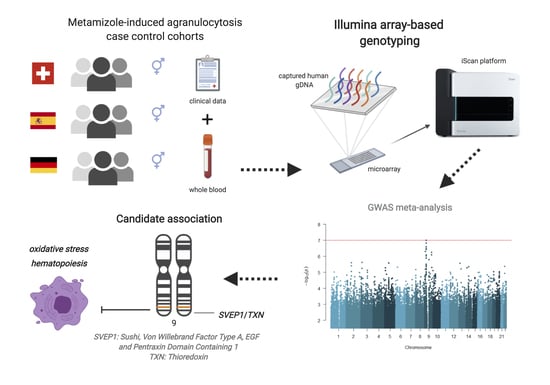
Genome-Wide Association Study of Metamizole-Induced Agranulocytosis in European Populations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Statement
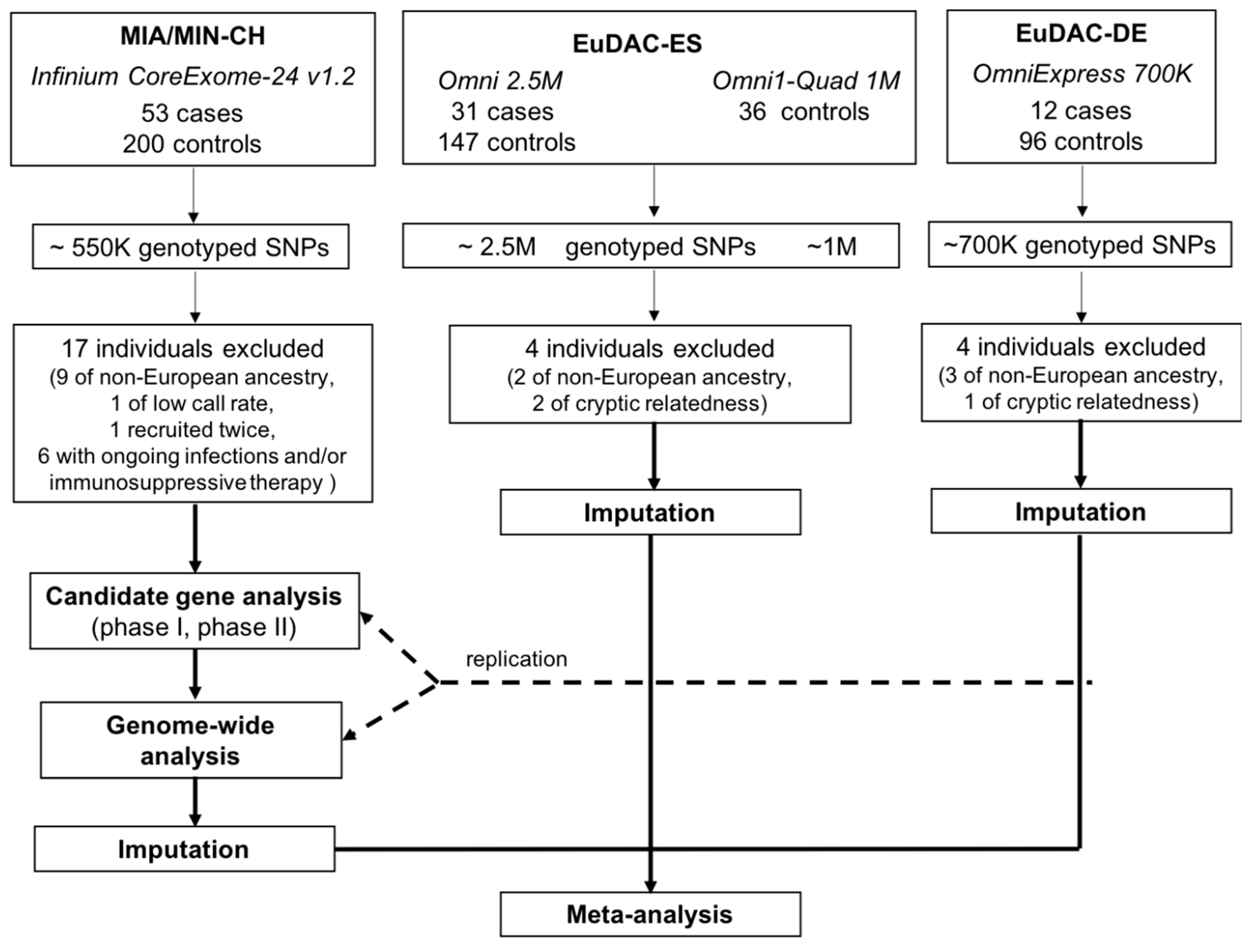
2.2. Study Design and Participants
2.3. Genotype Data and Quality Control
2.4. Multidimensional Scaling and Identification of Genetic Outliers
2.5. Imputation
2.6. Candidate Gene and Genome-Wide Association Analyses
3. Results
3.1. Cohort Characteristics
3.2. Association Analyses in the Discovery Cohort (MIA/MIN-CH and MIA-CH)
3.2.1. Candidate Gene Analyses
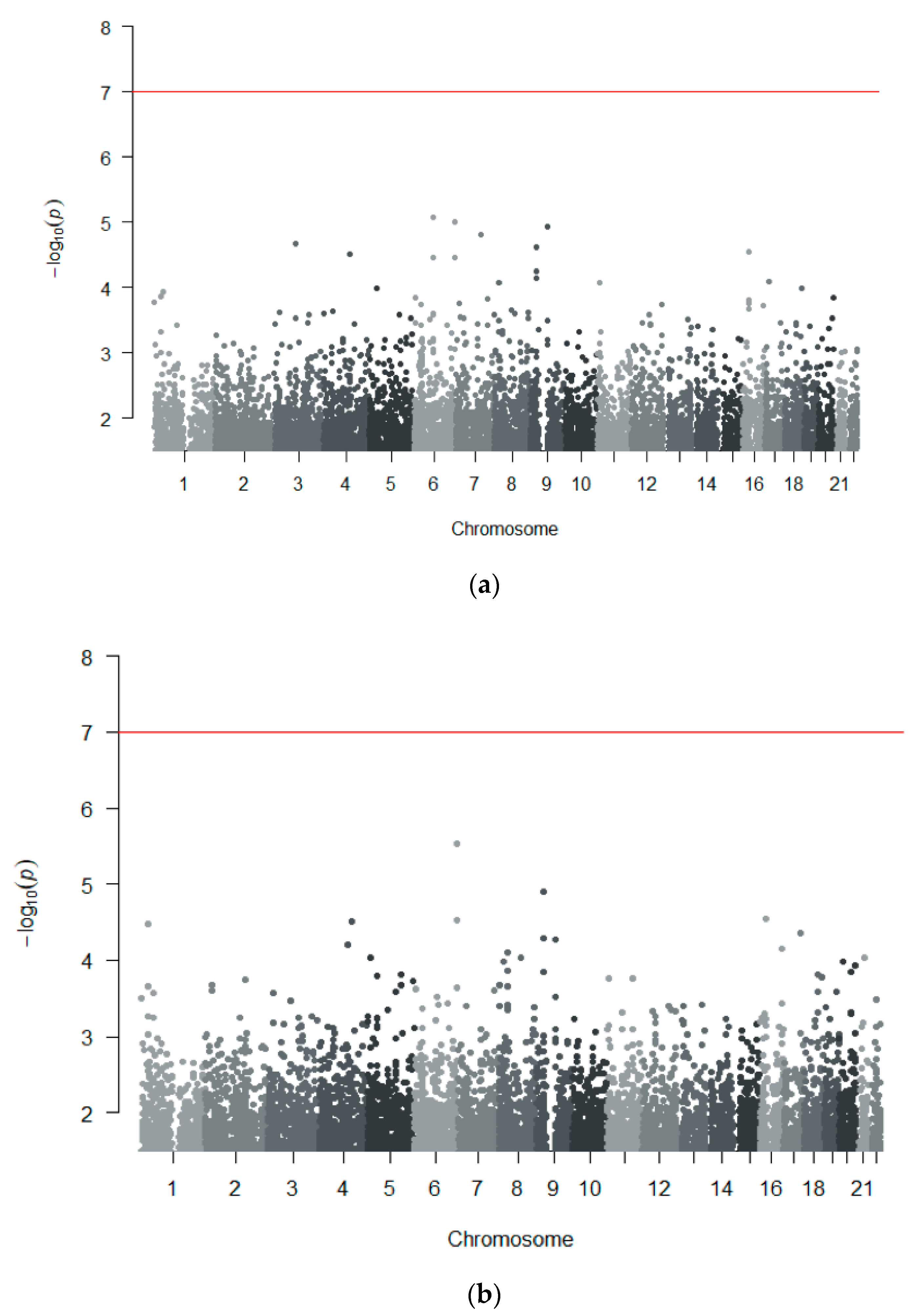
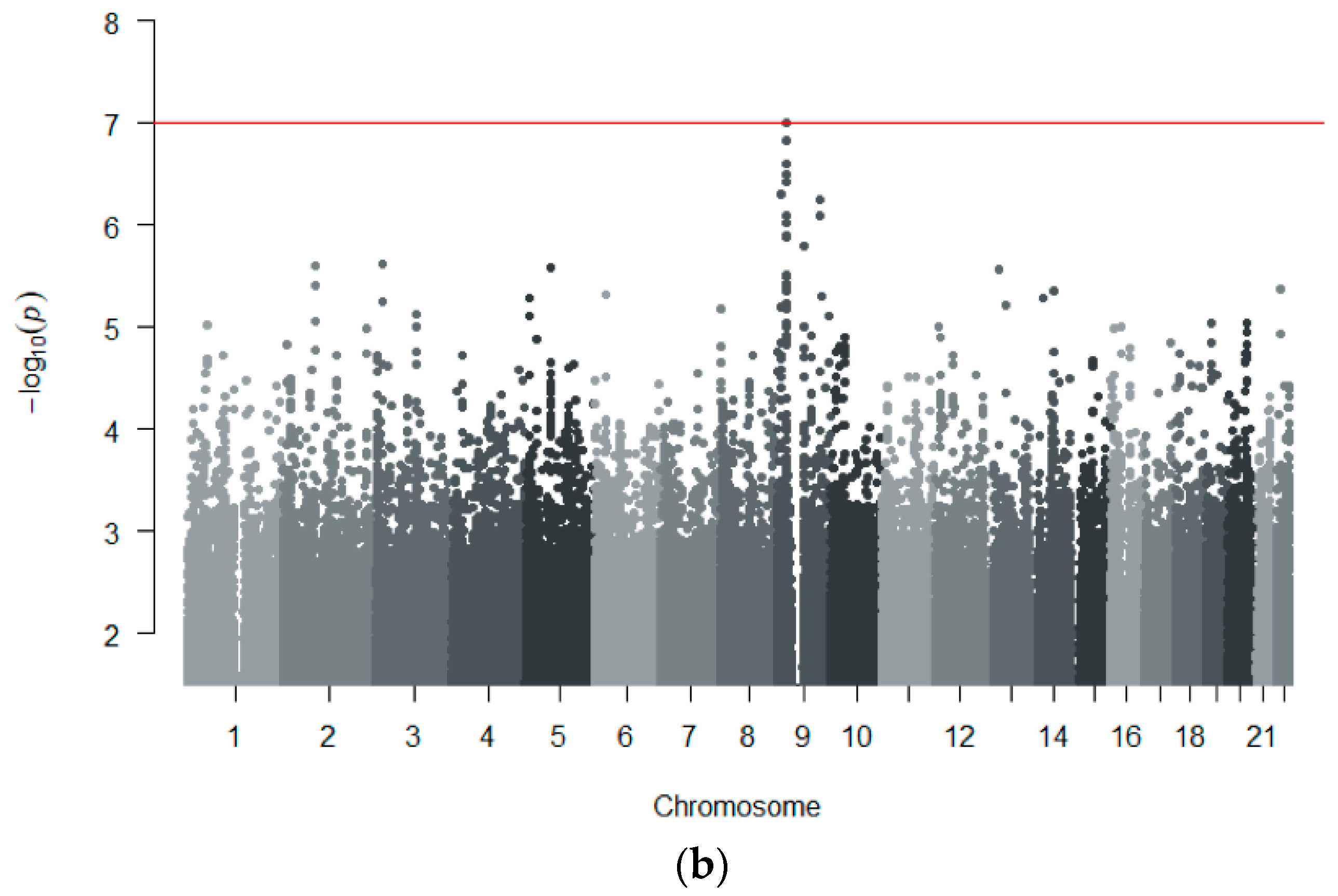
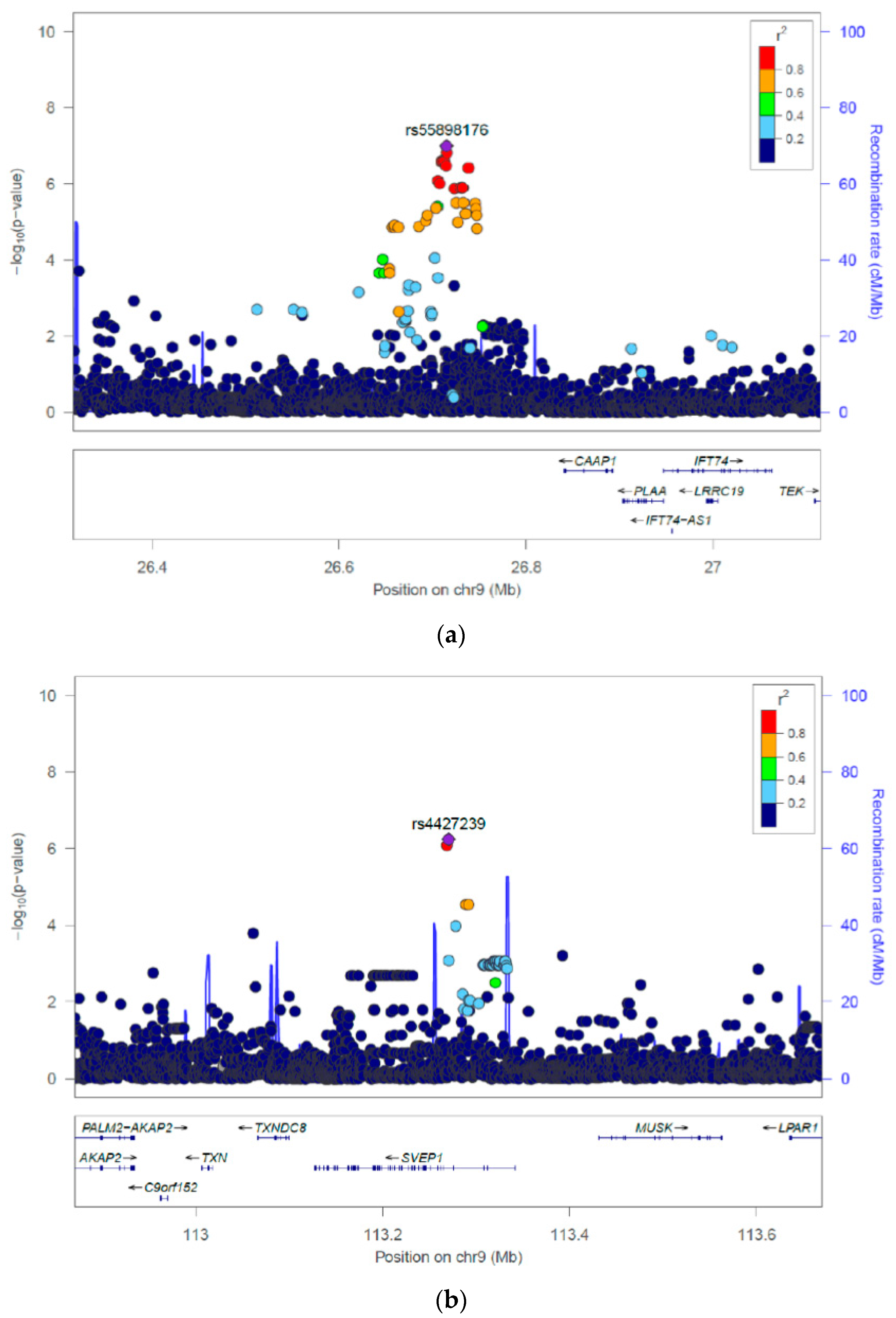
3.2.2. Genome-Wide Association Analyses
3.2.3. Replication in Independent Cohorts
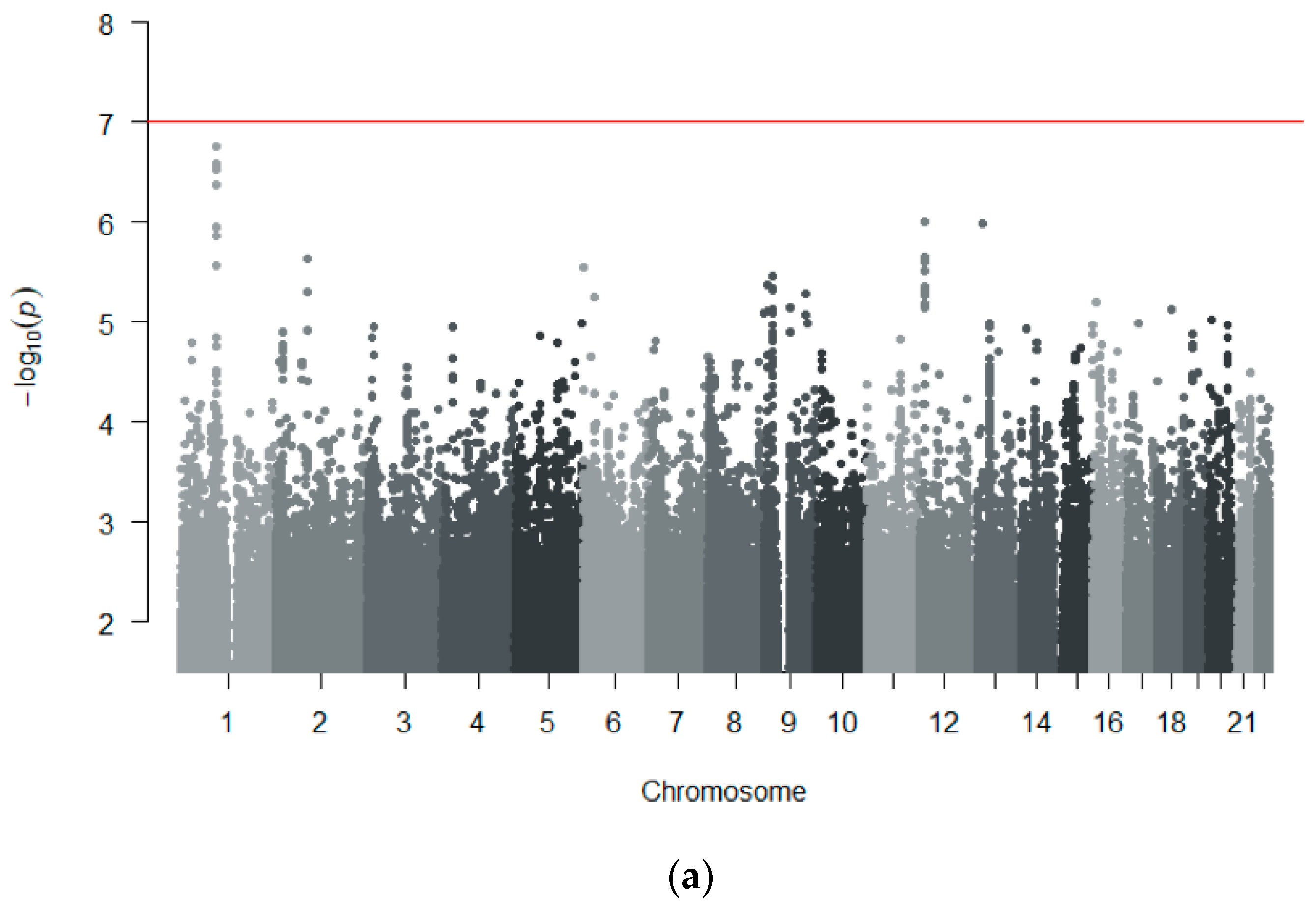
3.3. GWAS Meta-Analysis across Independent Cohorts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arellano, F.; Sacristán, J.A. Metamizole: Reassessment of its therapeutic role. Eur. J. Clin. Pharmacol. 1990, 38, 617–619. [Google Scholar] [CrossRef]
- Andrade, S.E.; Martinez, C.; Walker, A.M. Comparative safety evaluation of non-narcotic analgesics. J. Clin. Epidemiol. 1998, 51, 1357–1365. [Google Scholar] [CrossRef]
- Sánchez, S.; De la Lastra, C.A.; Ortiz, P.; Motilva, V.; Martín, M.J. Gastrointestinal tolerability of metamizol, acetaminophen, and diclofenac in subchronic treatment in rats. Dig. Dis. Sci. 2002, 47, 2791–2798. [Google Scholar] [CrossRef]
- Hedenmalm, K.; Spigset, O. Agranulocytosis and other blood dyscrasias associated with dipyrone (metamizole). Eur. J. Clin. Pharmacol. 2002, 58, 265–274. [Google Scholar] [CrossRef]
- Ibáñez, L.; Vidal, X.; Ballarín, E.; Laporte, J.R. Agranulocytosis associated with dipyrone (metamizol). Eur. J. Clin. Pharmacol. 2005, 60, 821–829. [Google Scholar] [CrossRef]
- Blaser, L.S.; Tramonti, A.; Egger, P.; Haschke, M.; Krähenbühl, S.; Rätz Bravo, A.E. Hematological safety of metamizole: Retrospective analysis of WHO and Swiss spontaneous safety reports. Eur. J. Clin. Pharmacol. 2015, 71, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Blaser, L.; Hassna, H.; Hofmann, S.; Holbro, A.; Haschke, M.; Rätz Bravo, A.; Zeller, A.; Krähenbühl, S.; Taegtmeyer, A. Leucopenia associated with metamizole: A case-control study. Swiss Med. Wkly. 2017, 147, w14438. [Google Scholar] [CrossRef] [PubMed]
- Andrès, E.; Maloisel, F. Idiosyncratic drug-induced agranulocytosis or acute neutropenia. Curr. Opin. Hematol. 2008, 15, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Van Der Klauw, M.M.; Goudsmit, R.; Halie, M.R.; Van’t Veer, M.B.; Herings, R.M.C.; Wilson, J.H.P.; Stricker, B.H.C. A population-based case-cohort study of drug-associated agranulocytosis. Arch. Intern. Med. 1999, 159, 369–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basak, G.W.; Drozd-Sokołowska, J.; Wiktor-Jedrzejczak, W. Update on the incidence of metamizole sodium-induced blood dyscrasias in Poland. J. Int. Med. Res. 2010, 38, 1374–1380. [Google Scholar] [CrossRef]
- Chan, T.Y.K.; Chan, A.W.K. Aminopyrine-induced blood dyscrasias-still a problem in many parts of the world. Pharmacoepidemiol. Drug Saf. 1996, 5, 215–219. [Google Scholar] [CrossRef]
- Stammschulte, T.; Ludwig, W.D.; Mühlbauer, B.; Bronder, E.; Gundert-Remy, U. Metamizole (dipyrone)-associated agranulocytosis. An analysis of German spontaneous reports 1990–2012. Eur. J. Clin. Pharmacol. 2015, 71, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Pearson, E.R. Insights from Genome-Wide Association Studies of Drug Response. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 299–310. [Google Scholar] [CrossRef]
- Daly, A.K. Genome-wide association studies in pharmacogenomics. Nat. Rev. Genet. 2010, 11, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Wadelius, M.; Eriksson, N.; Kreutz, R.; Bondon-Guitton, E.; Ibañez, L.; Carvajal, A.; Lucena, M.I.; Sancho Ponce, E.; Molokhia, M.; Martin, J.; et al. Sulfasalazine-Induced Agranulocytosis Is Associated With the Human Leukocyte Antigen Locus. Clin. Pharmacol. Ther. 2017, 103, 843–853. [Google Scholar] [CrossRef] [Green Version]
- Hallberg, P.; Eriksson, N.; Ibañez, L.; Bondon-Guitton, E.; Kreutz, R.; Carvajal, A.; Lucena, M.I.; Ponce, E.S.; Molokhia, M.; Martin, J.; et al. Genetic variants associated with antithyroid drug-induced agranulocytosis: A genome-wide association study in a European population. Lancet Diabetes Endocrinol. 2016, 4, 507–516. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, J.I.; Fredrik Jarskog, L.; Hilliard, C.; Alfirevic, A.; Duncan, L.; Fourches, D.; Huang, H.; Lek, M.; Neale, B.M.; Ripke, S.; et al. Clozapine-induced agranulocytosis is associated with rare HLA-DQB1 and HLA-B alleles. Nat. Commun. 2014, 5, 1–9. [Google Scholar] [CrossRef]
- Athanasiou, M.C.; Dettling, M.; Cascorbi, I.; Mosyagin, I.; Salisbury, B.A.; Pierz, K.A.; Zou, W.; Whalen, H.; Malhotra, A.K.; Lencz, T.; et al. Candidate gene analysis identifies a polymorphism in HLA-DQB1 associated with clozapine-induced agranulocytosis. J. Clin. Psychiatry 2011, 72, 458–463. [Google Scholar] [CrossRef]
- Pavlos, R.; Mallal, S.; Ostrov, D.; Buus, S.; Metushi, I.; Peters, B.; Phillips, E. T Cell–Mediated Hypersensitivity Reactions to Drugs. Annu. Rev. Med. 2015, 66, 439–454. [Google Scholar] [CrossRef] [Green Version]
- Rudin, D.; Lanzilotto, A.; Bachmann, F.; Housecroft, C.E.; Constable, E.C.; Drewe, J.; Haschke, M.; Krähenbühl, S. Non-immunological toxicological mechanisms of metamizole-associated neutropenia in HL60 cells. Biochem. Pharmacol. 2019, 163, 345–356. [Google Scholar] [CrossRef]
- Rudin, D.; Haschke, M.; Krahenbuhl, S. Mechanisms of cytotoxicity involved in metamizole-induced neutropenia. Clin. Toxicol. 2017, 55, 371–544. [Google Scholar] [CrossRef]
- Rudin, D.; Roos, N.J.; Duthaler, U.; Krähenbühl, S. Toxicity of metamizole on differentiating HL60 cells and human neutrophil granulocytes. Toxicology 2019, 426, 152254. [Google Scholar] [CrossRef] [PubMed]
- Rudin, D.; Spoendlin, J.; Cismaru, A.L.; Liakoni, E.; Bonadies, N.; Amstutz, U.; Meier, C.R.; Krähenbühl, S.; Haschke, M. Metamizole-associated neutropenia: Comparison of patients with neutropenia and metamizole-tolerant patients. Eur. J. Intern. Med. 2019, 68, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Igo, R.P.; Cooke Bailey, J.N.; Romm, J.; Haines, J.L.; Wiggs, J.L. Quality control for the illumina humanexome beadchip. Curr. Protoc. Hum. Genet. 2016, 90, 2–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coleman, J.R.I.; Euesden, J.; Patel, H.; Folarin, A.A.; Newhouse, S.; Breen, G. Quality control, imputation and analysis of genome-wide genotyping data from the Illumina HumanCoreExome microarray. Brief. Funct. Genom. 2016, 15, 298–304. [Google Scholar] [CrossRef] [Green Version]
- Turner, S.; Armstrong, L.L.; Bradford, Y.; Carlsony, C.S.; Crawford, D.C.; Crenshaw, A.T.; de Andrade, M.; Doheny, K.F.; Haines, J.L.; Hayes, G.; et al. Quality control procedures for genome-wide association studies. Curr. Protoc. Hum. Genet. 2011, 68, 1.19.1–1.19.18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, N.; Lucas, G.; Hysi, P. Big data challenges in bone research: Genome-wide association studies and next-generation sequencing. Bonekey Rep. 2015, 4, 635. [Google Scholar] [CrossRef]
- Marees, A.T.; de Kluiver, H.; Stringer, S.; Vorspan, F.; Curis, E.; Marie-Claire, C.; Derks, E.M. A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int. J. Methods Psychiatr. Res. 2018, 27, e1608. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Reddy, S.; Madireddy, S.; Nair, P. Association of Genetic Markers Contributing to Dyslexia Susceptibility in Indian Population. J. Neurol. Neurosci. 2016. [Google Scholar] [CrossRef]
- Graffelman, J.; Nelson, S.; Gogarten, S.M.; Weir, B.S. Exact inference for Hardy-Weinberg proportions with missing genotypes: Single and multiple imputation. G3 Genes Genomes Genet. 2015, 5, 2365–2373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyagawa, T.; Nishida, N.; Ohashi, J.; Kimura, R.; Fujimoto, A.; Kawashima, M.; Koike, A.; Sasaki, T.; Tanii, H.; Otowa, T.; et al. Appropriate data cleaning methods for genome-wide association study. J. Hum. Genet. 2008, 53, 886–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graffelman, J.; Jain, D.; Weir, B. A genome-wide study of Hardy–Weinberg equilibrium with next generation sequence data. Hum. Genet. 2017, 136, 727–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.X.; Yeung, J.M.Y.; Cherny, S.S.; Sham, P.C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 2012, 131, 747–756. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Zhi, D.; Zhang, K.; Gao, G.; Limdi, N.A.; Liu, N. Practical consideration of genotype imputation: Sample size, window size, reference choice, and untyped rate. Stat. Interface 2011, 4, 339–351. [Google Scholar] [PubMed]
- Li, Y.; Willer, C.J.; Ding, J.; Scheet, P.; Abecasis, G.R. MaCH: Using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010, 34, 816–834. [Google Scholar] [CrossRef] [Green Version]
- Marchini, J.; Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010, 11, 499–511. [Google Scholar] [CrossRef]
- Das, S.; Forer, L.; Schönherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, S.; Das, S.; Kretzschmar, W.; Delaneau, O.; Wood, A.; Teumer, A.; Kang, H.M.; Fuchsberger, C.; Danecek, P.; Sharp, K.; et al. A reference panel of 64,976 haplotypes for genotype imputation. bioRxiv 2015. [Google Scholar] [CrossRef]
- Freedman, M.L.; Reich, D.; Penney, K.L.; McDonald, G.J.; Mignault, A.A.; Patterson, N.; Gabriel, S.B.; Topol, E.J.; Smoller, J.W.; Pato, C.N.; et al. Assessing the impact of population stratification on genetic association studies. Nat. Genet. 2004, 36, 388–393. [Google Scholar] [CrossRef]
- Devlin, B.; Roeder, K. Genomic control for association studies. Biometrics 1999, 55, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Legge, S.E.; Hamshere, M.L.; Ripke, S.; Pardinas, A.F.; Goldstein, J.I.; Rees, E.; Richards, A.L.; Leonenko, G.; Jorskog, L.F.; Chambert, K.D.; et al. Genome-wide common and rare variant analysis provides novel insights into clozapine-associated neutropenia. Mol. Psychiatry 2017, 22, 1502–1508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, T.; Ikeda, M.; Mushiroda, T.; Ozeki, T.; Kondo, K.; Shimasaki, A.; Kawase, K.; Hashimoto, S.; Yamamori, H.; Yasuda, Y.; et al. Pharmacogenomic Study of Clozapine-Induced Agranulocytosis/Granulocytopenia in a Japanese Population. Biol. Psychiatry 2016, 80, 636–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.L.; Shih, S.R.; Wang, P.W.; Lin, Y.C.; Chu, C.C.; Lin, J.H.; Chen, S.C.; Chang, C.C.; Huang, T.S.; Tsai, K.S.; et al. Genetic determinants of antithyroid drug-induced agranulocytosis by human leukocyte antigen genotyping and genome-wide association study. Nat. Commun. 2015, 6, 7633. [Google Scholar] [CrossRef] [PubMed]
- Plantinga, T.S.; Arts, P.; Knarren, G.H.; Mulder, A.H.; Wakelkamp, I.M.; Hermus, A.R.; Joosten, L.A.; Netea, M.G.; Bisschop, P.H.; de Herder, W.W.; et al. Rare NOX3 Variants Confer Susceptibility to Agranulocytosis During Thyrostatic Treatment of Graves’ Disease. Clin. Pharmacol. Ther. 2017, 102, 1017–1024. [Google Scholar] [CrossRef]
- Ostrousky, O.; Meged, S.; Loewenthal, R.; Valevski, A.; Weizman, A.; Carp, H.; Gazit, E. NQO2 gene is associated with clozapine-induced agranulocytosis. Tissue Antigens 2003, 62, 483–491. [Google Scholar] [CrossRef]
- Van Der Weide, K.; Loovers, H.; Pondman, K.; Bogers, J.; Van Der Straaten, T.; Langemeijer, E.; Cohen, D.; Commandeur, J.; Van Der Weide, J. Genetic risk factors for clozapine-induced neutropenia and agranulocytosis in a Dutch psychiatric population. Pharm. J. 2017, 17, 471–478. [Google Scholar] [CrossRef]
- García-Martín, E.; Esguevillas, G.; Blanca-López, N.; García-Menaya, J.; Blanca, M.; Amo, G.; Canto, G.; Martínez, C.; Cordobés, C.; Agúndez, J.A.G. Genetic determinants of metamizole metabolism modify the risk of developing anaphylaxis. Pharmacogenet. Genom. 2015, 25, 462–464. [Google Scholar] [CrossRef]
- Skelly, D.A.; Squiers, G.T.; McLellan, M.A.; Bolisetty, M.T.; Robson, P.; Rosenthal, N.A.; Pinto, A.R. Single-Cell Transcriptional Profiling Reveals Cellular Diversity and Intercommunication in the Mouse Heart. Cell Rep. 2018, 22, 600–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panagiotou, O.A.; Ioannidis, J.P.A.; Hirschhorn, J.N.; Abecasis, G.R.; Frayling, T.M.; McCarthy, M.I.; Lindgren, C.M.; Beaty, T.H.; Eriksson, N.; Polychronakos, C.; et al. What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int. J. Epidemiol. 2012, 41, 273–286. [Google Scholar] [CrossRef]
- Willer, C.J.; Li, Y.; Abecasis, G.R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010, 26, 2190–2191. [Google Scholar] [CrossRef] [PubMed]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Zheng, J.; Zhang, Q.; Hou, P.; Zhu, F.; Yang, J.; Li, W.; Chen, P.; Liu, S.; Zhang, B.; et al. Association of HLA-B and HLA-DRB1 polymorphisms with antithyroid drug-induced agranulocytosis in a Han population from northern China. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Westra, H.J.; Peters, M.J.; Esko, T.; Yaghootkar, H.; Schurmann, C.; Kettunen, J.; Christiansen, M.W.; Fairfax, B.P.; Schramm, K.; Powell, J.E.; et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013, 45, 1238–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traynor, K. Suvorexant approved for insomnia. Am. J. Health Pharm. 2014, 71, 1524. [Google Scholar] [CrossRef] [PubMed]
- Lappalainen, T.; Sammeth, M.; Friedländer, M.R.; ’T Hoen, P.A.C.; Monlong, J.; Rivas, M.A.; Gonzàlez-Porta, M.; Kurbatova, N.; Griebel, T.; Ferreira, P.G.; et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013, 501, 506–511. [Google Scholar] [CrossRef]
- Ardlie, K.G.; DeLuca, D.S.; Segrè, A.V.; Sullivan, T.J.; Young, T.R.; Gelfand, E.T.; Trowbridge, C.A.; Maller, J.B.; Tukiainen, T.; Lek, M.; et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef] [Green Version]
- Andiappan, A.K.; Melchiotti, R.; Poh, T.Y.; Nah, M.; Puan, K.J.; Vigano, E.; Haase, D.; Yusof, N.; San Luis, B.; Lum, J.; et al. Genome-wide analysis of the genetic regulation of gene expression in human neutrophils. Nat. Commun. 2015, 6, 7971. [Google Scholar] [CrossRef] [Green Version]
- Carithers, L.J.; Moore, H.M. The Genotype-Tissue Expression (GTEx) Project. Biopreserv. Biobank. 2015, 13, 307–308. [Google Scholar] [CrossRef]
- Carvalho-Silva, D.; Pierleoni, A.; Pignatelli, M.; Ong, C.K.; Fumis, L.; Karamanis, N.; Carmona, M.; Faulconbridge, A.; Hercules, A.; McAuley, E.; et al. Open Targets Platform: New developments and updates two years on. Nucleic Acids Res. 2019, 47, D1056–D1065. [Google Scholar] [CrossRef]
- Daly, A. Human Leukocyte Antigen (HLA) Pharmacogenomic Tests: Potential and Pitfalls. Curr. Drug Metab. 2014, 15, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Johnston, A.; Uetrecht, J. Current understanding of the mechanisms of idiosyncratic drug-induced agranulocytosis. Expert Opin. Drug Metab. Toxicol. 2015, 11, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Tesfa, D.; Keisu, M.; Palmblad, J. Idiosyncratic drug-induced agranulocytosis: Possible mechanisms and management. Am. J. Hematol. 2009, 84, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Mallal, S.; Phillips, E.; Carosi, G.; Molina, J.-M.; Workman, C.; Tomažič, J.; Jägel-Guedes, E.; Rugina, S.; Kozyrev, O.; Cid, J.F.; et al. HLA-B*5701 Screening for Hypersensitivity to Abacavir. N. Engl. J. Med. 2008, 358, 568–579. [Google Scholar] [CrossRef] [Green Version]
- Hetherington, S.; Hughes, A.R.; Mosteller, M.; Shortino, D.; Baker, K.L.; Spreen, W.; Lai, E.; Davies, K.; Handley, A.; Dow, D.J.; et al. Genetic variations in HLA-B region and hypersensitivity reactions to abacavir. Lancet 2002, 359, 1121–1122. [Google Scholar] [CrossRef]
- Cheung, C.L.; Sing, C.W.; Tang, C.S.M.; Cheng, V.K.F.; Pirmohamed, M.; Choi, C.H.; Hung, C.S.; Lau, E.Y.F.; Lee, K.F.; Mak, M.W.H.; et al. HLA-B∗38:02:01 predicts carbimazole/methimazole-induced agranulocytosis. Clin. Pharmacol. Ther. 2016, 99, 555–561. [Google Scholar] [CrossRef]
- Spraggs, C.F.; Budde, L.R.; Briley, L.P.; Bing, N.; Cox, C.J.; King, K.S.; Whittaker, J.C.; Mooser, V.E.; Preston, A.J.; Stein, S.H.; et al. HLA-DQA1*02:01 is a major risk factor for lapatinib-induced hepatotoxicity in women with advanced breast cancer. J. Clin. Oncol. 2011, 29, 667–673. [Google Scholar] [CrossRef]
- Cismaru, A.L.; Grimm, L.; Rudin, D.; Ibañez, L.; Liakoni, E.; Bonadies, N.; Kreutz, R.; Hallberg, P.; Wadelius, M.; Haschke, M.; et al. High-Throughput Sequencing to Investigate Associations Between HLA Genes and Metamizole-Induced Agranulocytosis. Front. Genet. 2020, 11. [Google Scholar] [CrossRef]
- Astle, W.J.; Elding, H.; Jiang, T.; Allen, D.; Ruklisa, D.; Mann, A.L.; Mead, D.; Bouman, H.; Riveros-Mckay, F.; Kostadima, M.A.; et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell 2016, 167, 1415–1429.e19. [Google Scholar] [CrossRef] [Green Version]
- Kichaev, G.; Bhatia, G.; Loh, P.R.; Gazal, S.; Burch, K.; Freund, M.K.; Schoech, A.; Pasaniuc, B.; Price, A.L. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am. J. Hum. Genet. 2019, 104, 65–75. [Google Scholar] [CrossRef] [Green Version]
- Canela-Xandri, O.; Rawlik, K.; Tenesa, A. An atlas of genetic associations in UK Biobank. Nat. Genet. 2018, 50, 1593–1599. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, X.; Hu, J.; Lian, X.; Zhu, T.; Zhang, H.; Gu, N.; Li, J. PTPRO Promotes Oxidized Low-Density Lipoprotein Induced Oxidative Stress and Cell Apoptosis through Toll-Like Receptor 4/Nuclear Factor κb Pathway. Cell. Physiol. Biochem. 2017, 42, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Collet, J.F.; Messens, J. Structure, function, and mechanism of thioredoxin proteins. Antioxid. Redox Signal. 2010, 13, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, A.; Hamuro, J.; Nakamura, H.; Kondo, N.; Hirabayashi, Y.; Ishizaki-Koizumi, S.; Hirakawa, T.; Inoue, T.; Yodoi, J. Overexpression of human thioredoxin in transgenic mice controls oxidative stress and life span. Antioxid. Redox Signal. 2002, 4, 693–696. [Google Scholar] [CrossRef] [PubMed]
- Lillig, C.H.; Holmgren, A. Thioredoxin and related molecules-From biology to health and disease. Antioxid. Redox Signal. 2007, 9, 25–47. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Uhlen, M.; Karlsson, M.J.; Zhong, W.; Tebani, A.; Pou, C.; Mikes, J.; Lakshmikanth, T.; Forsström, B.; Edfors, F.; Odeberg, J.; et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 2019, 366, eaax9198. [Google Scholar] [CrossRef]
- Matsuzawa, A. Thioredoxin and redox signaling: Roles of the thioredoxin system in control of cell fate. Arch. Biochem. Biophys. 2017, 617, 101–105. [Google Scholar] [CrossRef]
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014, 66, 75–87. [Google Scholar] [CrossRef]
- Lu, J.; Holmgren, A. Thioredoxin system in cell death progression. Antioxid. Redox Signal. 2012, 17, 1738–1747. [Google Scholar] [CrossRef]
- Ravi, D.; Muniyappa, H.; Das, K.C. Endogenous thioredoxin is required for redox cycling of anthracyclines and p53-dependent apoptosis in cancer cells. J. Biol. Chem. 2005, 280, 40084–40096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Periasamy, P.; Tran, V.; O’Neill, H.C. Identification of genes which regulate stromadependent in vitro hematopoiesis. PLoS ONE 2018, 13, e0205583. [Google Scholar] [CrossRef] [PubMed]
- Sato-Nishiuchi, R.; Nakano, I.; Ozawa, A.; Sato, Y.; Takeichi, M.; Kiyozumi, D.; Yamazaki, K.; Yasunaga, T.; Futaki, S.; Sekiguchi, K. Polydom/SVEP1 is a ligand for integrin α9β1. J. Biol. Chem. 2012, 287, 25615–25630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schreiber, T.D.; Steinl, C.; Essl, M.; Abele, H.; Geiger, K.; Müller, C.A.; Aicher, W.K.; Klein, G. The integrin α9β1 on hematopoietic stem and progenitor cells: Involvement in cell adhesion, proliferation and differentiation. Haematologica 2009, 94, 1493–1501. [Google Scholar] [CrossRef]
- Chen, C.; Huang, X.; Atakilit, A.; Zhu, Q.S.; Corey, S.J.; Sheppard, D. The Integrin α9β1 Contributes to Granulopoiesis by Enhancing Granulocyte Colony-Stimulating Factor Receptor Signaling. Immunity 2006, 25, 895–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Johansson, E.; Miller, M.L.; Jänicke, R.U.; Ferguson, D.J.; Plas, D.; Meller, J.; Anderson, M.W. Identification of a conserved anti-apoptotic protein that modulates the mitochondrial apoptosis pathway. PLoS ONE 2011, 6, e25284. [Google Scholar] [CrossRef]
- Wright, G.E.B.; Amstutz, U.; Drögemöller, B.I.; Shih, J.; Rassekh, S.R.; Hayden, M.R.; Carleton, B.C.; Ross, C.J.D.; Visscher, H.; Aminkeng, F.; et al. Pharmacogenomics of Vincristine-Induced Peripheral Neuropathy Implicates Pharmacokinetic and Inherited Neuropathy Genes. Clin. Pharmacol. Ther. 2019, 105, 402–410. [Google Scholar] [CrossRef] [Green Version]
- Usami, Y.; Wu, Y.; Göttlinger, H.G. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 2015, 526, 218–223. [Google Scholar] [CrossRef] [Green Version]
- Regev, A.; Teichmann, S.A.; Lander, E.S.; Amit, I.; Benoist, C.; Birney, E.; Bodenmiller, B.; Campbell, P.; Carninci, P.; Clatworthy, M.; et al. The human cell atlas. Elife 2017, 6. [Google Scholar] [CrossRef]
- Popescu, D.M.; Botting, R.A.; Stephenson, E.; Green, K.; Webb, S.; Jardine, L.; Calderbank, E.F.; Polanski, K.; Goh, I.; Efremova, M.; et al. Decoding human fetal liver haematopoiesis. Nature 2019, 574, 365–371. [Google Scholar] [CrossRef]
- Setty, M.; Kiseliovas, V.; Levine, J.; Gayoso, A.; Mazutis, L.; Pe’er, D. Palantir characterizes cell fate continuities in human hematopoiesis. bioRxiv 2018, 385328. [Google Scholar] [CrossRef]
- Daly, A.K. Using genome-wide association studies to identify genes important in serious adverse drug reactions. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Diouf, B.; Crews, K.R.; Lew, G.; Pei, D.; Cheng, C.; Bao, J.; Zheng, J.J.; Yang, W.; Fan, Y.; Wheeler, H.E.; et al. Association of an inherited genetic variant with vincristine-related peripheral neuropathy in children with acute lymphoblastic leukemia. JAMA J. Am. Med. Assoc. 2015, 313, 815–823. [Google Scholar] [CrossRef]
- Aminkeng, F.; Bhavsar, A.P.; Visscher, H.; Rassekh, S.R.; Li, Y.; Lee, J.W.; Brunham, L.R.; Caron, H.N.; Van Dalen, E.C.; Kremer, L.C.; et al. A coding variant in RARG confers susceptibility to anthracycline-induced cardiotoxicity in childhood cancer. Nat. Genet. 2015, 47, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Kowalec, K.; Wright, G.E.B.; Drögemöller, B.I.; Aminkeng, F.; Bhavsar, A.P.; Kingwell, E.; Yoshida, E.M.; Traboulsee, A.; Marrie, R.A.; Kremenchutzky, M.; et al. Common variation near IRF6 is associated with IFN-β-induced liver injury in multiple sclerosis. Nat. Genet. 2018, 50, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.R.; Bacanu, S.A.; Mosteller, M.; Li, L.; Bowman, C.E.; Roses, A.D.; Lai, E.H.; Ehm, M.G. Genome-wide approaches to identify pharmacogenetic contributions to adverse drug reactions. Pharm. J. 2009, 9, 23–33. [Google Scholar] [CrossRef] [Green Version]
| Cohort | MIA/MIN-CH | EuDAC-DE | EuDAC-ES | |||
|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | |
| N = 45 | N = 191 | N = 12 | N = 92 | N = 29 | N = 181 | |
| ANC < 500/uL | 30 (67) | - | 12 | - | 29 | - |
| Sex, male (%) | 13 (42) | 17 (45) | 4 (33) | 41 (44.5) | 6 (21) | 87 (48) |
| Age, years (%) | ||||||
| <25 | 7 (23) | 1 (3) | 2 (16.6) | NA | 3 (10.3) | NA |
| 25–44 | 11 (35) | 6 (16) | 6 (50) | NA | 6 (21) | NA |
| 45–64 | 10 (32) | 15 (39) | 2 (16.6) | NA | 12 (41.4) | NA |
| 65–74 | 3 (10) | 9 (24) | 2 (16.6) | NA | 5 (17) | NA |
| >74 | - | 7 (18) | - | NA | 3 (10.3) | NA |
| BMI, median (range) | 24 (19–47) | 28 (16–39) | NA | NA | NA | NA |
| Latency time * a/treatment duration b, days | 17 (1–204) | 25 (1–5297) | 33.5 (4–9855) | NA | 11.5 (1–235) | NA |
| CHR | SNP | Alleles (Minor/Major) | BP | MAF Cases MIA/MIN-CH| EuDAC-ES| EuDAC-DE | MAF Controls MIA/MIN-CH| EuDAC-ES| EuDAC-DE | OR [95%] | p-Value | Gene Region | HetISq |
|---|---|---|---|---|---|---|---|---|---|
| 1 | rs11583606 | T/C | 92349247 | 0.10| 014| 0.083 | 0.023| 0.025| 0.027 | 7.0 [3.37–14.5] | 1.72 × 10−7 | TGFBR3 | 0 |
| 1 | rs149072800 | C/T | 92445720 | 0.089| 0.12| 0.083 | 0.020| 0.019| 0.022 | 7.81 [3.57–17.1] | 2.66 × 10−7 | BRDT | 0 |
| 1 | rs146378328 | G/A | 92528047 | 0.089| 0.10| 0.083 | 0.018| 0.019| 0.021 | 8.31 [3.69–18.7] | 2.93 × 10−7 | EPHX4 | 0 |
| 1 | rs75499485 | G/A | 92486274 | 0.089| 0.10| 0.083 | 0.020| 0.019| 0.021 | 7.96 [3.56–17.8] | 4.23 × 10−7 | EPHX4 | 0 |
| 12 | rs112917452 | C/A | 15638858 | 0.067| 0.15| 0.042 | 0.016| 0.027| 0.016 | 7.30 [3.29–16.20] | 9.92 × 10−7 | PTPRO | 0 |
| 12 | rs118135416 | A/G | 15638914 | 0.067| 0.15| 0.042 | 0.016| 0.027| 0.016 | 7.30 [3.29–16.20] | 9.92 × 10−7 | PTPRO | 0 |
| 12 | rs7135120 | T/C | 15626920 | 0.067| 0.15| 0.042 | 0.016| 0.027| 0.016 | 7.30 [3.29–16.20] | 9.92 × 10−7 | PTPRO | 0 |
| 12 | rs143843248 | T/C | 15633812 | 0.067| 0.15| 0.041 | 0.016| 0.027| 0.016 | 7.30 [3.29–16.20] | 9.92 × 10−7 | PTPRO | 0 |
| 13 | rs73163933 | A/G | 33968020 | 0.12| 0.086| 0.21 | 0.036| 0.027| 0.054 | 5.36 [2.73–10.5] | 1.01 × 10−6 | STARD13 | 0 |
| 1 | rs78201766 | G/A | 92379078 | 0.10| 0.14| 0.083 | 0.026| 0.030| 0.027 | 5.65 [2.81–11.34] | 1.12 × 10−6 | TGFBR3 | 0 |
| CHR | SNP | Alleles (Minor/Major) | BP | MAF Cases MIA/MIN-CH| EuDAC-ES| EuDAC-DE | MAF Controls MIA/MIN-CH| EuDAC-ES| EuDAC-DE | OR [95%] | p-Value | Gene Region | HetISq |
|---|---|---|---|---|---|---|---|---|---|
| 9 | rs55898176 | T/C | 26715294 | 0.27| 0.24| 0.25 | 0.065| 0.12| 0.081 | 4.01 [2.41–6.68] | 1.01 × 10−7 | - | 21.7 |
| 9 | rs112223975 | C/G | 26715828 | 0.27| 0.24| 0.25 | 0.065| 0.12| 0.081 | 3.89 [2.34–6.48] | 1.50 × 10−7 | - | 28.6 |
| 9 | rs11790418 | G/A | 26713012 | 0.27| 0.24| 0.25 | 0.065| 0.12| 0.098 | 3.81 [2.29–6.35] | 2.54 × 10−7 | - | 29.3 |
| 9 | rs1434481 | G/C | 26711134 | 0.27| 0.24| 0.25 | 0.065| 0.11| 0.098 | 3.81 [2.29–6.35] | 2.54 × 10−7 | - | 29.3 |
| 9 | rs28475568 | G/C | 26709933 | 0.27| 0.24| 0.25 | 0.065| 0.11| 0.098 | 3.81 [2.29–6.35] | 2.54 × 10−7 | - | 29.3 |
| 9 | rs28649995 | A/G | 26709912 | 0.27| 0.24| 0.25 | 0.065| 0.11| 0.098 | 3.81 [2.29–6.35] | 2.54 × 10−7 | - | 14.6 |
| 9 | rs56285046 | A/G | 26714950 | 0.28| 0.24| 0.25 | 0.073| 0.12| 0.092 | 3.70 [2.27–6.05] | 3.25 × 10−7 | - | 0 |
| 9 | rs77949268 | A/G | 26738366 | 0.27| 0.26| 0.25 | 0.078| 0.12| 0.10 | 3.59 [2.20–5.87] | 3.74 × 10−7 | - | 0 |
| 9 | rs4427239 | A/G | 113270601 | 0.16| 0.15| 0.041 | 0.029| 0.041| 0.016 | 5.47 [2.81–10.65] | 5.75 × 10−7 | SVEP1 | 0 |
| 9 | rs10759436 | C/T | 113268650 | 0.15| 0.15| 0.041 | 0.029| 0.041| 0.016 | 5.81 [2.92–11.54] | 8.13 × 10−7 | SVEP1 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cismaru, A.L.; Rudin, D.; Ibañez, L.; Liakoni, E.; Bonadies, N.; Kreutz, R.; Carvajal, A.; Lucena, M.I.; Martin, J.; Sancho Ponce, E.; et al. Genome-Wide Association Study of Metamizole-Induced Agranulocytosis in European Populations. Genes 2020, 11, 1275. https://doi.org/10.3390/genes11111275
Cismaru AL, Rudin D, Ibañez L, Liakoni E, Bonadies N, Kreutz R, Carvajal A, Lucena MI, Martin J, Sancho Ponce E, et al. Genome-Wide Association Study of Metamizole-Induced Agranulocytosis in European Populations. Genes. 2020; 11(11):1275. https://doi.org/10.3390/genes11111275
Chicago/Turabian StyleCismaru, Anca Liliana, Deborah Rudin, Luisa Ibañez, Evangelia Liakoni, Nicolas Bonadies, Reinhold Kreutz, Alfonso Carvajal, Maria Isabel Lucena, Javier Martin, Esther Sancho Ponce, and et al. 2020. "Genome-Wide Association Study of Metamizole-Induced Agranulocytosis in European Populations" Genes 11, no. 11: 1275. https://doi.org/10.3390/genes11111275
APA StyleCismaru, A. L., Rudin, D., Ibañez, L., Liakoni, E., Bonadies, N., Kreutz, R., Carvajal, A., Lucena, M. I., Martin, J., Sancho Ponce, E., Molokhia, M., Eriksson, N., EuDAC collaborators, Krähenbühl, S., Largiadèr, C. R., Haschke, M., Hallberg, P., Wadelius, M., & Amstutz, U. (2020). Genome-Wide Association Study of Metamizole-Induced Agranulocytosis in European Populations. Genes, 11(11), 1275. https://doi.org/10.3390/genes11111275