Unlocking Survival Mechanisms for Metal and Oxidative Stress in the Extremely Acidophilic, Halotolerant Acidihalobacter Genus
Abstract
1. Introduction
2. Materials and Methods
Genome Annotation and Comparisons
3. Results and Discussion
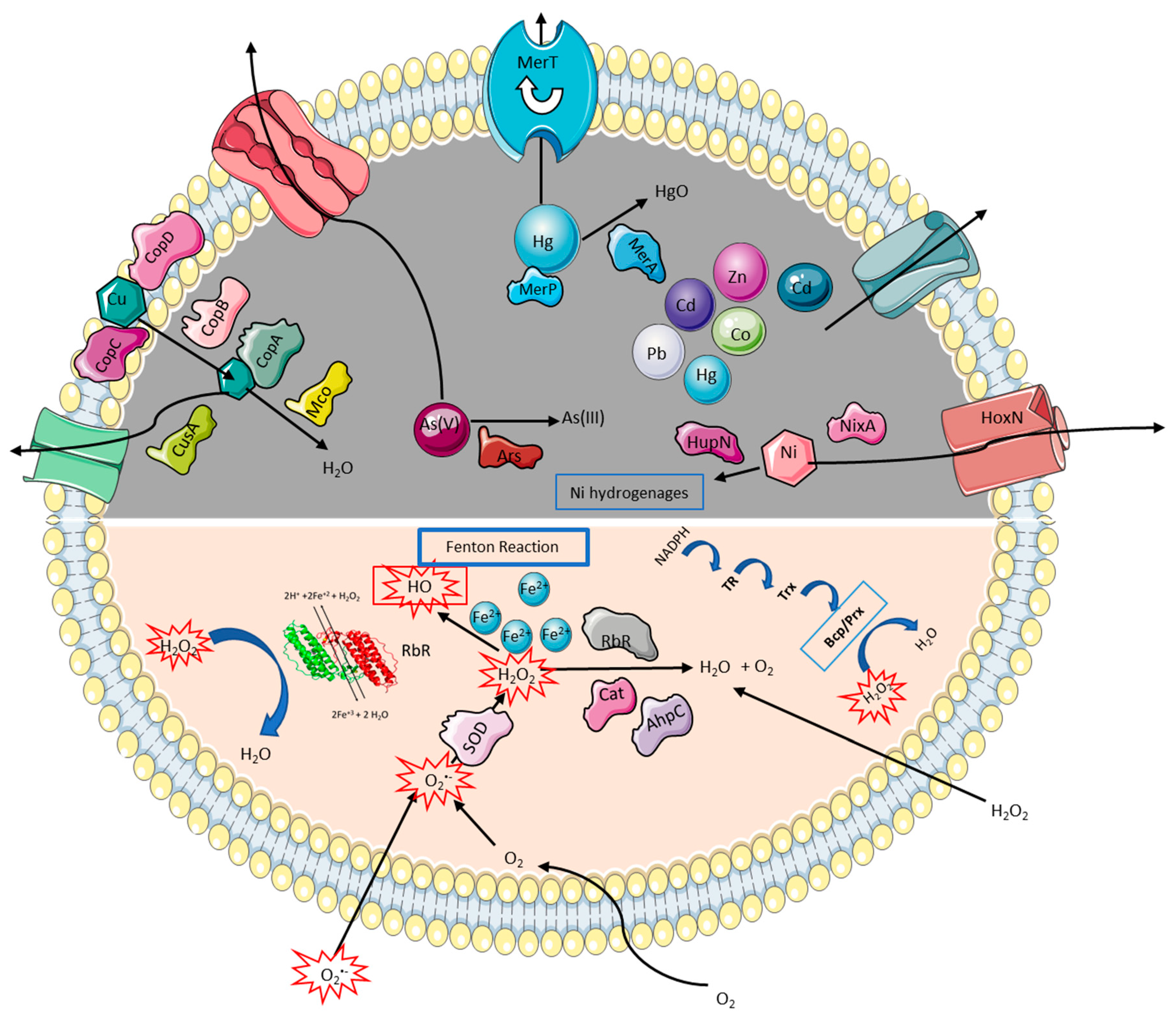
3.1. Strategies to Cope with High Metal and Metalloid Concentrations
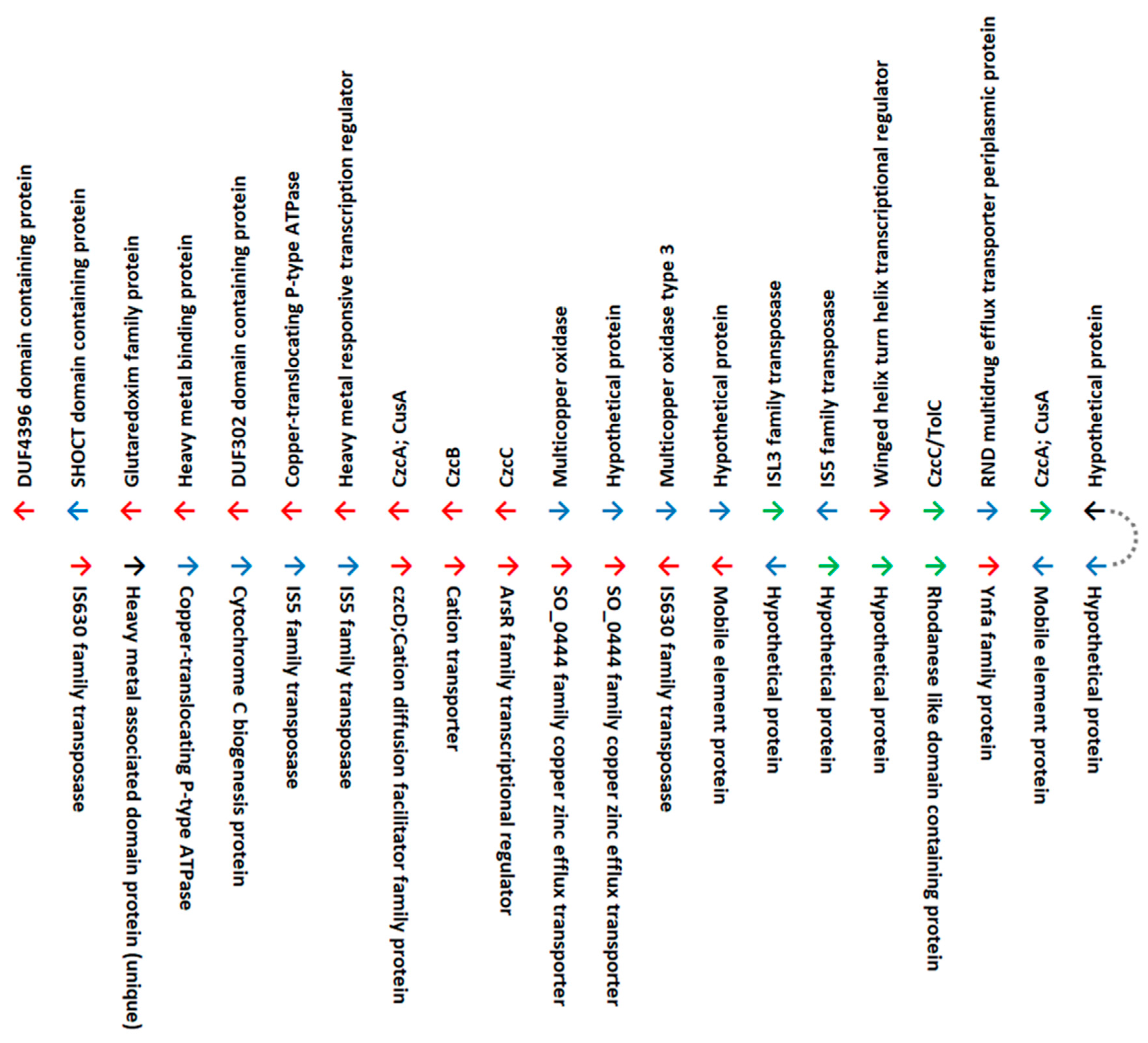
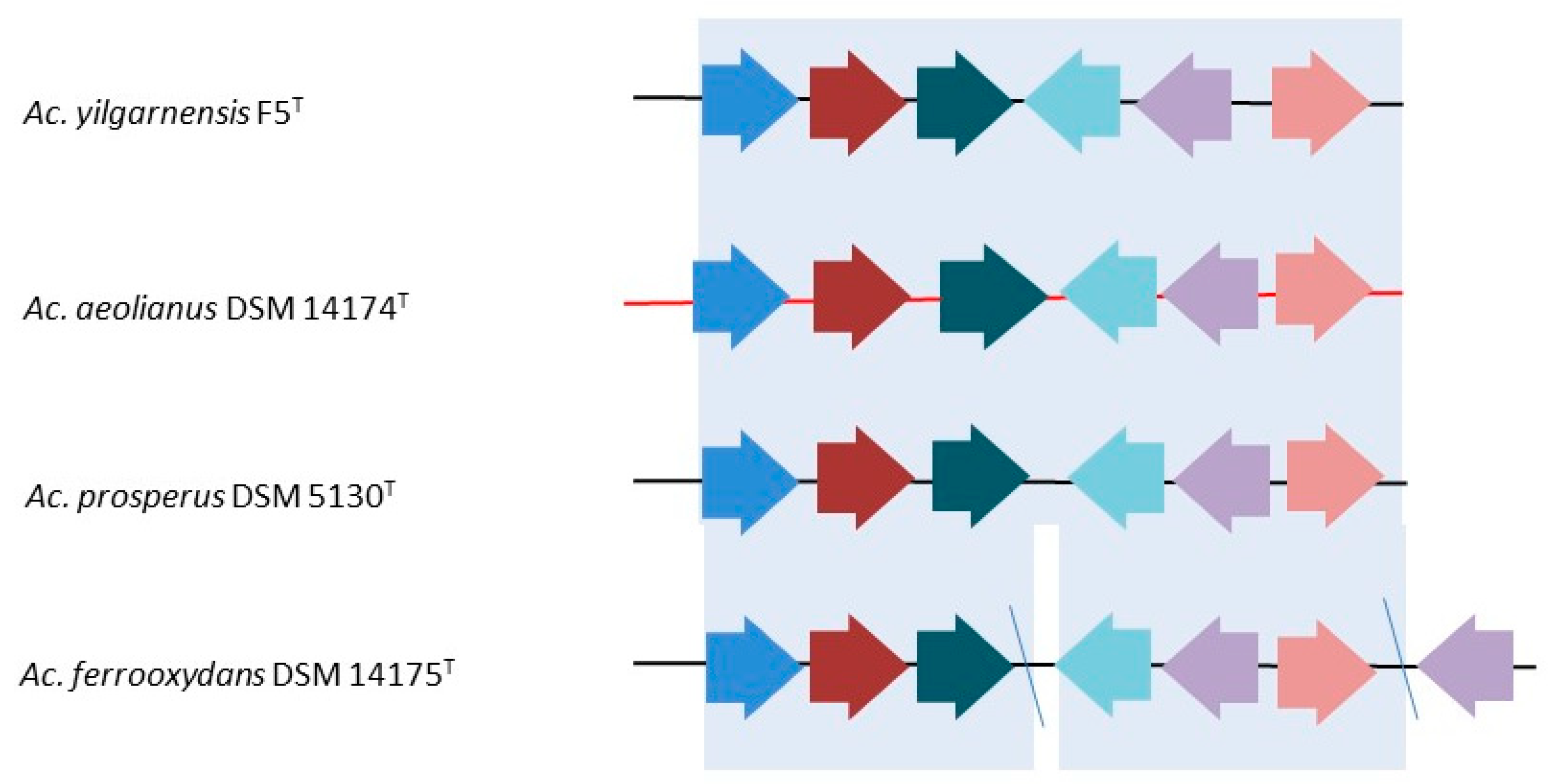
3.1.1. Copper
3.1.2. Other Divalent Heavy Metals and Metalloids
3.1.3. The poly-P Mechanism
3.2. Strategies to Tolerate Oxidative Stress
3.2.1. Rubrerythrin and Neighborhood Genes
3.2.2. Catalase and Alkyl Hydroperoxide Reductase
3.2.3. Other Mechanisms of Oxidative Stress Tolerance
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Khaleque, H.N.; González, C.; Kaksonen, A.H.; Boxall, N.J.; Holmes, D.S.; Watkin, E.L. Genome-based classification of two halotolerant extreme acidophiles, Acidihalobacter prosperus V6 (= DSM 14174= JCM 32253) and ‘Acidihalobacter ferrooxidans’ V8 (= DSM 14175= JCM 32254) as two new species, Acidihalobacter aeolianus sp. nov. and Acidihalobacter ferrooxydans sp. nov., respectively. Int. J. Syst. Evol. Microbiol. 2019, 69, 1557–1565. [Google Scholar] [PubMed]
- Ossandon, F.J.; Cárdenas, J.P.; Corbett, M.; Quatrini, R.; Holmes, D.S.; Watkin, E.L.J.; Guo, M.-Y.; Huo, D.-Q.; Ghai, R.; Rodriguez-Valera, F.; et al. Draft Genome Sequence of the Iron-Oxidizing, Acidophilic, and Halotolerant “Thiobacillus prosperus” Type Strain DSM 5130. Genome Announc. 2014, 2, e01045-14. [Google Scholar] [CrossRef] [PubMed]
- Khaleque, H.N.; González, C.; Johnson, D.B.; Kaksonen, A.H.; Holmes, D.S.; Watkin, E.L.J. Genome-based classification of Acidihalobacter prosperus F5 (=DSM 105917=JCM 32255) as Acidihalobacter yilgarnensis sp. nov. Int. J. Syst. Evol. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Khaleque, H.N.; Kaksonen, A.H.; Boxall, N.J.; Watkin, E.L. Chloride ion tolerance and pyrite bioleaching capabilities of pure and mixed halotolerant, acidophilic iron- and sulfur-oxidizing cultures. Miner. Eng. 2018, 120, 87–93. [Google Scholar] [CrossRef]
- Khaleque, H.N.; González, C.; Shafique, R.; Kaksonen, A.H.; Holmes, D.S.; Watkin, E.L.J. Uncovering the Mechanisms of Halotolerance in the Extremely Acidophilic Members of the Acidihalobacter Genus Through Comparative Genome Analysis. Front. Microbiol. 2019, 10, 155. [Google Scholar] [CrossRef] [PubMed]
- Khaleque, H.N.; Corbett, M.K.; Ramsay, J.P.; Kaksonen, A.H.; Boxall, N.J.; Watkin, E.L. Complete genome sequence of Acidihalobacter prosperus strain F5, an extremely acidophilic, iron- and sulfur-oxidizing halophile with potential industrial applicability in saline water bioleaching of chalcopyrite. J. Biotechnol. 2017, 262, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Dopson, M.; Baker-Austin, C.; Koppineedi, P.R.; Bond, P.L. Growth in sulfidic mineral environments: Metal resistance mechanisms in acidophilic micro-organisms. Microbiology 2003, 149, 1959–1970. [Google Scholar] [CrossRef]
- Bruins, M.R.; Kapil, S.; Oehme, F.W. Microbial Resistance to Metals in the Environment. Ecotoxicol. Environ. Saf. 2000, 45, 198–207. [Google Scholar] [CrossRef]
- Nies, D.H. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 1999, 51, 730–750. [Google Scholar] [CrossRef]
- Ferrer, A.; Orellana, O.; Levicán, G. Oxidative Stress and Metal Tolerance in Extreme Acidophiles. Acidophiles Life Extrem. Acidic Environ. 2016, 2016, 63–76. [Google Scholar] [CrossRef]
- Watkin, E.L.; Keeling, S.E.; Perrot, F.A.; Shiers, D.W.; Palmer, M.-L.; Watling, H.R. Metals tolerance in moderately thermophilic isolates from a spent copper sulfide heap, closely related to Acidithiobacillus caldus, Acidimicrobium ferrooxidans and Sulfobacillus thermosulfidooxidans. J. Ind. Microbiol. Biotechnol. 2009, 36, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A. Pathways of Oxidative Damage. Annu. Rev. Microbiol. 2003, 57, 395–418. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A. Cellular Defenses against Superoxide and Hydrogen Peroxide. Annu. Rev. Biochem. 2008, 77, 755–776. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, A.; Rivera, J.; Zapata, C.; Norambuena, J.; Sandoval, Á.; Chávez, R.; Orellana, O.; Levicán, G. Cobalamin Protection against Oxidative Stress in the Acidophilic Iron-oxidizing Bacterium Leptospirillum Group II CF-1. Front. Microbiol. 2016, 7, 748. [Google Scholar] [CrossRef] [PubMed]
- Schoonen, M.A.; Cohn, C.A.; Roemer, E.; Laffers, R.; Simon, S.R.; O’Riordan, T. Mineral-Induced Formation of Reactive Oxygen Species. Rev. Miner. Geochem. 2006, 64, 179–221. [Google Scholar] [CrossRef]
- Schoonen, M.A.; Harrington, A.D.; Laffers, R.; Strongin, D.R. Role of hydrogen peroxide and hydroxyl radical in pyrite oxidation by molecular oxygen. Geochim. Cosmochim. Acta 2010, 74, 4971–4987. [Google Scholar] [CrossRef]
- Jones, G.C.; Van Hille, R.P.; Harrison, S.T.L. Reactive oxygen species generated in the presence of fine pyrite particles and its implication in thermophilic mineral bioleaching. Appl. Microbiol. Biotechnol. 2012, 97, 2735–2742. [Google Scholar] [CrossRef]
- Kehrer, J.P. The Haber–Weiss reaction and mechanisms of toxicity. Toxicology 2000, 149, 43–50. [Google Scholar] [CrossRef]
- Cárdenas, J.P.; Moya, F.; Covarrubias, P.C.; Shmaryahu, A.; Levicán, G.; Holmes, D.S.; Quatrini, R. Comparative genomics of the oxidative stress response in bioleaching microorganisms. Hydrometallurgy 2012, 127, 162–167. [Google Scholar] [CrossRef]
- Fridovich, I. The biology of oxygen radicals. Science 1978, 201, 875–880. [Google Scholar] [CrossRef]
- Nordberg, J.; Arnér, E.S. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic. Biol. Med. 2001, 31, 1287–1312. [Google Scholar] [CrossRef]
- González, D.; Álamos, P.; Rivero, M.; Orellana, O.; Norambuena, J.; Chávez, R.; Levicán, G. Deciphering the Role of Multiple Thioredoxin Fold Proteins of Leptospirillum sp. in Oxidative Stress Tolerance. Int. J. Mol. Sci. 2020, 21, 1880. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, H.; Wu, W.; Zhang, X.; Gu, T.; Zhu, M.; Tan, W. Oxidative Stress Induced by Metal Ions in Bioleaching of LiCoO2 by an Acidophilic Microbial Consortium. Front. Microbiol. 2020, 10, 3058. [Google Scholar] [CrossRef] [PubMed]
- Bellenberg, S.; Huynh, D.; Poetsch, A.; Sand, W.; Vera, M. Proteomics Reveal Enhanced Oxidative Stress Responses and Metabolic Adaptation in Acidithiobacillus ferrooxidans Biofilm Cells on Pyrite. Front. Microbiol. 2019, 10, 592. [Google Scholar] [CrossRef]
- West, M.D.; Clarke, J.D.; Laing, J.H.; Willson, D.; Waldie, J.M.; Murphy, G.M.; Thomas, M.; Mann, G. Testing technologies and strategies for exploration in Australian Mars analogues: A review. Planet. Space Sci. 2010, 58, 658–670. [Google Scholar] [CrossRef]
- Bowen, B.B.; Benison, K.C.; Story, S.; Grotzinger, J.P.; Milliken, R.E. Early Diagenesis by Modern Acid Brines in Western Australia and Implications for the History of Sedimentary Modification on Mars. Sediment. Geol. Mars 2012, 102, 229–252. [Google Scholar] [CrossRef]
- Quatrini, R.; Escudero, L.V.; Moya-Beltrán, A.; Galleguillos, P.A.; Issotta, F.; Acosta, M.; Cárdenas, J.P.; Nuñez, H.; Salinas, K.; Holmes, D.S.; et al. Draft genome sequence of Acidithiobacillus thiooxidans CLST isolated from the acidic hypersaline Gorbea salt flat in northern Chile. Stand. Genom. Sci. 2017, 12, 84. [Google Scholar] [CrossRef]
- Benison, K.C. The Physical and Chemical Sedimentology of Two High-Altitude Acid Salars in Chile: Sedimentary Processes in an Extreme Environment. J. Sediment. Res. 2019, 89, 147–167. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 1–15. [Google Scholar] [CrossRef]
- Riadi, G.; Medina-Moenne, C.; Holmes, D.S. TnpPred: A Web Service for the Robust Prediction of Prokaryotic Transposases. Comp. Funct. Genom. 2012, 2012, 1–5. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Schneider, T.D.; Stephens, R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Li, X.; Zhu, Y.-G.; Shaban, B.; Bruxner, T.J.C.; Bond, P.L.; Huang, L. Assessing the genetic diversity of Cu resistance in mine tailings through high-throughput recovery of full-length copA genes. Sci. Rep. 2015, 5, srep13258. [Google Scholar] [CrossRef] [PubMed]
- Völlmecke, C.; Drees, S.L.; Reimann, J.; Albers, S.-V.; Lübben, M. The ATPases CopA and CopB both contribute to copper resistance of the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiology 2012, 158, 1622–1633. [Google Scholar] [CrossRef] [PubMed]
- Falagán, C.; Johnson, D.B. The significance of pH in dictating the relative toxicities of chloride and copper to acidophilic bacteria. Res. Microbiol. 2018, 169, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Navarro, C.A.; Von Bernath, D.; Jerez, C.A. Heavy Metal Resistance Strategies of Acidophilic Bacteria and Their Acquisition: Importance for Biomining and Bioremediation. Biol. Res. 2013, 46, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Rowland, J.L.; Niederweis, M. A Multicopper Oxidase Is Required for Copper Resistance in Mycobacterium tuberculosis. J. Bacteriol. 2013, 195, 3724–3733. [Google Scholar] [CrossRef] [PubMed]
- Wen, Q.; Liu, X.-M.; Wang, H.; Lin, J. A versatile and efficient markerless gene disruption system forAcidithiobacillus thiooxidans: Application for characterizing a copper tolerance related multicopper oxidase gene. Environ. Microbiol. 2014, 16, 3499–3514. [Google Scholar] [CrossRef]
- Grass, G.; Rensing, C. CueO Is a Multi-copper Oxidase That Confers Copper Tolerance in Escherichia coli. Biochem. Biophys. Res. Commun. 2001, 286, 902–908. [Google Scholar] [CrossRef]
- Quintanar, L.; Stoj, C.; Taylor, A.B.; Hart, P.J.; Kosman, D.J.; Solomon, E.I. Shall we dance? How a multicopper oxidase chooses its electron transfer partner. Acc. Chem. Res. 2007, 40, 445–452. [Google Scholar] [CrossRef]
- Solomon, E.I.; Sundaram, U.M.; Machonkin, T.E. Multicopper Oxidases and Oxygenases. Chem. Rev. 1996, 96, 2563–2606. [Google Scholar] [CrossRef]
- Nies, D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Boyd, E.S.; Barkay, T. The Mercury Resistance Operon: From an Origin in a Geothermal Environment to an Efficient Detoxification Machine. Front. Microbiol. 2012, 3, 349. [Google Scholar] [CrossRef] [PubMed]
- Khaleque, H.N.; Shafique, R.; Kaksonen, A.H.; Boxall, N.J.; Watkin, E.L. Quantitative proteomics using SWATH-MS identifies mechanisms of chloride tolerance in the halophilic acidophile Acidihalobacter prosperus DSM 14174. Res. Microbiol. 2018, 169, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Dopson, M.; Holmes, D.S. Metal resistance in acidophilic microorganisms and its significance for biotechnologies. Appl. Microbiol. Biotechnol. 2014, 98, 8133–8144. [Google Scholar] [CrossRef] [PubMed]
- Fulkerson, J.F.; Mobley, H.L.T. Membrane Topology of the NixA Nickel Transporter ofHelicobacter pylori: Two Nickel Transport-Specific Motifs within Transmembrane Helices II and III. J. Bacteriol. 2000, 182, 1722–1730. [Google Scholar] [CrossRef]
- Degen, O.; Kobayashi, M.; Shimizu, S.; Eitinger, T. Selective transport of divalent cations by transition metal permeases: The Alcaligenes eutrophus HoxN and the Rhodococcus rhodochrous NhlF. Arch. Microbiol. 1999, 171, 139–145. [Google Scholar] [CrossRef]
- Bartels, F.; Fernández, S.; Holtel, A.; Timmis, K.N.; De Lorenzo, V. The Essential HupB and HupN Proteins ofPseudomonas putidaProvide Redundant and Nonspecific DNA-bending Functions. J. Biol. Chem. 2001, 276, 16641–16648. [Google Scholar] [CrossRef]
- Soboh, B.; Pinske, C.; Kuhns, M.; Waclawek, M.; Ihling, C.; Trchounian, A.; Trchounian, A.; Sinz, A.; Sawers, R.G. The respiratory molybdo-selenoprotein formate dehydrogenases of Escherichia coli have hydrogen: Benzyl viologen oxidoreductase activity. BMC Microbiol. 2011, 11, 173. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Dopson, M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 2007, 15, 165–171. [Google Scholar] [CrossRef]
- Heijerick, D.; De Schamphelaere, K.; Janssen, C. Biotic ligand model development predicting Zn toxicity to the alga Pseudokirchneriella subcapitata: Possibilities and limitations. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2002, 133, 207–218. [Google Scholar] [CrossRef]
- Ullrich, S.R.; Poehlein, A.; Tischler, J.S.; González, C.; Ossandon, F.J.; Daniel, R.; Holmes, D.S.; Schlömann, M.; Mühling, M. Genome Analysis of the Biotechnologically Relevant Acidophilic Iron Oxidising Strain JA12 Indicates Phylogenetic and Metabolic Diversity within the Novel Genus “Ferrovum”. PLoS ONE 2016, 11, e0146832. [Google Scholar] [CrossRef]
- Orell, A.; Navarro, C.A.; Arancibia, R.; Mobarec, J.C.; Jerez, C.A. Life in blue: Copper resistance mechanisms of bacteria and Archaea used in industrial biomining of minerals. Biotechnol. Adv. 2010, 28, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Orell, A.; Navarro, C.A.; Rivero, M.; Aguilar, J.S.; Jerez, C.A. Inorganic polyphosphates in extremophiles and their possible functions. Extremophiles 2012, 16, 573–583. [Google Scholar] [CrossRef]
- Rao, N.N.; Gómez-García, M.R.; Kornberg, A. Inorganic Polyphosphate: Essential for Growth and Survival. Annu. Rev. Biochem. 2009, 78, 605–647. [Google Scholar] [CrossRef] [PubMed]
- Keasling, J.D. Regulation of Intracellular Toxic Metals and Other Cations by Hydrolysis of Polyphosphate. Ann. N. Y. Acad. Sci. 1997, 829, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Orell, A.; Remonsellez, F.; Arancibia, R.; Jerez, C.A. Molecular Characterization of Copper and Cadmium Resistance Determinants in the Biomining Thermoacidophilic ArchaeonSulfolobus metallicus. Archaea 2013, 2013, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, S.; Jerez, C.A. Copper Ions Stimulate Polyphosphate Degradation and Phosphate Efflux in Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2004, 70, 5177–5182. [Google Scholar] [CrossRef] [PubMed]
- Navarro, C.A.; Orellana, L.H.; Mauriaca, C.; Jerez, C.A. Transcriptional and Functional Studies of Acidithiobacillus ferrooxidans Genes Related to Survival in the Presence of Copper. Appl. Environ. Microbiol. 2009, 75, 6102–6109. [Google Scholar] [CrossRef]
- Fathollahzadeh, H.; Hackett, M.J.; Khaleque, H.N.; Eksteen, J.; Kaksonen, A.H.; Watkin, E.L.J. Better together: Potential of co-culture microorganisms to enhance bioleaching of rare earth elements from monazite. Bioresour. Technol. Rep. 2018, 3, 109–118. [Google Scholar] [CrossRef]
- Imlay, J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef]
- Rivera Araya, J.I. Characterization of NaCl Tolerance Mechanism and Its Relation with the Antioxidant Mechanisms in the Acidophilic Bacterium Leptospirillum ferriphilum DSM 14647. Ph.D. Thesis, Universidad de Chile, Santiago, Chile, 2019. [Google Scholar]
- Andrews, S.C. The Ferritin-like superfamily: Evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochim. Biophys. Acta (BBA) Gen. Subj. 2010, 1800, 691–705. [Google Scholar] [CrossRef]
- Cardenas, J.P.; Quatrini, R.; Holmes, D.S. Aerobic Lineage of the Oxidative Stress Response Protein Rubrerythrin Emerged in an Ancient Microaerobic, (Hyper)Thermophilic Environment. Front. Microbiol. 2016, 7, 1822. [Google Scholar] [CrossRef] [PubMed]
- Pedone, E.; Fiorentino, G.; Bartolucci, S.; Limauro, D. Enzymatic Antioxidant Signatures in Hyperthermophilic Archaea. Antioxidants 2020, 9, 703. [Google Scholar] [CrossRef] [PubMed]
- Coulter, E.D.; Shenvi, N.V.; Beharry, Z.M.; Smith, J.J.; Prickril, B.C.; Kurtz, N.M. Rubrerythrin-catalyzed substrate oxidation by dioxygen and hydrogen peroxide. Inorg. Chim. Acta 2000, 297, 231–241. [Google Scholar] [CrossRef]
- Weinberg, M.V.; Jenney, F.E.; Cui, X.; Adams, M.W.W. Rubrerythrin from the Hyperthermophilic Archaeon Pyrococcus furiosus is a Rubredoxin-Dependent, Iron-Containing Peroxidase. J. Bacteriol. 2004, 186, 7888–7895. [Google Scholar] [CrossRef]
- Mishra, S.; Imlay, J. Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch. Biochem. Biophys. 2012, 525, 145–160. [Google Scholar] [CrossRef]
- Martins, M.C.; Romão, C.V.; Folgosa, F.; Borges, P.T.; Frazão, C.; Teixeira, M. How superoxide reductases and flavodiiron proteins combat oxidative stress in anaerobes. Free Radic. Biol. Med. 2019, 140, 36–60. [Google Scholar] [CrossRef]
- Nóbrega, C.S.; Pauleta, S.R. Reduction of hydrogen peroxide in gram-negative bacteria—Bacterial peroxidases. Adv. Microb. Physiol. 2019, 74, 415–464. [Google Scholar] [CrossRef]
- Soito, L.; Williamson, C.; Knutson, S.T.; Fetrow, J.S.; Poole, L.B.; Nelson, K.J. PREX: PeroxiRedoxin classification indEX, a database of subfamily assignments across the diverse peroxiredoxin family. Nucleic Acids Res. 2010, 39, D332–D337. [Google Scholar] [CrossRef]
- Maaty, W.S.; Wiedenheft, B.; Tarlykov, P.; Schaff, N.; Heinemann, J.; Robison-Cox, J.; Valenzuela, J.; Dougherty, A.; Blum, P.; Lawrence, C.M.; et al. Something Old, Something New, Something Borrowed; How the Thermoacidophilic Archaeon Sulfolobus solfataricus Responds to Oxidative Stress. PLoS ONE 2009, 4, e6964. [Google Scholar] [CrossRef]
- Poynton, R.A.; Hampton, M.B. Peroxiredoxins as biomarkers of oxidative stress. Biochim. Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 906–912. [Google Scholar] [CrossRef]
- Jeong, W.; Cha, M.-K.; Kim, I.-H. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/alkyl hydroperoxide peroxidase C (AhpC) family. J. Biol. Chem. 2000, 275, 2924–2930. [Google Scholar] [CrossRef] [PubMed]
- Wakita, M.; Masuda, S.; Motohashi, K.; Hisabori, T.; Ohta, H.; Takamiya, K.-I. The Significance of Type II and PrxQ Peroxiredoxins for Antioxidative Stress Response in the Purple Bacterium Rhodobacter sphaeroides. J. Biol. Chem. 2007, 282, 27792–27801. [Google Scholar] [CrossRef] [PubMed]
- Clarke, D.; Mackay, C.L.; Campopiano, D.J.; Langridge-Smith, P.; Brown, A.R. Interrogating the Molecular Details of the Peroxiredoxin Activity of the Escherichia coli Bacterioferritin Comigratory Protein Using High-Resolution Mass Spectrometry. Biochemistry 2009, 48, 3904–3914. [Google Scholar] [CrossRef]
- Hall, A.; Nelson, K.; Poole, L.B.; Karplus, P.A. Structure-based Insights into the Catalytic Power and Conformational Dexterity of Peroxiredoxins. Antioxid. Redox Signal. 2011, 15, 795–815. [Google Scholar] [CrossRef] [PubMed]
- Nelson, K.J.; Knutson, S.T.; Soito, L.; Klomsiri, C.; Poole, L.B.; Fetrow, J.S. Analysis of the peroxiredoxin family: Using active-site structure and sequence information for global classification and residue analysis. Proteins Struct. Funct. Bioinform. 2011, 79, 947–964. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.G. Overview on Peroxiredoxin. Mol. Cells 2016, 39, 1–5. [Google Scholar] [CrossRef]
- Braida, W.; Ong, S.K. Decomposition of Nitrite under Various pH and Aeration Conditions. Water Air Soil Pollut. 2000, 118, 13–26. [Google Scholar] [CrossRef]
- Winterbourn, C.C. Biological chemistry of superoxide radicals. ChemTexts 2020, 6, 1–13. [Google Scholar] [CrossRef]
- Parimelzaghan, A.; Anbarasu, A.; Ramaiah, S. Gene Network Analysis of Metallo Beta Lactamase Family Proteins Indicates the Role of Gene Partners in Antibiotic Resistance and Reveals Important Drug Targets. J. Cell. Biochem. 2016, 117, 1330–1339. [Google Scholar] [CrossRef]
- Panyushkina, A.E.; Matyushkina, D.; Pobeguts, O. Understanding Stress Response to High-Arsenic Gold-Bearing Sulfide Concentrate in Extremely Metal-Resistant Acidophile Sulfobacillus thermotolerans. Microorganisms 2020, 8, 1076. [Google Scholar] [CrossRef]
- Walsh, T.R.; Toleman, M.A.; Poirel, L.; Nordmann, P. Metallo-β-Lactamases: The Quiet before the Storm? Clin. Microbiol. Rev. 2005, 18, 306–325. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Maisonneuve, E. Bacterial Persistence and Toxin-Antitoxin Loci. Annu. Rev. Microbiol. 2012, 66, 103–123. [Google Scholar] [CrossRef]
- Fléchard, M.; Gilot, P. Physiological impact of transposable elements encoding DDE transposases in the environmental adaptation of Streptococcus agalactiae. Microbiology 2014, 160, 1298–1315. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.S.; Mountain, S.; Beeri, K.; Adams, M.D. Assessment of Insertion Sequence Mobilization as an Adaptive Response to Oxidative Stress in Acinetobacter baumannii Using IS-seq. J. Bacteriol. 2017, 199. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.; Vulić, M.; Min, J.; Yen, T.-J.; Schumacher, M.A.; Brennan, R.G.; Lewis, K. Regulation of the Escherichia coli HipBA toxin-antitoxin system by proteolysis. PLoS ONE 2012, 7, e39185. [Google Scholar] [CrossRef]
- Virtanen, P.; Wäneskog, M.; Koskiniemi, S. Class II contact-dependent growth inhibition (CDI) systems allow for broad-range cross-species toxin delivery within the Enterobacteriaceae family. Mol. Microbiol. 2019, 111, 1109–1125. [Google Scholar] [CrossRef] [PubMed]
- Cañas-Duarte, S.J.; Perez-Lopez, M.I.; Herrfurth, C.; Sun, L.; Contreras, L.M.; Feussner, I.; Leidy, C.; Riaño-Pachón, D.M.; Restrepo, S.; Pedraza, J.M. An integrative approach points to membrane composition as a key factor in E. coli persistence. bioRxiv 2020. [Google Scholar] [CrossRef]
- Johnson, L.A.; Hug, L.A. Distribution of reactive oxygen species defense mechanisms across domain bacteria. Free Radic. Biol. Med. 2019, 140, 93–102. [Google Scholar] [CrossRef]
- Christel, S.; Herold, M.; Bellenberg, S.; El Hajjami, M.; Buetti-Dinh, A.; Pivkin, I.V.; Sand, W.; Wilmes, P.; Poetsch, A.; Dopson, M. Multi-omics Reveals the Lifestyle of the Acidophilic, Mineral-Oxidizing Model Species Leptospirillum ferriphilum T. Appl. Environ. Microbiol. 2017, 84, e02091-17. [Google Scholar] [CrossRef]
- Kojima, H.; Watanabe, T.; Fukui, M. Sulfuricaulis limicola gen. nov., sp. nov., a sulfur oxidizer isolated from a lake. Int. J. Syst. Evol. Microbiol. 2016, 66, 266–270. [Google Scholar] [CrossRef]
- Terrón-González, L.; Martín-Cabello, G.; Ferrer, M.; Santero, E. Functional Metagenomics of a Biostimulated Petroleum-Contaminated Soil Reveals an Extraordinary Diversity of Extradiol Dioxygenases. Appl. Environ. Microbiol. 2016, 82, 2467–2478. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.I.; Hwang, C.Y.; Cho, B.C. Nitratireductor aquimarinus sp. nov., isolated from a culture of the diatom Skeletonema costatum, and emended description of the genus Nitratireductor. Int. J. Syst. Evol. Microbiol. 2011, 61, 2676–2681. [Google Scholar] [CrossRef] [PubMed]
- Ou, D.; Huang, H.; Bai, R.; Li, Q.; Wang, Y.; Yin, Y. Nitratireductor aestuarii sp. nov., a marine alphaproteobacterium isolated from an estuary. Int. J. Syst. Evol. Microbiol. 2017, 67, 1637–1642. [Google Scholar] [CrossRef] [PubMed]
- Lemire, J.; AlHasawi, A.; Appanna, V.D.; Tharmalingam, S. Metabolic defence against oxidative stress: The road less travelled so far. J. Appl. Microbiol. 2017, 123, 798–809. [Google Scholar] [CrossRef]
- Cosgrove, K.; Coutts, G.; Jonsson, I.-M.; Tarkowski, A.; Kokai-Kun, J.F.; Mond, J.J.; Foster, S.J. Catalase (KatA) and Alkyl Hydroperoxide Reductase (AhpC) Have Compensatory Roles in Peroxide Stress Resistance and Are Required for Survival, Persistence, and Nasal Colonization in Staphylococcus aureus. J. Bacteriol. 2006, 189, 1025–1035. [Google Scholar] [CrossRef]
- Pagán-Ramos, E.; Song, J.; McFalone, M.; Mudd, M.H.; Deretic, V. Oxidative Stress Response and Characterization of the oxyR-ahpC and furA-katG Loci in Mycobacterium marinum. J. Bacteriol. 1998, 180, 4856–4864. [Google Scholar] [CrossRef]
- Tralau, T.; Vuilleumier, S.; Thibault, C.; Campbell, B.J.; Hart, C.A.; Kertesz, M.A. Transcriptomic Analysis of the Sulfate Starvation Response of Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 6743–6750. [Google Scholar] [CrossRef]
- Park, C.; Shin, B.; Park, W. Protective Role of Bacterial Alkanesulfonate Monooxygenase under Oxidative Stress. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef]
- Meyer, Y.; Buchanan, B.B.; Vignols, F.; Reichheld, J.-P. Thioredoxins and Glutaredoxins: Unifying Elements in Redox Biology. Annu. Rev. Genet. 2009, 43, 335–367. [Google Scholar] [CrossRef]
- Zeller, T.; Klug, G. Thioredoxins in bacteria: Functions in oxidative stress response and regulation of thioredoxin genes. Naturwissenschaften 2006, 93, 259–266. [Google Scholar] [CrossRef]
- D’Autréaux, B.; Toledano, M.B. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Hammel, K.E.; Cornwell, K.L.; Buchanan, B.B. Ferredoxin/flavoprotein-linked pathway for the reduction of thioredoxin. Proc. Natl. Acad. Sci. USA 1983, 80, 3681–3685. [Google Scholar] [CrossRef] [PubMed]
- Kashima, Y. Alkyl Hydroperoxide Reductase Dependent on Thioredoxin-Like Protein from Pyrococcus horikoshii. J. Biochem. 2003, 134, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Sarin, R.; Sharma, Y.D. Thioredoxin system in obligate anaerobe Desulfovibrio desulfuricans: Identification and characterization of a novel thioredoxin 2. Gene 2006, 376, 107–115. [Google Scholar] [CrossRef]
- Hernandez, H.; Jaquez, O.A.; Hamill, M.J.; Elliott, S.J.; Drennan, C.L. Thioredoxin Reductase from Thermoplasma acidophilum: A New Twist on Redox Regulation†,‡. Biochemistry 2008, 47, 9728–9737. [Google Scholar] [CrossRef]
- Hosoya-Matsuda, N.; Inoue, K.; Hisabori, T. Roles of Thioredoxins in the Obligate Anaerobic Green Sulfur Photosynthetic Bacterium Chlorobaculum tepidum. Mol. Plant 2009, 2, 336–343. [Google Scholar] [CrossRef]
- Pieulle, L.; Stocker, P.; Vinay, M.; Nouailler, M.; Vita, N.; Brasseur, G.; Garcin, E.B.; Sebban-Kreuzer, C.; Dolla, A. Study of the Thiol/Disulfide Redox Systems of the Anaerobe Desulfovibrio vulgaris Points Out Pyruvate:Ferredoxin Oxidoreductase as a New Target for Thioredoxin 1. J. Biol. Chem. 2011, 286, 7812–7821. [Google Scholar] [CrossRef]
- McCarver, A.C.; Lessner, D.J. Molecular characterization of the thioredoxin system from Methanosarcina acetivorans. FEBS J. 2014, 281, 4598–4611. [Google Scholar] [CrossRef]
- Norambuena, J.; Flores, R.; Cardenas, J.P.; Quatrini, R.; Chávez, R.; Levicán, G. Thiol/Disulfide System Plays a Crucial Role in Redox Protection in the Acidophilic Iron-Oxidizing Bacterium Leptospirillum ferriphilum. PLoS ONE 2012, 7, e44576. [Google Scholar] [CrossRef]
- Cha, M.-K.; Kim, H.-K.; Kim, I.-H.; Johnston, J.A.; Wang, L.-M.; Hanson, E.P.; Sun, X.-J.; White, M.F.; Oakes, S.A.; Pierce, J.H.; et al. Thioredoxin-linked “Thiol Peroxidase” from Periplasmic Space of Escherichia coli. J. Biol. Chem. 1995, 270, 28635–28641. [Google Scholar] [CrossRef]
- Atack, J.M.; Harvey, P.; Jones, M.A.; Kelly, D.J. The Campylobacter jejuni Thiol Peroxidases Tpx and Bcp Both Contribute to Aerotolerance and Peroxide-Mediated Stress Resistance but Have Distinct Substrate Specificities. J. Bacteriol. 2008, 190, 5279–5290. [Google Scholar] [CrossRef] [PubMed]
- Ruddock, L.W.; Klappa, P. Oxidative stress: Protein folding with a novel redox switch. Curr. Biol. 1999, 9, R400–R402. [Google Scholar] [CrossRef][Green Version]
- Carmel-Harel, O.; Storz, G. Roles of the Glutathione- and Thioredoxin-Dependent Reduction Systems in the Escherichia coli and Saccharomyces cerevisiae Responses to Oxidative Stress. Annu. Rev. Microbiol. 2000, 54, 439–461. [Google Scholar] [CrossRef] [PubMed]
- Ritz, D.; Beckwith, J. Roles of Thiol-Redox Pathways in Bacteria. Annu. Rev. Microbiol. 2001, 55, 21–48. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.D.; Meshnick, S.R.; Eaton, J.W. Superoxide dismutase-rich bacteria. Paradoxical increase in oxidant toxicity. J. Biol. Chem. 1987, 262, 3640–3645. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khaleque, H.N.; Fathollazadeh, H.; González, C.; Shafique, R.; Kaksonen, A.H.; Holmes, D.S.; Watkin, E.L.J. Unlocking Survival Mechanisms for Metal and Oxidative Stress in the Extremely Acidophilic, Halotolerant Acidihalobacter Genus. Genes 2020, 11, 1392. https://doi.org/10.3390/genes11121392
Khaleque HN, Fathollazadeh H, González C, Shafique R, Kaksonen AH, Holmes DS, Watkin ELJ. Unlocking Survival Mechanisms for Metal and Oxidative Stress in the Extremely Acidophilic, Halotolerant Acidihalobacter Genus. Genes. 2020; 11(12):1392. https://doi.org/10.3390/genes11121392
Chicago/Turabian StyleKhaleque, Himel Nahreen, Homayoun Fathollazadeh, Carolina González, Raihan Shafique, Anna H. Kaksonen, David S. Holmes, and Elizabeth L.J. Watkin. 2020. "Unlocking Survival Mechanisms for Metal and Oxidative Stress in the Extremely Acidophilic, Halotolerant Acidihalobacter Genus" Genes 11, no. 12: 1392. https://doi.org/10.3390/genes11121392
APA StyleKhaleque, H. N., Fathollazadeh, H., González, C., Shafique, R., Kaksonen, A. H., Holmes, D. S., & Watkin, E. L. J. (2020). Unlocking Survival Mechanisms for Metal and Oxidative Stress in the Extremely Acidophilic, Halotolerant Acidihalobacter Genus. Genes, 11(12), 1392. https://doi.org/10.3390/genes11121392







