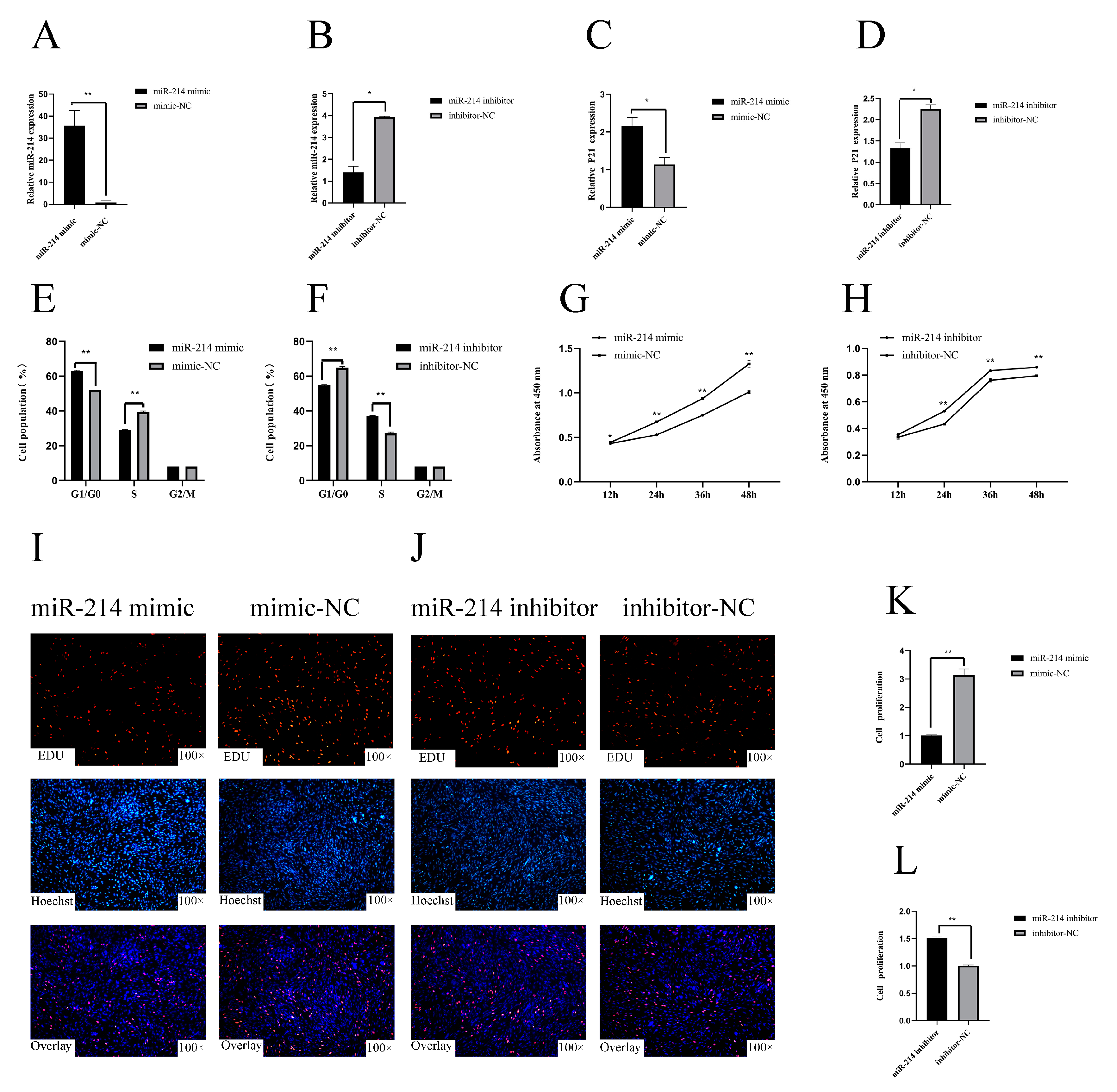
3.1. miR-214 Inhibits the Proliferation of Chicken Myoblasts
3.2. miR-214 Promotes the Differentiation of Chicken Myoblasts
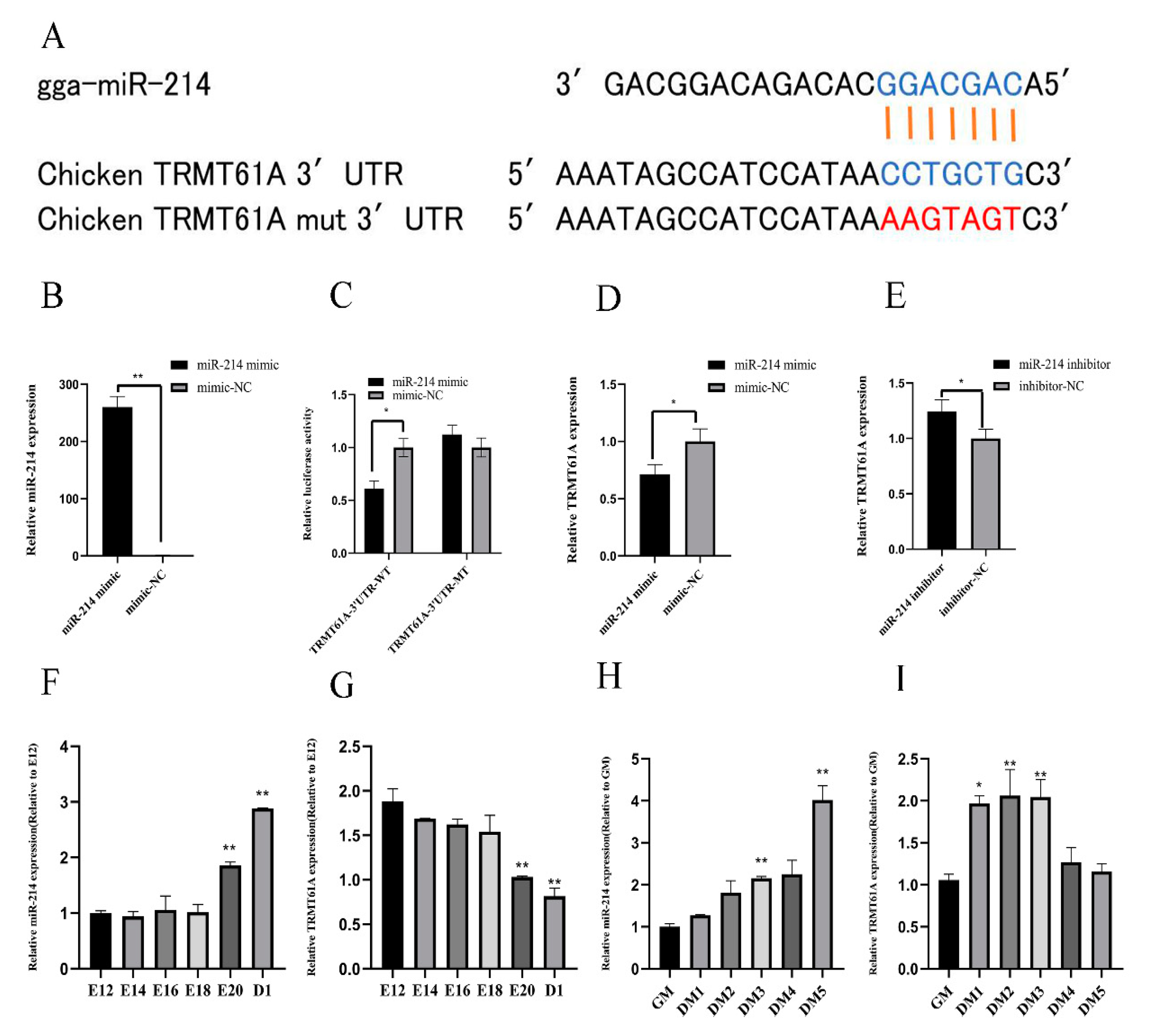
3.3. miR-214 Targets TRMT61A and Negatively Regulates TRMT61A
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pavlath, G.K.; Horsley, V. Cell fusion in skeletal muscle—Central role of NFATC2 in regulating muscle cell size. Cell Cycle 2003, 2, 419–422. [Google Scholar] [CrossRef]
- Koomkrong, N.; Theerawatanasirikul, S.; Boonkaewwan, C.; Jaturasitha, S.; Kayan, A. Breed-Related Number and Size of Muscle Fibres and Their Response to Carcass Quality in Chickens. Ital. J. Anim. Sci. 2015, 14, 4145. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Wu, H.; Ye, Y.; Li, Z.; Hao, S.; Kong, L.; Zheng, X.; Lin, S.; Nie, Q.; Zhang, X. The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation. Cell Death Dis. 2014, 5, e1347. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Luo, W.; Abdalla, B.A.; Ouyang, H.; Yu, J.; Hu, F.; Nie, Q.; Zhang, X. miRNA-223 upregulated by MYOD inhibits myoblast proliferation by repressing IGF2 and facilitates myoblast differentiation by inhibiting ZEB1. Cell Death Dis. 2017, 8, e3094. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.S.; Yang, X.H.; Wang, X.D.; Wang, Y.L.; Zhou, B.; Song, Z.S. MiR-214 regulate gastric cancer cell proliferation, migration and invasion by targeting PTEN. Cancer Cell Int. 2013, 13, 68. [Google Scholar] [CrossRef]
- Xia, H.; Ooi, L.L.P.J.; Hui, K.M. MiR-214 Targets β-Catenin Pathway to Suppress Invasion, Stem-Like Traits and Recurrence of Human Hepatocellular Carcinoma. PLoS ONE 2012, 7, e44206. [Google Scholar] [CrossRef]
- Flynt, A.S.; Li, N.; Thatcher, E.J.; Solnica-Krezel, L.; Patton, J.G. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat. Genet. 2007, 39, 259–263. [Google Scholar] [CrossRef]
- Liu, J.; Luo, X.J.; Xiong, A.W.; Zhang, Z.D.; Yue, S.; Zhu, M.S.; Cheng, S.Y. MicroRNA-214 Promotes Myogenic Differentiation by Facilitating Exit from Mitosis via Down-regulation of Proto-oncogene N-ras. J. Biol. Chem. 2010, 285, 26599–26607. [Google Scholar] [CrossRef]
- Li, T.; Zhang, G.; Wu, P.; Duan, L.; Li, G.; Liu, Q.; Wang, J. Dissection of Myogenic Differentiation Signatures in Chickens by RNA-Seq Analysis. Genes 2018, 9, 34. [Google Scholar] [CrossRef]
- Schwartz, S. M1A within cytoplasmic mRNAs at single nucleotide resolution: A reconciled transcriptome-wide map. RNA 2018, 24, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Ma, M.; Chen, B.; Li, Z.; Abdalla, B.A.; Nie, Q.; Zhang, X. MiR-16-5p targets SESN1 to regulate the p53 signaling pathway, affecting myoblast proliferation and apoptosis, and is involved in myoblast differentiation. Cell Death Dis. 2018, 9, 367. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Tan, W.-H.; Liu, W.; Jin, Y.-X.; Liu, Q. Effects of miR-214 on cervical cancer cell proliferation, apoptosis and invasion via modulating PI3K/AKT/mTOR signal pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 1891–1898. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Li, L.; Wang, J.; Liu, Z. The effect of miR-214 on HepG2 cell proliferation and cell cycle. Chin. Public Health 2017, 33, 614–617. [Google Scholar]
- El-Deiry, W.S.; Tokino, T.; Velculescu, V.E.; Levy, D.B.; Parsons, R.; Trent, J.M.; Lin, D.; Mercer, W.E.; Kinzler, K.W.; Vogelstein, B.E.; et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993, 75, 817–825. [Google Scholar] [CrossRef]
- Liu, L.; Cai, X.; Liu, E.; Tian, X.; Tian, C. MicroRNA-18a promotes proliferation and metastasis in hepatocellular carcinoma via targeting KLF4. Oncotarget 2017, 8, 68263. [Google Scholar] [CrossRef]
- Speidel, D. Transcription-independent p53 apoptosis: An alternative route to death. Trends Cell Biol. 2010, 20, 14–24. [Google Scholar] [CrossRef]
- Juan, A.H.; Kumar, R.M.; Marx, J.G.; Young, R.A.; Sartorelli, V. Mir-214-Dependent Regulation of the Polycomb Protein Ezh2 in Skeletal Muscle and Embryonic Stem Cells. Mol. Cell 2009, 36, 61–74. [Google Scholar] [CrossRef]
- Lee, T.I.; Jenner, R.G.; Boyer, L.A.; Guenther, M.G.; Levine, S.S.; Kumar, R.M.; Chevalier, B.; Johnstone, S.E.; Cole, M.F.; Isono, K.; et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 2006, 125, 301–313. [Google Scholar] [CrossRef]
- Caretti, G.; Di Padova, M.; Micales, B.; Lyons, G.E.; Sartorelli, V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004, 18, 2627–2638. [Google Scholar] [CrossRef]
- Huang, W.; Guo, L.; Zhao, M.; Zhang, D.; Xu, H.; Nie, Q. The Inhibition on MDFIC and PI3K/AKT Pathway Caused by miR-146b-3p Triggers Suppression of Myoblast Proliferation and Differentiation and Promotion of Apoptosis. Cells 2019, 8, 656. [Google Scholar] [CrossRef] [PubMed]
- Safra, M.; Sas-Chen, A.; Nir, R.; Winkler, R.; Nachshon, A.; Bar-Yaacov, D.; Erlacher, M.; Rossmanith, W.; Stern-Ginossar, N.; Schwartz, S. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 2017, 551, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, Q.; Kaboli, P.J.; Shen, J.; Li, M.; Wu, X.; Yin, J.; Zhang, H.; Wu, Y.; Lin, L.; et al. m1A Regulated Genes Modulate PI3K/AKT/mTOR and ErbB Pathways in Gastrointestinal Cancer. Transl. Oncol. 2019, 12, 1323–1333. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Primer Sequences (5′ to 3′) | Product Size (bp) | Annealing Temperature (°C) | Accession Number |
|---|---|---|---|---|
| TRMT61A | F: GTGGGAAGCCATTGGACAT | 266 | 57 | XM_421386.6 |
| R: CTGGGCTACCTTGGTTTGAT | ||||
| p21 | F: CCCGTAGACCACGAGCAGAT | 102 | 61 | NM_204396.1 |
| R: CGTCTCGGTCTCGAAGTTGA | ||||
| MYOD | GCTACTACACGGAATCACCAAAT | 200 | 53 | NM_204214.2 |
| R: CTGGGCTCCACTGTCACTCA | ||||
| MYHC | F: CTCCTCACGCTTTGGTAA | 213 | 53 | NM_001319304.1 |
| R: TGATAGTCGTATGGGTTGGT | ||||
| MYOG | F: CGGAGGCTGAAGAAGGTGAA | 320 | 53 | NM_204184.1 |
| R: CGGTCCTCTGCCTGGTCAT | ||||
| β-actin | F: CAGCCATCTTTCTTGGGTAT | 169 | 60 | NM_205518.1 |
| R: CTGTGATCTCCTTCTGCATCC |
| Gene Name | Primer Sequences (5′ to 3′) | Annealing Temperature (°C) |
|---|---|---|
| gga-miR-214 | F: CGCGACAGCAGGCACAGAC | 60 |
| R: AGTGCAGGGTCCGAGGTATT | ||
| U6 | F: GTCACTTCTGGTGGCGGTAA | 60 |
| R: GTTCAGTAGAGGGTCAAA | ||
| Stem loop primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTGCCT |
| Sequence Name | Sequence Information |
|---|---|
| miR-214 mimic | ACAGCAGGCACAGACAGGCAG |
| GCCUGUCUGUGCCUGCUGUUU | |
| Mimic NC | UUCUCCGAACGUGUCACGUTT |
| ACGUGACACGUUCGGAGAATT | |
| miR-214 inhibitor | CUGCCUGUCUGUGCCUGCUGU |
| Inhibitor NC | CAGUACUUUUGUGUAGUACAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, Y.; Wu, Y.; Yin, X.; Li, T.; Chen, F.; Wu, P.; Zhang, S.; Wang, J.; Zhang, G. MicroRNA-214 Inhibits Chicken Myoblasts Proliferation, Promotes Their Differentiation, and Targets the TRMT61A Gene. Genes 2020, 11, 1400. https://doi.org/10.3390/genes11121400
Duan Y, Wu Y, Yin X, Li T, Chen F, Wu P, Zhang S, Wang J, Zhang G. MicroRNA-214 Inhibits Chicken Myoblasts Proliferation, Promotes Their Differentiation, and Targets the TRMT61A Gene. Genes. 2020; 11(12):1400. https://doi.org/10.3390/genes11121400
Chicago/Turabian StyleDuan, Yanjun, Yulin Wu, Xuemei Yin, Tingting Li, Fuxiang Chen, Pengfei Wu, Shanshan Zhang, Jinyu Wang, and Genxi Zhang. 2020. "MicroRNA-214 Inhibits Chicken Myoblasts Proliferation, Promotes Their Differentiation, and Targets the TRMT61A Gene" Genes 11, no. 12: 1400. https://doi.org/10.3390/genes11121400
APA StyleDuan, Y., Wu, Y., Yin, X., Li, T., Chen, F., Wu, P., Zhang, S., Wang, J., & Zhang, G. (2020). MicroRNA-214 Inhibits Chicken Myoblasts Proliferation, Promotes Their Differentiation, and Targets the TRMT61A Gene. Genes, 11(12), 1400. https://doi.org/10.3390/genes11121400






