Vertical and Horizontal Transmission of ESBL Plasmid from Escherichia coli O104:H4
Abstract
1. Introduction
2. Materials and Methods
2.1. Standard Microbiology Procedures
2.2. Transposon-Insertion Sequencing
2.3. Transposon-Insertion Sequencing Data Analysis
2.4. Electrophoretic Mobility Shift Assay
2.5. Microscopy
3. Results
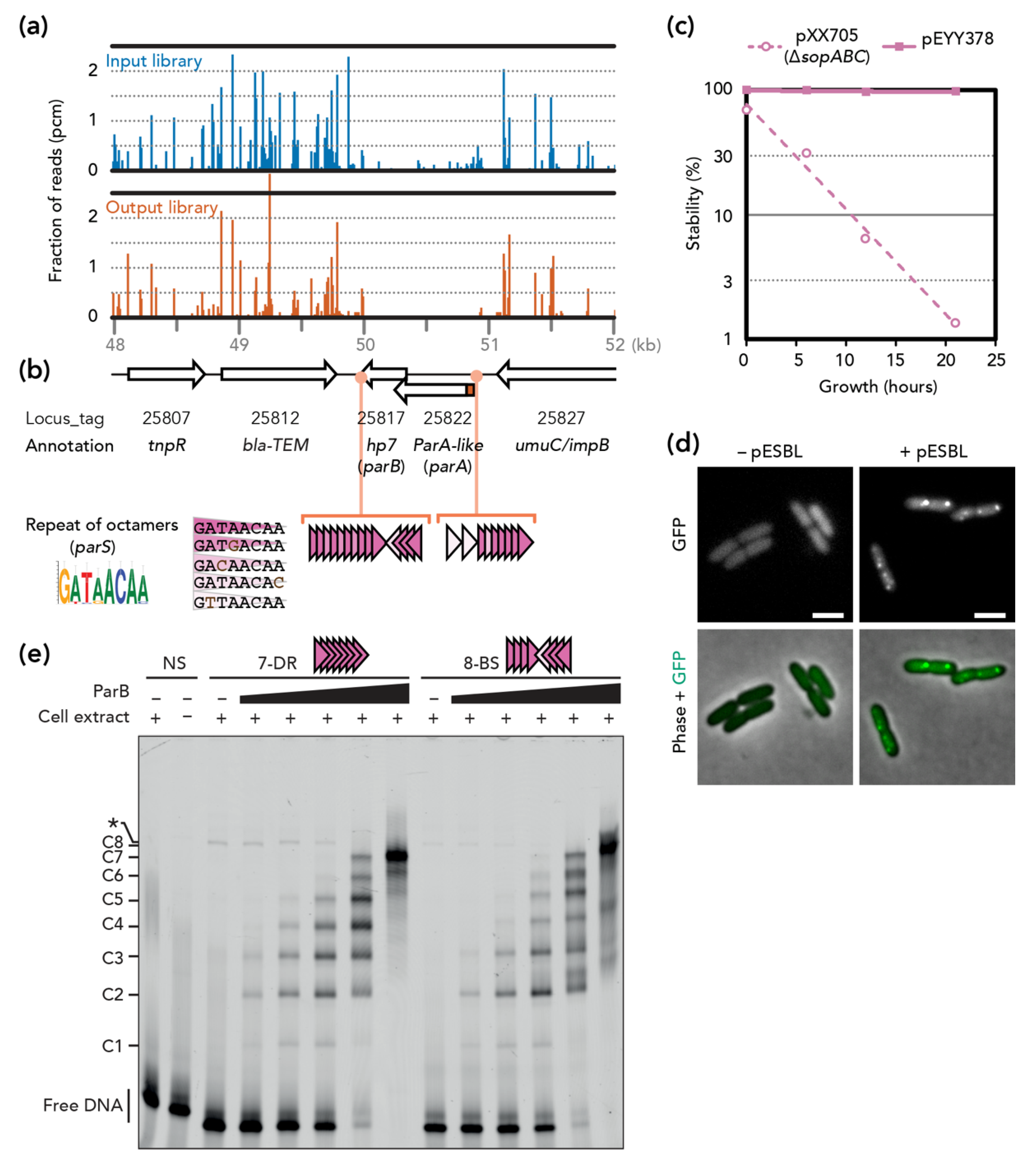
3.1. Transposon-Insertion Sequencing
3.2. pESBL Par Locus
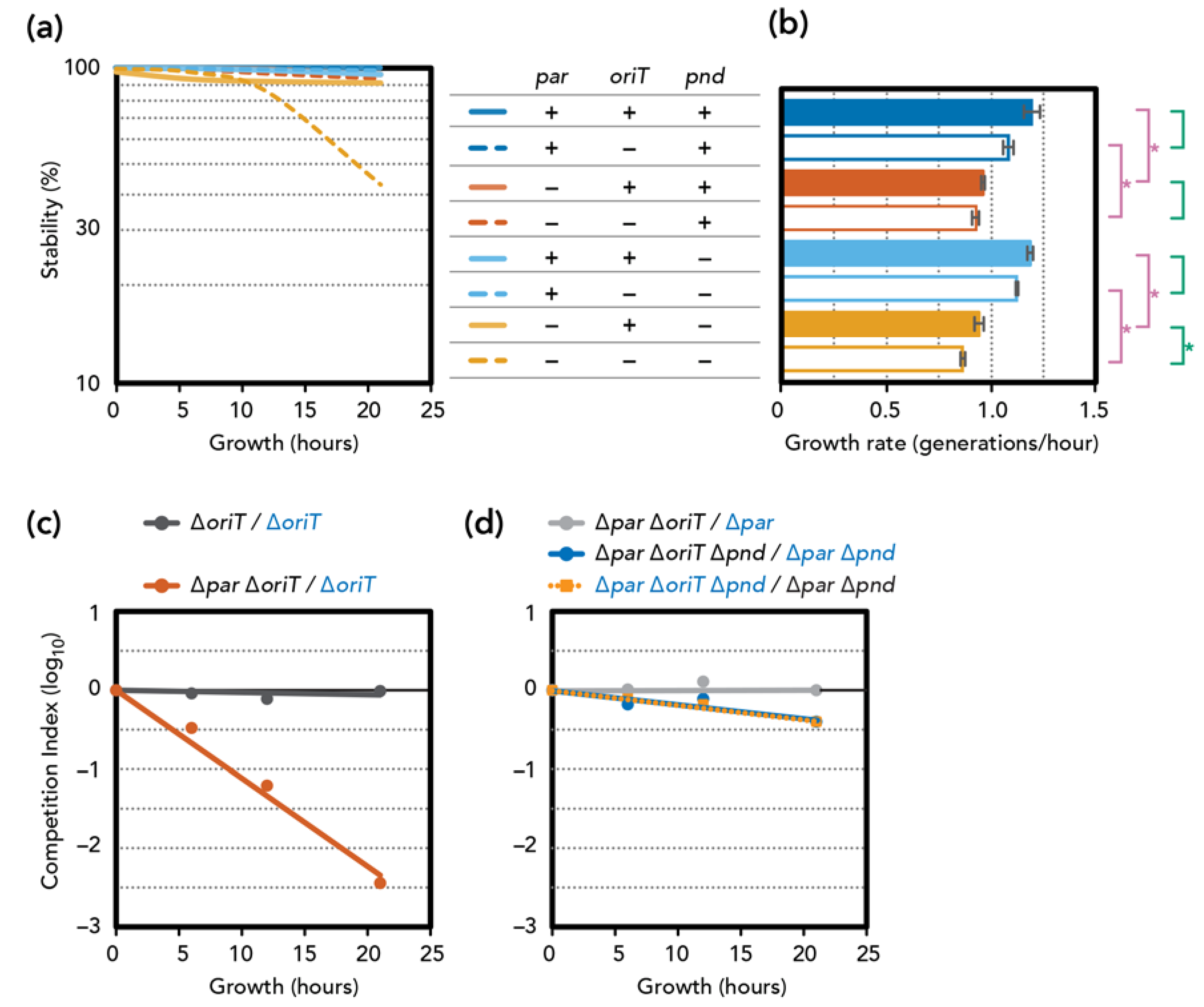
3.3. Stability and Fitness ∆par Mutants
3.4. Involvement of Plasmid Transfer
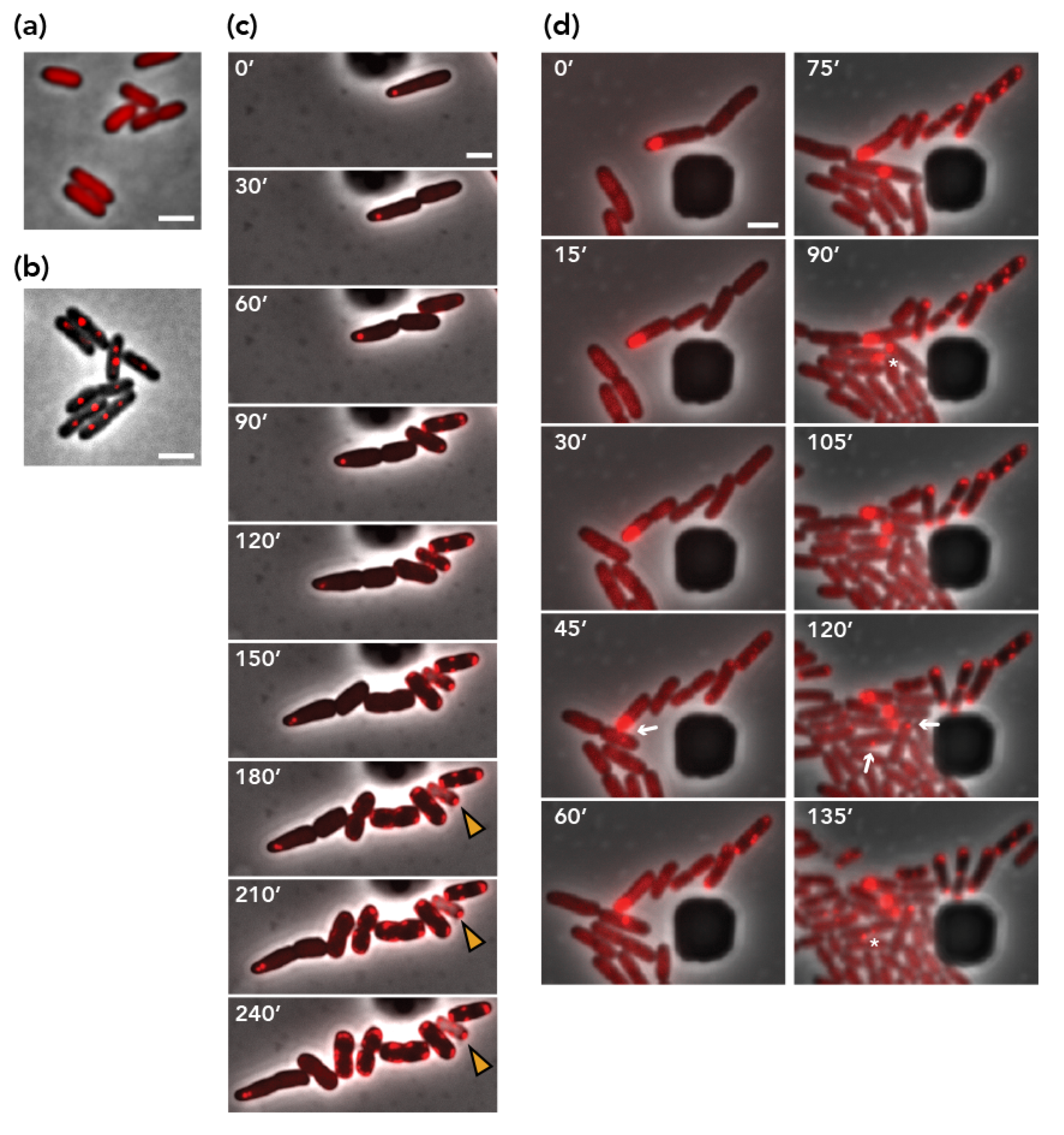
3.5. Visualization of Plasmid Re-Transfer after Mis-Segregation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smillie, C.; Garcillán-Barcia, M.P.; Francia, M.V.; Rocha, E.P.C.; de la Cruz, F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010, 74, 434–452. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Howard, M.; Szardenings, F. Pushing and pulling in prokaryotic DNA segregation. Cell 2010, 141, 927–942. [Google Scholar] [CrossRef]
- Salje, J. Plasmid segregation: How to survive as an extra piece of DNA. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 296–317. [Google Scholar] [CrossRef]
- Bouet, J.-Y.; Funnell, B.E. Plasmid Localization and Partition in Enterobacteriaceae. EcoSal Plus 2019, 8. [Google Scholar] [CrossRef]
- Gerdes, K.; Møller-Jensen, J.; Bugge Jensen, R. Plasmid and chromosome partitioning: Surprises from phylogeny. Mol. Microbiol. 2000, 37, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Livny, J.; Yamaichi, Y.; Waldor, M.K. Distribution of centromere-like parS sites in bacteria: Insights from comparative genomics. J. Bacteriol. 2007, 189, 8693–8703. [Google Scholar] [CrossRef] [PubMed]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Brodersen, D.E.; Mitarai, N.; Gerdes, K. Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol. Cell 2018, 70, 768–784. [Google Scholar] [CrossRef]
- Pandey, D.P.; Gerdes, K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005, 33, 966–976. [Google Scholar] [CrossRef]
- Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Type II Toxin-Antitoxin Systems: Evolution and Revolutions. J. Bacteriol. 2020, 202. [Google Scholar] [CrossRef]
- Rohde, H.; Qin, J.; Cui, Y.; Li, D.; Loman, N.J.; Hentschke, M.; Chen, W.; Pu, F.; Peng, Y.; Li, J.; et al. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N. Engl. J. Med. 2011, 365, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Rasko, D.A.; Webster, D.R.; Sahl, J.W.; Bashir, A.; Boisen, N.; Scheutz, F.; Paxinos, E.E.; Sebra, R.; Chin, C.-S.S.; Iliopoulos, D.; et al. Origins of the E. coli Strain Causing an Outbreak of Hemolytic-Uremic Syndrome in Germany. N. Engl. J. Med. 2011, 365, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Munera, D.; Ritchie, J.M.; Hatzios, S.K.; Bronson, R.; Fang, G.; Schadt, E.E.; Davis, B.M.; Waldor, M.K. Autotransporters but not pAA are critical for rabbit colonization by Shiga toxin-producing Escherichia coli O104:H4. Nat. Commun. 2014, 5, 3080. [Google Scholar] [CrossRef] [PubMed]
- Yamaichi, Y.; Chao, M.C.; Sasabe, J.; Clark, L.; Davis, B.M.; Yamamoto, N.; Mori, H.; Kurokawa, K.; Waldor, M.K. High-resolution genetic analysis of the requirements for horizontal transmission of the ESBL plasmid from Escherichia coli O104:H4. Nucleic Acids Res. 2015, 43, 348–360. [Google Scholar] [CrossRef] [PubMed]
- Hamm, K.; Barth, S.A.; Stalb, S.; Geue, L.; Liebler-Tenorio, E.; Teifke, J.P.; Lange, E.; Tauscher, K.; Kotterba, G.; Bielaszewska, M.; et al. Experimental Infection of Calves with Escherichia coli O104:H4 outbreak strain. Sci. Rep. 2016, 6, 32812. [Google Scholar] [CrossRef]
- Giles, M.; Cawthraw, S.A.; AbuOun, M.; Thomas, C.M.; Munera, D.; Waldor, M.K.; La Ragione, R.M.; Ritchie, J.M. Host-specific differences in the contribution of an ESBL IncI1 plasmid to intestinal colonization by Escherichia coli O104:H4. J. Antimicrob. Chemother. 2018, 73, 1579–1585. [Google Scholar] [CrossRef]
- Cain, A.K.; Barquist, L.; Goodman, A.L.; Paulsen, I.T.; Parkhill, J.; van Opijnen, T. A decade of advances in transposon-insertion sequencing. Nat. Rev. Genet. 2020, 21, 526–540. [Google Scholar] [CrossRef]
- Hancock, S.J.; Phan, M.-D.D.; Peters, K.M.; Forde, B.M.; Chong, T.M.; Yin, W.-F.F.; Chan, K.-G.G.; Paterson, D.L.; Walsh, T.R.; Beatson, S.A.; et al. Identification of IncA/C Plasmid Replication and Maintenance Genes and Development of a Plasmid Multilocus Sequence Typing Scheme. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Poidevin, M.; Sato, M.; Altinoglu, I.; Delaplace, M.; Sato, C.; Yamaichi, Y. Mutation in ESBL Plasmid from Escherichia coli O104:H4 Leads Autoagglutination and Enhanced Plasmid Dissemination. Front. Microbiol. 2018, 9, 130. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Yamaichi, Y.; Fogel, M.A.; McLeod, S.M.; Hui, M.P.; Waldor, M.K. Distinct centromere-like parS sites on the two chromosomes of Vibrio spp. J. Bacteriol. 2007, 189, 5314–5324. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, E.; Paly, E.; Barre, F.-X. High-Resolution Whole-Genome Analysis of Sister-Chromatid Contacts. Mol. Cell 2020, 79, 857–869. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef]
- Pillet, F.; Sanchez, A.; Lane, D.; Anton Leberre, V.; Bouet, J.-Y. Centromere binding specificity in assembly of the F plasmid partition complex. Nucleic Acids Res. 2011, 39, 7477–7486. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.D.; Stephens, R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef] [PubMed]
- Niki, H.; Hiraga, S. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell 1997, 90, 951–957. [Google Scholar] [CrossRef]
- Yamaichi, Y.; Niki, H. Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 14656–14661. [Google Scholar] [CrossRef]
- Bartosik, A.A.; Lasocki, K.; Mierzejewska, J.; Thomas, C.M.; Jagura-Burdzy, G. ParB of Pseudomonas aeruginosa: Interactions with its partner ParA and its target parS and specific effects on bacterial growth. J. Bacteriol. 2004, 186, 6983–6998. [Google Scholar] [CrossRef] [PubMed]
- Dubarry, N.; Pasta, F.; Lane, D. ParABS systems of the four replicons of Burkholderia cenocepacia: New chromosome centromeres confer partition specificity. J. Bacteriol. 2006, 188, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H.J.; Ottesen, J.R.; Youngren, B.; Austin, S.J.; Hansen, F.G. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol. Microbiol. 2006, 62, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Thanbichler, M.; Shapiro, L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 2006, 126, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Poulsen, L.K.; Thisted, T.; Nielsen, A.K.; Martinussen, J.; Andreasen, P.H. The hok killer gene family in gram-negative bacteria. New Biol. 1990, 2, 946–956. [Google Scholar]
- Nielsen, A.K.; Gerdes, K. Mechanism of post-segregational killing by hok-homologue pnd of plasmid R483: Two translational control elements in the pnd mRNA. J. Mol. Biol. 1995, 249, 270–282. [Google Scholar] [CrossRef]
- Sampei, G.-I.; Furuya, N.; Tachibana, K.; Saitou, Y.; Suzuki, T.; Mizobuchi, K.; Komano, T. Complete genome sequence of the incompatibility group I1 plasmid R64. Plasmid 2010, 64, 92–103. [Google Scholar] [CrossRef]
- Brouwer, M.S.M.; Bossers, A.; Harders, F.; van Essen-Zandbergen, A.; Mevius, D.J.; Smith, H.E. Complete Genome Sequences of IncI1 Plasmids Carrying Extended-Spectrum β-Lactamase Genes. Genome Announc. 2014, 2, e00859-14. [Google Scholar] [CrossRef]
- Nolivos, S.; Cayron, J.; Dedieu, A.; Page, A.; Delolme, F.; Lesterlin, C. Role of AcrAB-TolC multidrug efflux pump in drug-resistance acquisition by plasmid transfer. Science 2019, 364, 778–782. [Google Scholar] [CrossRef]
- Lam, M.M.C.; Wyres, K.L.; Wick, R.R.; Judd, L.M.; Fostervold, A.; Holt, K.E.; Löhr, I.H. Convergence of virulence and MDR in a single plasmid vector in MDR Klebsiella pneumoniae ST15. J. Antimicrob. Chemother. 2019, 74, 1218–1222. [Google Scholar] [CrossRef]
- Li, R.; Cheng, J.; Dong, H.; Li, L.; Liu, W.; Zhang, C.; Feng, X.; Qin, S. Emergence of a novel conjugative hybrid virulence multidrug-resistant plasmid in extensively drug-resistant Klebsiella pneumoniae ST15. Int. J. Antimicrob. Agents 2020, 55, 105952. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, M.; Austin, S. Prevalence and significance of plasmid maintenance functions in the virulence plasmids of pathogenic bacteria. Infect. Immun. 2011, 79, 2502–2509. [Google Scholar] [CrossRef] [PubMed]
- Ebersbach, G.; Gerdes, K. The double par locus of virulence factor pB171: DNA segregation is correlated with oscillation of ParA. Proc. Natl. Acad. Sci. USA 2001, 98, 15078–15083. [Google Scholar] [CrossRef] [PubMed]
- Ringgaard, S.; Ebersbach, G.; Borch, J.; Gerdes, K. Regulatory cross-talk in the double par locus of plasmid pB171. J. Biol. Chem. 2007, 282, 3134–3145. [Google Scholar] [CrossRef] [PubMed]
- Pratto, F.; Suzuki, Y.; Takeyasu, K.; Alonso, J.C. Single-molecule analysis of proteinxDNA complexes formed during partition of newly replicated plasmid molecules in Streptococcus pyogenes. J. Biol. Chem. 2009, 284, 30298–30306. [Google Scholar] [CrossRef]
- Gilks, W.R.; Audit, B.; de Angelis, D.; Tsoka, S.; Ouzounis, C.A. Percolation of annotation errors through hierarchically structured protein sequence databases. Math. Biosci. 2005, 193, 223–234. [Google Scholar] [CrossRef]
- Orlek, A.; Phan, H.; Sheppard, A.E.; Doumith, M.; Ellington, M.; Peto, T.; Crook, D.; Walker, A.S.; Woodford, N.; Anjum, M.F.; et al. A curated dataset of complete Enterobacteriaceae plasmids compiled from the NCBI nucleotide database. Data Brief 2017, 12, 423–426. [Google Scholar] [CrossRef]
- Orlek, A.; Phan, H.; Sheppard, A.E.; Doumith, M.; Ellington, M.; Peto, T.; Crook, D.; Walker, A.S.; Woodford, N.; Anjum, M.F.; et al. Ordering the mob: Insights into replicon and MOB typing schemes from analysis of a curated dataset of publicly available plasmids. Plasmid 2017, 91, 42–52. [Google Scholar] [CrossRef]
- Soh, Y.-M.; Davidson, I.F.; Zamuner, S.; Basquin, J.; Bock, F.P.; Taschner, M.; Veening, J.-W.; De Los Rios, P.; Peters, J.-M.; Gruber, S. Self-organization of parS centromeres by the ParB CTP hydrolase. Science 2019, 366, 1129–1133. [Google Scholar] [CrossRef]
- Osorio-Valeriano, M.; Altegoer, F.; Steinchen, W.; Urban, S.; Liu, Y.; Bange, G.; Thanbichler, M. ParB-type DNA Segregation Proteins Are CTP-Dependent Molecular Switches. Cell 2019, 179, 1512–1524.e15. [Google Scholar] [CrossRef]
- Youngren, B.; Radnedge, L.; Hu, P.; Garcia, E.; Austin, S. A plasmid partition system of the P1-P7par family from the pMT1 virulence plasmid of Yersinia pestis. J. Bacteriol. 2000, 182, 3924–3928. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ebersbach, G.; Gerdes, K. Bacterial mitosis: Partitioning protein ParA oscillates in spiral-shaped structures and positions plasmids at mid-cell. Mol. Microbiol. 2004, 52, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Lamothe, R.; Tran, T.; Meas, D.; Lee, L.; Li, A.M.; Sherratt, D.J.; Tolmasky, M.E. High-copy bacterial plasmids diffuse in the nucleoid-free space, replicate stochastically and are randomly partitioned at cell division. Nucleic Acids Res. 2014, 42, 1042–1051. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, A.; Cattoni, D.I.; Guilhas, B.; Mathieu-Demazière, C.; Oudjedi, L.; Fiche, J.-B.B.; Rech, J.; Abrahamsson, S.; Murray, H.; Bouet, J.-Y.Y.; et al. Bacterial partition complexes segregate within the volume of the nucleoid. Nat. Commun. 2016, 7, 12107. [Google Scholar] [CrossRef]
- Guynet, C.; Cuevas, A.; Moncalián, G.; de la Cruz, F. The stb operon balances the requirements for vegetative stability and conjugative transfer of plasmid R388. PLoS Genet 2011, 7, e1002073. [Google Scholar] [CrossRef]
- Frost, L.S.; Ippen-Ihler, K.; Skurray, R.A. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 1994, 58, 162–210. [Google Scholar] [CrossRef]
- Marrero, J.; Waldor, M.K. Interactions between inner membrane proteins in donor and recipient cells limit conjugal DNA transfer. Dev. Cell 2005, 8, 963–970. [Google Scholar] [CrossRef][Green Version]
- Sakuma, T.; Tazumi, S.; Furuya, N.; Komano, T. ExcA proteins of IncI1 plasmid R64 and IncIγ plasmid R621a recognize different segments of their cognate TraY proteins in entry exclusion. Plasmid 2013, 69, 138–145. [Google Scholar] [CrossRef]
- Fernandez-Lopez, R.; de la Cruz, F. Rebooting the genome: The role of negative feedback in horizontal gene transfer. Mob. Genet. Elements 2014, 4, 1–6. [Google Scholar] [CrossRef][Green Version]
- Stocker, B.A.D.A.; Smith, S.M.; Ozeki, H. High Infectivity of Salmonella typhimurium newly infected by the colI factor. J. Gen. Microbiol. 1963, 30, 201–221. [Google Scholar] [CrossRef][Green Version]
- Watanabe, T. Episome-mediated transfer of drug resistance in enterobacteriaceae. Vi. High-frequency resistance transfer system in Escherichia coli. J. Bacteriol. 1963, 85, 788–794. [Google Scholar] [CrossRef] [PubMed]
- Demarre, G.; Guérout, A.-M.M.; Matsumoto-Mashimo, C.; Rowe-Magnus, D.A.; Marlière, P.; Mazel, D. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 2005, 156, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Yamaichi, Y.; Fogel, M.A.; Waldor, M.K. par genes and the pathology of chromosome loss in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2007, 104, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef]
- Milton, D.L.; O’Toole, R.; Horstedt, P.; Wolf-Watz, H. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 1996, 178, 1310–1319. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daniel, S.; Goldlust, K.; Quebre, V.; Shen, M.; Lesterlin, C.; Bouet, J.-Y.; Yamaichi, Y. Vertical and Horizontal Transmission of ESBL Plasmid from Escherichia coli O104:H4. Genes 2020, 11, 1207. https://doi.org/10.3390/genes11101207
Daniel S, Goldlust K, Quebre V, Shen M, Lesterlin C, Bouet J-Y, Yamaichi Y. Vertical and Horizontal Transmission of ESBL Plasmid from Escherichia coli O104:H4. Genes. 2020; 11(10):1207. https://doi.org/10.3390/genes11101207
Chicago/Turabian StyleDaniel, Sandra, Kelly Goldlust, Valentin Quebre, Minjia Shen, Christian Lesterlin, Jean-Yves Bouet, and Yoshiharu Yamaichi. 2020. "Vertical and Horizontal Transmission of ESBL Plasmid from Escherichia coli O104:H4" Genes 11, no. 10: 1207. https://doi.org/10.3390/genes11101207
APA StyleDaniel, S., Goldlust, K., Quebre, V., Shen, M., Lesterlin, C., Bouet, J.-Y., & Yamaichi, Y. (2020). Vertical and Horizontal Transmission of ESBL Plasmid from Escherichia coli O104:H4. Genes, 11(10), 1207. https://doi.org/10.3390/genes11101207





