bta-miR-23a Regulates the Myogenic Differentiation of Fetal Bovine Skeletal Muscle-Derived Progenitor Cells by Targeting MDFIC Gene
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Ethics Statement
2.2. Cell Culture and Treatment
2.3. RNA Extration and Real-Time PCR Assay
2.4. Immunofluorescence Assay
2.5. Western Blotting
2.6. RNA Oligonucleotides, Plasmids Construction and Cell Transfection
2.7. Bioinformatic Analyses
2.8. Dual-Luciferase Reporter Assay
2.9. Statistical Analysis
3. Results
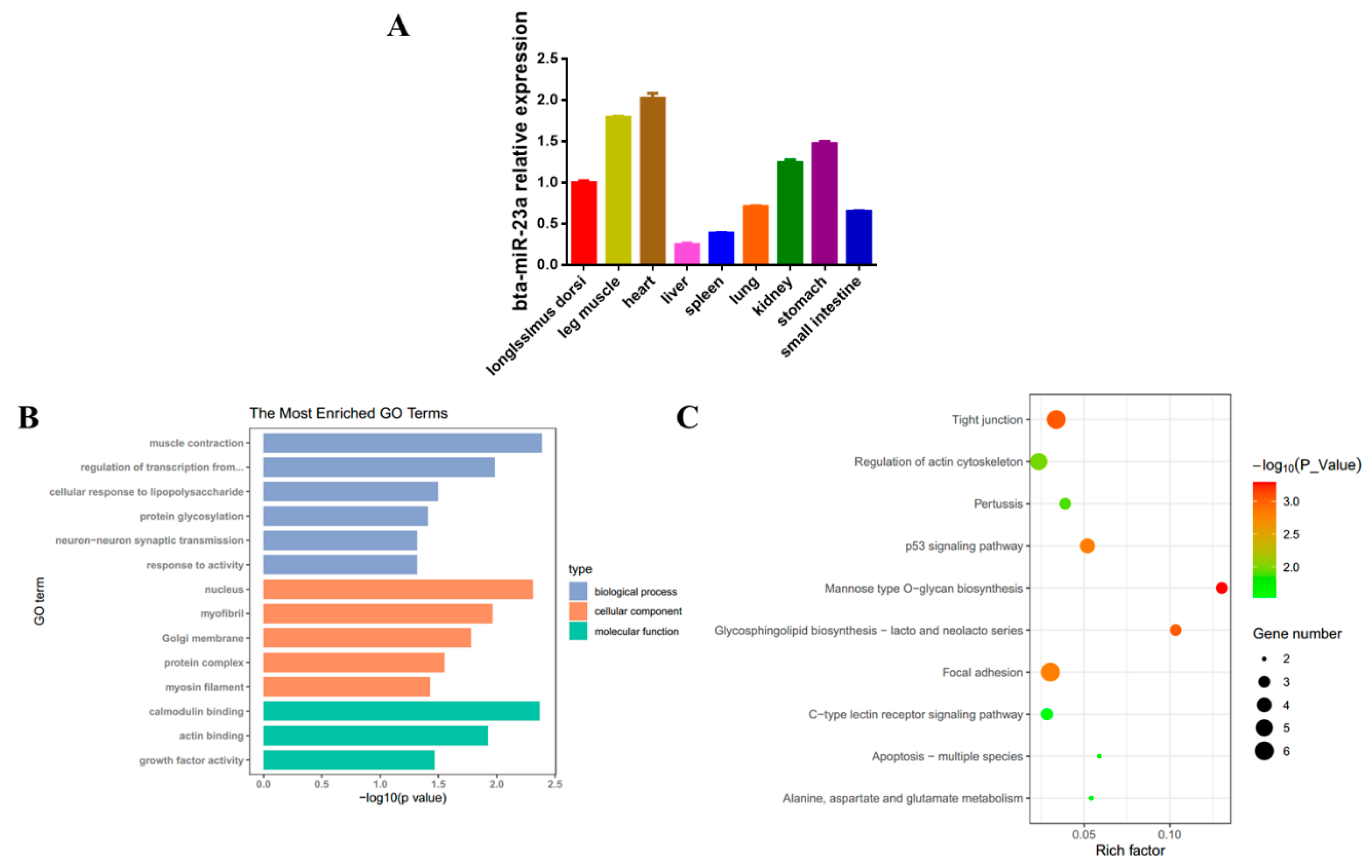
3.1. bta-miR-23a Expression Profile and Its Targets Analysis
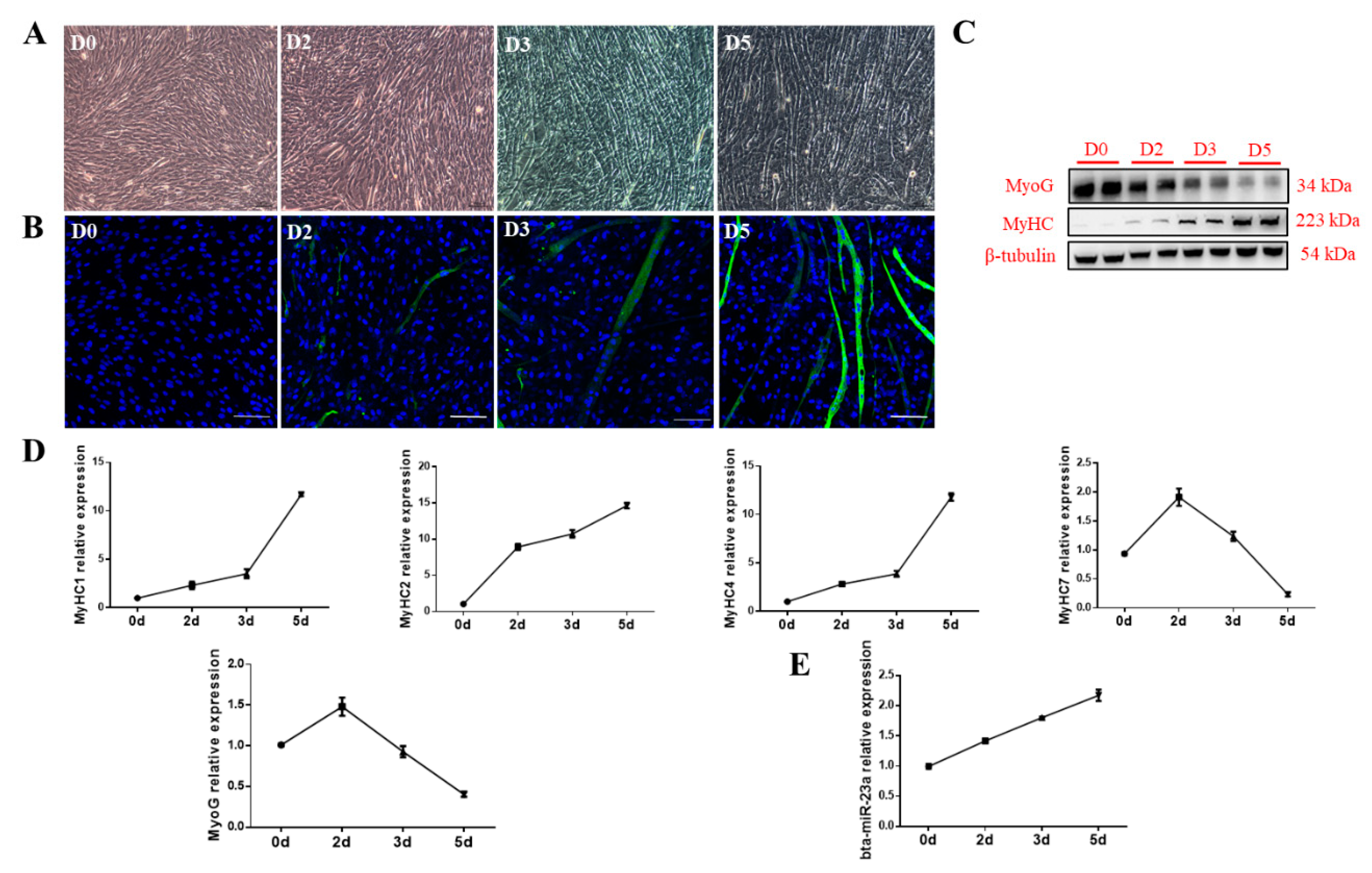
3.2. bta-miR-23a Is Upregulated During MD of PDGFRα− bPCs
3.3. bta-miR-23a Promotes the MD of PDGFRα− bPCs
3.4. MDFIC Is A Promising Target Gene of bta-miR-23a
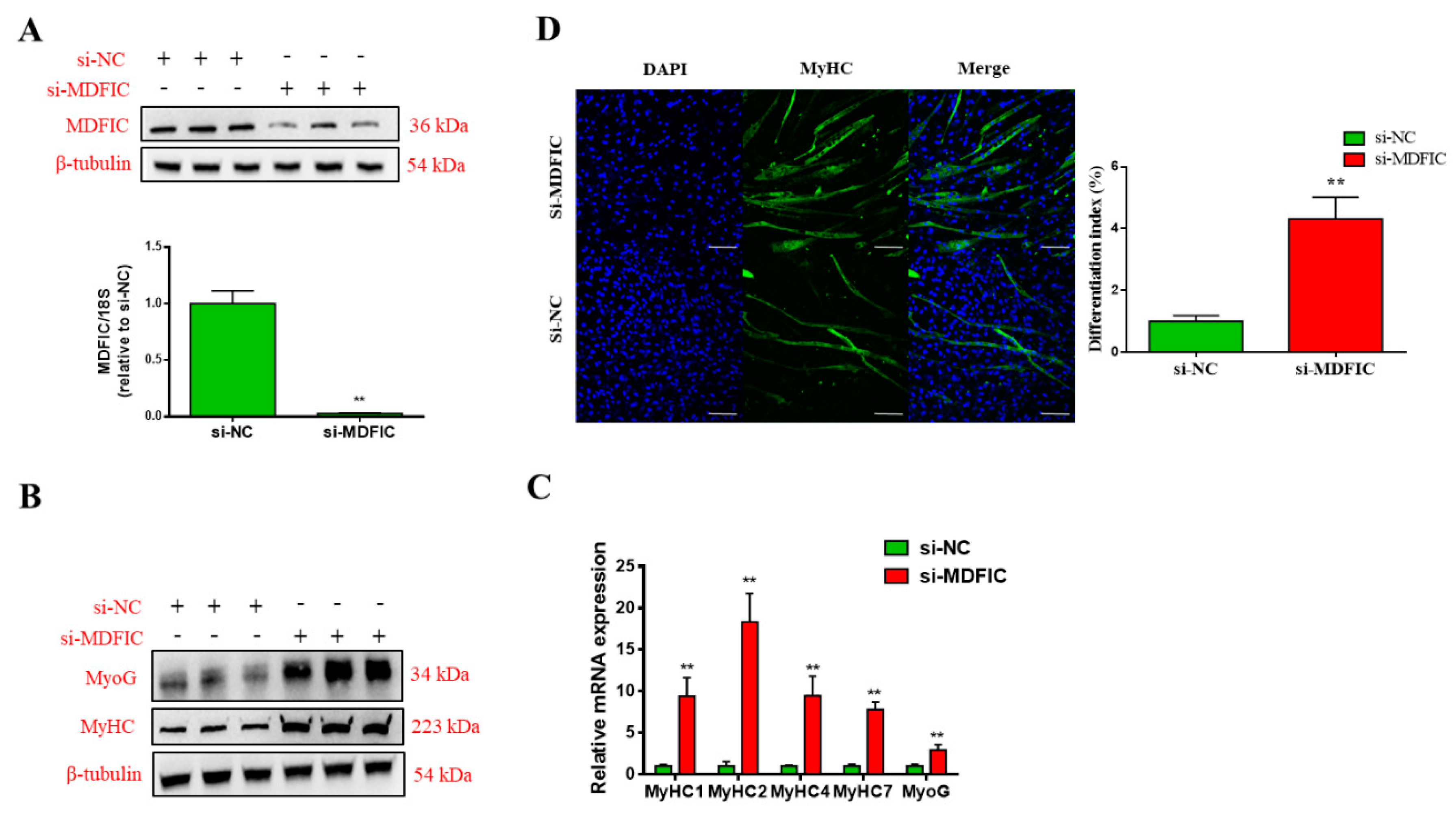
3.5. Knockdown of MDFIC Promotes MD
3.6. Overexpression of MDFIC Rescues the bta-miR-23a-Induced Effects
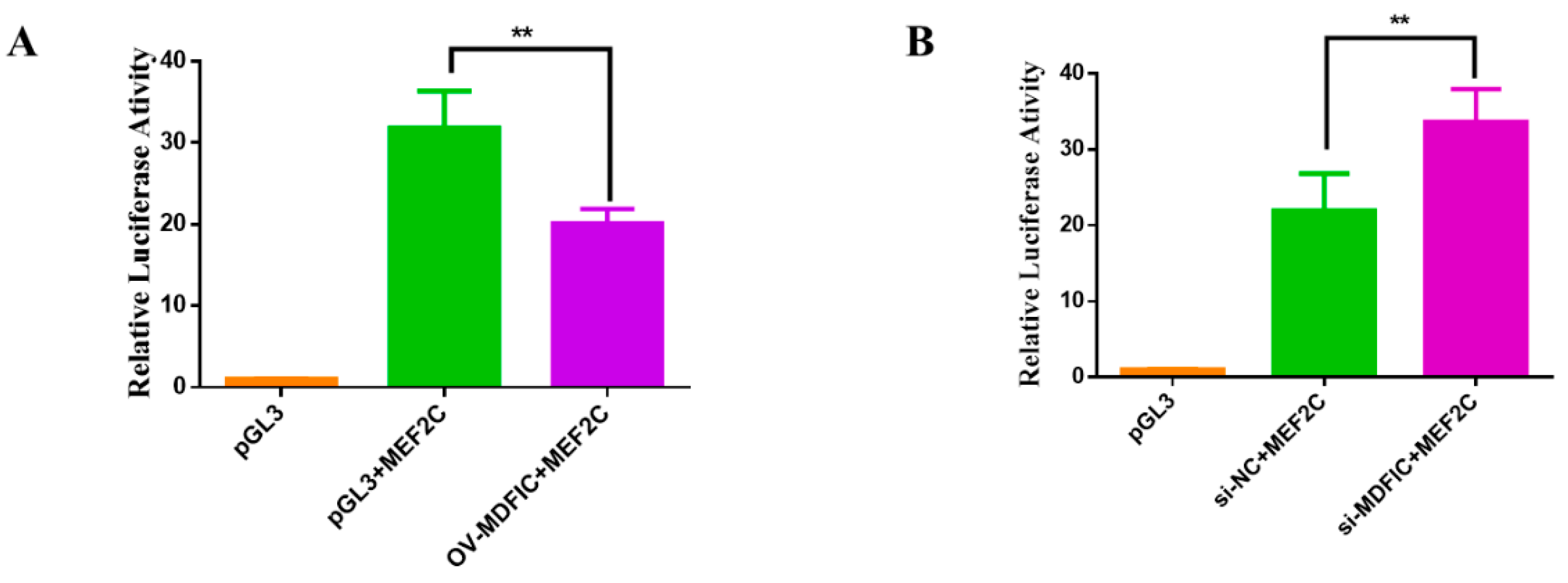
3.7. MDFIC Regulate the Transcription Activity of MEF2C
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cossu, G.; Borello, U. Wnt signaling and the activation of myogenesis in mammals. EMBO J. 1999, 18, 6867–6872. [Google Scholar] [CrossRef] [PubMed]
- Stickland, N.C. A quantitative study of muscle development in the bovine foetus (Bos indicus). Anat. Histol. Embryol. 1978, 7, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Tong, J.; Zhao, J.; Underwood, K.R.; Zhu, M.; Ford, S.P.; Nathanielsz, P.W. Fetal programming of skeletal muscle development in ruminant animals. J. Anim. Sci. 2010, 88, E51–E60. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Yan, X.; Tong, J.F.; Zhao, J.X.; Zhu, M.J. Maternal Obesity, Inflammation, and Fetal Skeletal Muscle Development. Biol. Reprod. 2010, 82, 4–12. [Google Scholar] [CrossRef]
- Wang, Y.K.; Schnegelsberg, P.N.J.; Dausman, J.; Jaenisch, R. Functional redundancy of the muscle-specific transcription factors Myf5 and myogenin. Nature 1996, 379, 823–825. [Google Scholar] [CrossRef]
- Bober, E.; Lyons, G.E.; Braun, T.; Cossu, G.; Buckingham, M.; Arnold, H.H. The muscle regulatory gene, Myf-6, has a biphasic pattern of expression during early mouse development. J. Cell Biol. 1991, 113, 1255–1265. [Google Scholar] [CrossRef]
- Sassoon, D.; Lyons, G.; Wright, W.E.; Lin, V.; Lassar, A.; Weintraub, H.; Buckingham, M. Expression of two myogenic regulatory factors myogenin and MyoDl during mouse embryogenesis. Nature 1989, 341, 303–307. [Google Scholar] [CrossRef]
- Ying, F.; Zhang, L.; Bu, G.; Xiong, Y.; Zuo, B. Muscle fiber-type conversion in the transgenic pigs with overexpression of PGC1α gene in muscle. Biochem. Biophys. Res. Commun. 2016, 480, 669–674. [Google Scholar] [CrossRef]
- Zhao, R.-Q.; Yang, X.-J.; Xu, Q.-F.; Wei, X.-H.; Xia, D.; Chen, J. Expression of GHR and PGC-lα in association with changes of MyHC isof orm types in longissimus muscle of Erhualian and Large White pigs (Sus scrofa) during postnatal growth. Anim. Sci. 2016, 79, 203–211. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Dai, Y.; Wang, Y.M.; Zhang, W.R.; Liu, X.F.; Li, X.; Ding, X.B.; Guo, H. The role of microRNA-1 and microRNA-206 in the proliferation and differentiation of bovine skeletal muscle satellite cells. Vitr. Cell Dev. Biol. Anim. 2016, 52, 27–34. [Google Scholar] [CrossRef]
- Wang, Y.M.; Ding, X.B.; Dai, Y.; Liu, X.F.; Guo, H.; Zhang, Y. Identification and bioinformatics analysis of miRNAs involved in bovine skeletal muscle satellite cell myogenic differentiation. Mol. Cell. Biochem. 2015, 404, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.W.; Sun, X.F.; Tong, H.L.; Wang, Y.H.; Li, S.F.; Yan, Y.Q.; Li, G.P. Effect of differentiation on microRNA expression in bovine skeletal muscle satellite cells by deep sequencing. Cell. Mol. Biol. Lett. 2016, 21, 8. [Google Scholar] [CrossRef]
- Wu, J.Y.; He, D.D.; Yue, B.L.; Zhang, C.L.; Fang, X.T.; Chen, H. miR-101-1 expression pattern in Qinchuan cattle and its role in the regulation of cell differentiation. Gene 2017, 636, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Miretti, S.; Martignani, E.; Accornero, P.; Baratta, M. Functional effect of mir-27b on myostatin expression: A relationship in piedmontese cattle with double-muscled phenotype. BMC Genom. 2013, 14, 194. [Google Scholar] [CrossRef] [PubMed]
- Song, C.C.; Yang, Z.X.; Dong, D.; Xu, J.W.; Wang, J.; Li, H.; Huang, Y.Z.; Lan, X.Y.; Lei, C.Z.; Ma, Y.; et al. miR-483 inhibits bovine myoblast cell proliferation and differentiation via IGF1/PI3K/AKT signal pathway. J. Cell. Physiol. 2019, 234, 9839–9848. [Google Scholar] [CrossRef]
- Wada, S.; Kato, Y.; Okutsu, M.; Miyaki, S.; Suzuki, K.; Yan, Z.; Schiaffino, S.; Asahara, H.; Ushida, T.; Akimoto, T. Translational suppression of atrophic regulators by microRNA-23a integrates resistance to skeletal muscle atrophy. J. Biol. Chem. 2011, 286, 38456–38465. [Google Scholar] [CrossRef]
- Yong, F.L.; Wang, C.W.; Roslani, A.C.; Law, C.W. The involvement of miR-23a/APAF1 regulation axis in colorectal cancer. Int. J. Mol. Sci. 2014, 15, 11713–11729. [Google Scholar] [CrossRef]
- Shang, J.; Yang, F.; Wang, Y.; Wang, Y.; Xue, G.; Mei, Q.; Wang, F.; Sun, S. MicroRNA-23a antisense enhances 5-fluorouracil chemosensitivity through APAF-1/caspase-9 apoptotic pathway in colorectal cancer cells. J. Cell. Biochem. 2014, 115, 772–784. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, J.; Hui, B.; Sun, W.; Li, B.; Shi, F.; Che, S.; Chai, L.; Song, L. Pristimerin enhances the effect of cisplatin by inhibiting the miR-23a/Akt/GSK3β signaling pathway and suppressing autophagy in lung cancer cells. Int. J. Mol. Med. 2019, 43, 1382–1394. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhu, A.; Tian, L.; Xin, Y.; Liu, X.; Peng, Y.; Zhang, J.; Miao, Y.; Wei, J. miR-23a suppresses pancreatic cancer cell progression by inhibiting PLK-1 expression. Mol. Med. Rep. 2018, 18, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Dai, W.; Sang, A.; Yang, X.; Gao, C. Upregulation of microRNA-23a/b promotes tumor progression and confers poor prognosis in patients with gastric cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 8833–8840. [Google Scholar] [PubMed]
- Wang, N.; Zhu, M.; Tsao, S.W.; Man, K.; Zhang, Z.; Feng, Y. MiR-23a-mediated inhibition of topoisomerase 1 expression potentiates cell response to etoposide in human hepatocellular carcinoma. Mol. Cancer 2013, 12, 119. [Google Scholar] [CrossRef] [PubMed]
- Thebault, S.; Gachon, F.; Lemasson, I.; Devaux, C.; Mesnard, J.M. Molecular cloning of a novel human I-mfa domain-containing protein that differently regulates human T-cell leukemia virus type I and HIV-1 expression. J. Biol. Chem. 2000, 275, 4848–4857. [Google Scholar] [CrossRef]
- Thebault, S.; Mesnard, J.M. How the sequestration of a protein interferes with its mechanism of action: Example of a new family of proteins characterized by a particular cysteine-rich carboxy-terminal domain involved in gene expression regulation. Curr. Protein Pept. Sci. 2001, 2, 155–167. [Google Scholar] [CrossRef]
- Chen, C.M.A.; Kraut, N.; Groudine, M.; Weintraub, H. I-mf, a Novel Myogenic Repressor, Interacts with Members of the MyoD Family. Cell 1996, 86, 731–741. [Google Scholar] [CrossRef]
- Kraut, N.; Snider, L.; Chen, C.M.A.; Tapscott, S.J.; Groudine, M. Requirement of the mouse I-mfa gene for placental development and skeletal patterning. Embo J. 1998, 17, 6276–6288. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, X.; Michal, J.J.; Ding, B.; Li, R.; Jiang, Z. Genome wide screening of candidate genes for improving piglet birth weight using high and low estimated breeding value populations. Int. J. Biol. Sci. 2014, 10, 236–244. [Google Scholar] [CrossRef]
- Guan, L.; Hu, X.; Liu, L.; Xing, Y.S.; Zhou, Z.K.; Liang, X.W.; Yang, Q.Y.; Jin, S.Y.; Bao, J.S.; Gao, H.J.; et al. bta-miR-23a involves in adipogenesis of progenitor cells derived from fetal bovine skeletal muscle. Sci. Rep.-UK 2017, 7, 43716. [Google Scholar] [CrossRef]
- Xing, Y.S.H.X.; Ren, L.; Wang, Y.H.; Xing, L.Y.; Li, J.Y.; Zhang, L.P. Identification of microRNAs Involved in Myogenic Differentiation of Bovine Fetal Skeletal Muscle Derived Myoblasts. Acta Vet. Zootech. Sin. 2018, 49, 1134–1144. [Google Scholar]
- Wang, D.Z.; Valdez, M.R.; McAnally, J.; Richardson, J.; Olson, E.N. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Dev. (Camb. Engl.) 2001, 128, 4623–4633. [Google Scholar]
- Ma, M.; Dai, J.; Tang, H.; Xu, T.; Yu, S.; Si, L.; Cui, C.; Sheng, X.; Chi, Z.; Mao, L.; et al. MicroRNA-23a-3p Inhibits Mucosal Melanoma Growth and Progression through Targeting Adenylate Cyclase 1 and Attenuating cAMP and MAPK Pathways. Theranostics 2019, 9, 945–960. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, R.J.; Bai, X.X.; Liu, W. MicroRNA-23a depletion promotes apoptosis of ovarian cancer stem cell and inhibits cell migration by targeting DLG2. Cancer Biol. Ther. 2019, 1–15. [Google Scholar] [CrossRef]
- Gao, P.; Tchernyshyov, I.; Chang, T.C.; Lee, Y.S.; Kita, K.; Ochi, T.; Zeller, K.I.; De Marzo, A.M.; Van Eyk, J.E.; Mendell, J.T.; et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef]
- Hadjimichael, C.; Nikolaou, C.; Papamatheakis, J.; Kretsovali, A. MicroRNAs for Fine-Tuning of Mouse Embryonic Stem Cell Fate Decision through Regulation of TGF-β Signaling. Stem Cell Rep. 2016, 6, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Chen, Y.S.; Guo, L.J.; Jiang, T.J.; Lin, Z.Y. miR-23a/b regulates the balance between osteoblast and adipocyte differentiation in bone marrow mesenchymal stem cells. Bone Res. 2016, 4, 16022. [Google Scholar] [CrossRef]
- Hassan, M.Q.; Gordon, J.A.; Beloti, M.M.; Croce, C.M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc. Natl. Acad. Sci. USA 2010, 107, 19879–19884. [Google Scholar] [CrossRef]
- Ying, Z.; Rong-Lin, X.; Jonathan, G.; Kimberly, L.B.; Stein, J.L.; Lian, J.B.; Van Wijnen, A.J.; Stein, G.S. Control of mesenchymal lineage progression by microRNAs targeting skeletal gene regulators Trps1 and Runx2. J. Biol. Chem. 2012, 287, 21926–21935. [Google Scholar]
- Tanay, G.; Julieta, A.; Jeannette, N.; Hannes, E.; Christian, S.; Cedric, M.; Thomas, L.; Imane, M.; Leslie, S.; Martina, D. MicroRNAs establish robustness and adaptability of a critical gene network to regulate progenitor fate decisions during cortical neurogenesis. Cell Rep. 2014, 7, 1779–1788. [Google Scholar]
- Ubaldo, G.; Valerio, D.C.; Pasquale, C.; Camilla, T.; Antonella, C.; Marcella, M.; Pietro, L.; Stefano, B.; Irene, B.; Emanuele, C. Mir-23a and mir-125b regulate neural stem/progenitor cell proliferation by targeting Musashi1. RNA Biol. 2014, 11, 1105–1112. [Google Scholar]
- Murata, M.; Nonaka, H.; Komatsu, S.; Goto, M.; Mai, M.; Yamada, S.; Lin, I.C.; Yamashita, S.; Tachibana, H. Delphinidin Prevents Muscle Atrophy and Upregulates miR-23a Expression. J. Agric. Food Chem. 2016, 65, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Zhiqiang, L.; Iram, M.; Kun, W.; Jianqin, J.; Jie, G.; Pei-Feng, L. miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 2009, 106, 12103–12108. [Google Scholar]
- Mercatelli, N.; Fittipaldi, S.; Paola, E.D.; Dimauro, I.; Paronetto, M.P.; Jackson, M.J.; Caporossi, D. MiR-23-TrxR1 as a novel molecular axis in skeletal muscle differentiation. Sci. Rep. 2017, 7, 7219. [Google Scholar] [CrossRef]
- Wang, L.; Chen, X.; Zheng, Y.; Li, F.; Lu, Z.; Chen, C.; Liu, J.; Wang, Y.; Peng, Y.; Shen, Z.; et al. MiR-23a inhibits myogenic differentiation through down regulation of fast myosin heavy chain isoforms. Exp. Cell Res. 2012, 318, 2324–2334. [Google Scholar] [CrossRef]
- Snider, L.; Thirlwell, H.; Miller, J.R.; Moon, R.T.; Groudine, M.; Tapscott, S.J. Inhibition of Tcf3 binding by I-mfa domain proteins. Mol. Cell. Biol. 2001, 21, 1866–1873. [Google Scholar] [CrossRef]
- Weijun, P.; Yingying, J.; Tao, H.; Jiyong, W.; Donglei, T.; Xiaoqing, G.; Lin, L. Beta-catenin relieves I-mfa-mediated suppression of LEF-1 in mammalian cells. J. Cell Sci. 2006, 119, 4850. [Google Scholar]




| Name | Primer Sequence (5’–3’) | Annealing Temperature | Accession Number |
|---|---|---|---|
| Myogenin (MyoG) | F: CAAATCCACTCCCTGAAA | 60 °C | NM_001111325.1 |
| R: GCATAGGAAGAGATGAACA | |||
| Myosin heavy chain 1 (MyHC1) | F: GGGAAACTGGCTTCTGCTGAT | 60 °C | NM_174117.1 |
| R: TGGGTTGGTGGTGATTAGGAG | |||
| Myosin heavy chain 2 (MyHC2) | F: GTCAAAGGGACTATCCAGAGCAG | 60 °C | NM_001166227.1 |
| R: AGAAGAGGCCCGAGTAGGTGT | |||
| Myosin heavy chain 4 (MyHC4) | F: CTCCTAATCACCACCAACCCATA | 60 °C | XM_002695806.5 |
| R: TGTCAGCAACTTCAGTGCCATC | |||
| Myosin heavy chain 7 (MyHC7) | F: AAGACAGTGACCGTGAAGGAGG | 60 °C | NM_174727.1 |
| R: GGTTGATGGTGACGCAGAAGA | |||
| MyoD family inhibitor domain containing (MDFIC) | F: TGAGGAGGAAATAAGCAAGATAA | 60 °C | NM_001101102.1 |
| R: CAGGATACAGTGGACACAGCAGT | |||
| bta-miR-23a | F: ATCACATTGCCAGGGATTTCC | 65 °C | NR_031347.1 |
| 18s | F: GTAACCCGTTGAACCCCATT | 60 °C | NM_001025002.1 |
| R: CCATCCAATCGGTAGTAGCG | |||
| U6 | F: GCTTCGGCAGCACATATACTAAAAT | 65 °C | NM_001075477.2 |
| Name | Primer Sequence (5’–3’) | Annealing Temperature | Accession Number |
|---|---|---|---|
| Psi-CHECK-MDFIC-W | F: ACTCTCGAGACTTTTCCTTTGTTGTCTATTC | 60 °C | NM_001101102.1 |
| R: ATAGCGGCCGCAGAGACCAAATCTGTAACTGTA | |||
| Psi-CHECK-MDFIC-Mut | F: AAAACAAAAGTAGCAAATATTTTCCATATG | 60 °C | NM_001101102.1 |
| R: CATATGGAAAATATTTGCTACTTTTGTTTT | |||
| pBI-CMV3 | F: TCCTTCCTAAATCTCCAGAGGATCATAATCAGCCATAC | 68 °C | |
| R: GATGATGATGATCGTCGACAAGCTTATCGATGC | |||
| MDFIC | F: TGTCGACGATCATCATCATCATCATCACATGTCCGGCGCGGGCGAAG | 60 °C | NM_001101102.1 |
| R: CTCTGGAGATTTAGGAAGGAAAACAAATCCCGCAGCACTCCATGC | |||
| pGL3-basic | F: GGGCTCGAGATCTGCGATCTAAGTAAG | 60 °C | |
| R: GGGCTAGCACGCGTAAGAGC | |||
| MEF2C | F: TCTTACGCGTGCTAGCCCTCACTTAGTATTAAAAATAGTTTGATTTTAAAAGTA GAAAGGTCATATATGAAAAACATAATAAAGTCCAGGTAAAGAAATACCTGATAG | 68 °C | NM_001046113.1 |
| R: AGATCGCAGATCTCGAGCCCTCTATGAAGACCCAGGCTTTCCCCCCTTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, X.; Xing, Y.; Ren, L.; Wang, Y.; Li, Q.; Yang, Q.; Du, M.; Xu, L.; Willems, L.; Li, J.; et al. bta-miR-23a Regulates the Myogenic Differentiation of Fetal Bovine Skeletal Muscle-Derived Progenitor Cells by Targeting MDFIC Gene. Genes 2020, 11, 1232. https://doi.org/10.3390/genes11101232
Hu X, Xing Y, Ren L, Wang Y, Li Q, Yang Q, Du M, Xu L, Willems L, Li J, et al. bta-miR-23a Regulates the Myogenic Differentiation of Fetal Bovine Skeletal Muscle-Derived Progenitor Cells by Targeting MDFIC Gene. Genes. 2020; 11(10):1232. https://doi.org/10.3390/genes11101232
Chicago/Turabian StyleHu, Xin, Yishen Xing, Ling Ren, Yahui Wang, Qian Li, Qiyuan Yang, Min Du, Lingyang Xu, Luc Willems, Junya Li, and et al. 2020. "bta-miR-23a Regulates the Myogenic Differentiation of Fetal Bovine Skeletal Muscle-Derived Progenitor Cells by Targeting MDFIC Gene" Genes 11, no. 10: 1232. https://doi.org/10.3390/genes11101232
APA StyleHu, X., Xing, Y., Ren, L., Wang, Y., Li, Q., Yang, Q., Du, M., Xu, L., Willems, L., Li, J., & Zhang, L. (2020). bta-miR-23a Regulates the Myogenic Differentiation of Fetal Bovine Skeletal Muscle-Derived Progenitor Cells by Targeting MDFIC Gene. Genes, 11(10), 1232. https://doi.org/10.3390/genes11101232





