Comparative Genomics Analysis of Lactobacillus mucosae from Different Niches
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and General Genome Features
2.2. The Average Nucleotide Identity (ANI) Values and Phylogenetic Analyses
2.3. Pan-Genome and Core-Genome Analysis
2.4. Genotype/Phenotype Association Applied to Carbohydrate Metabolism
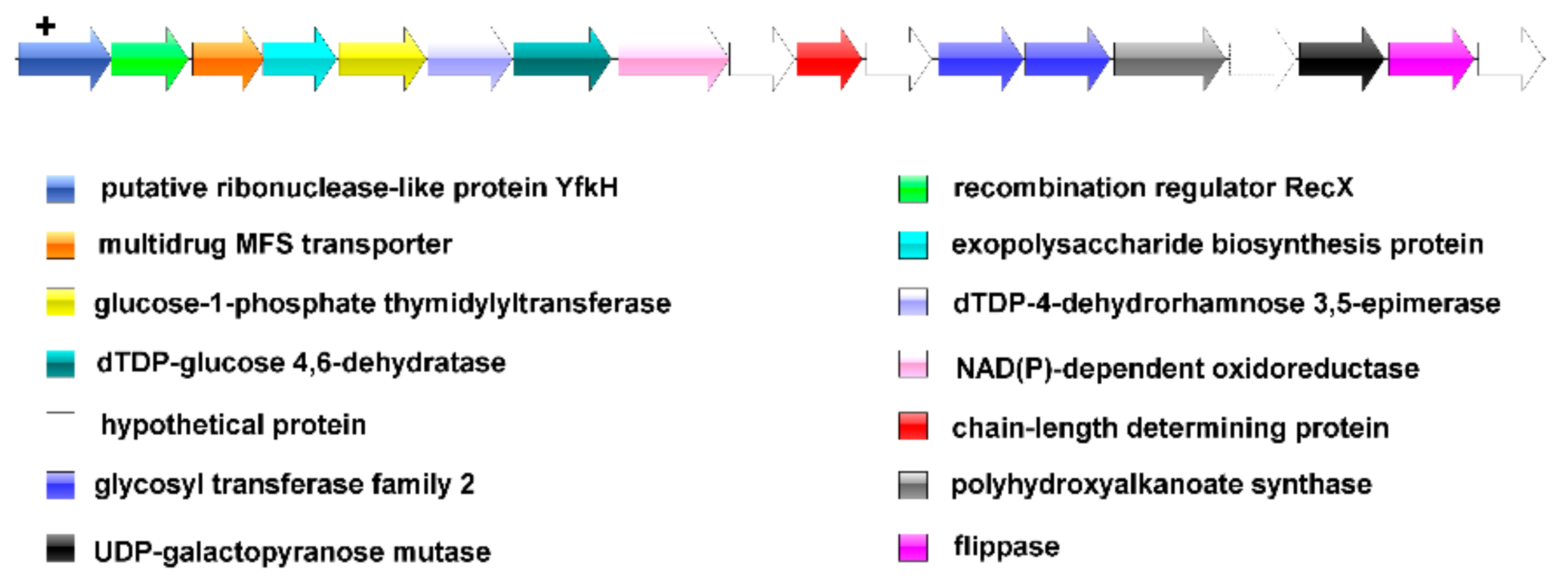
2.5. Prediction of the EPS Gene Operon
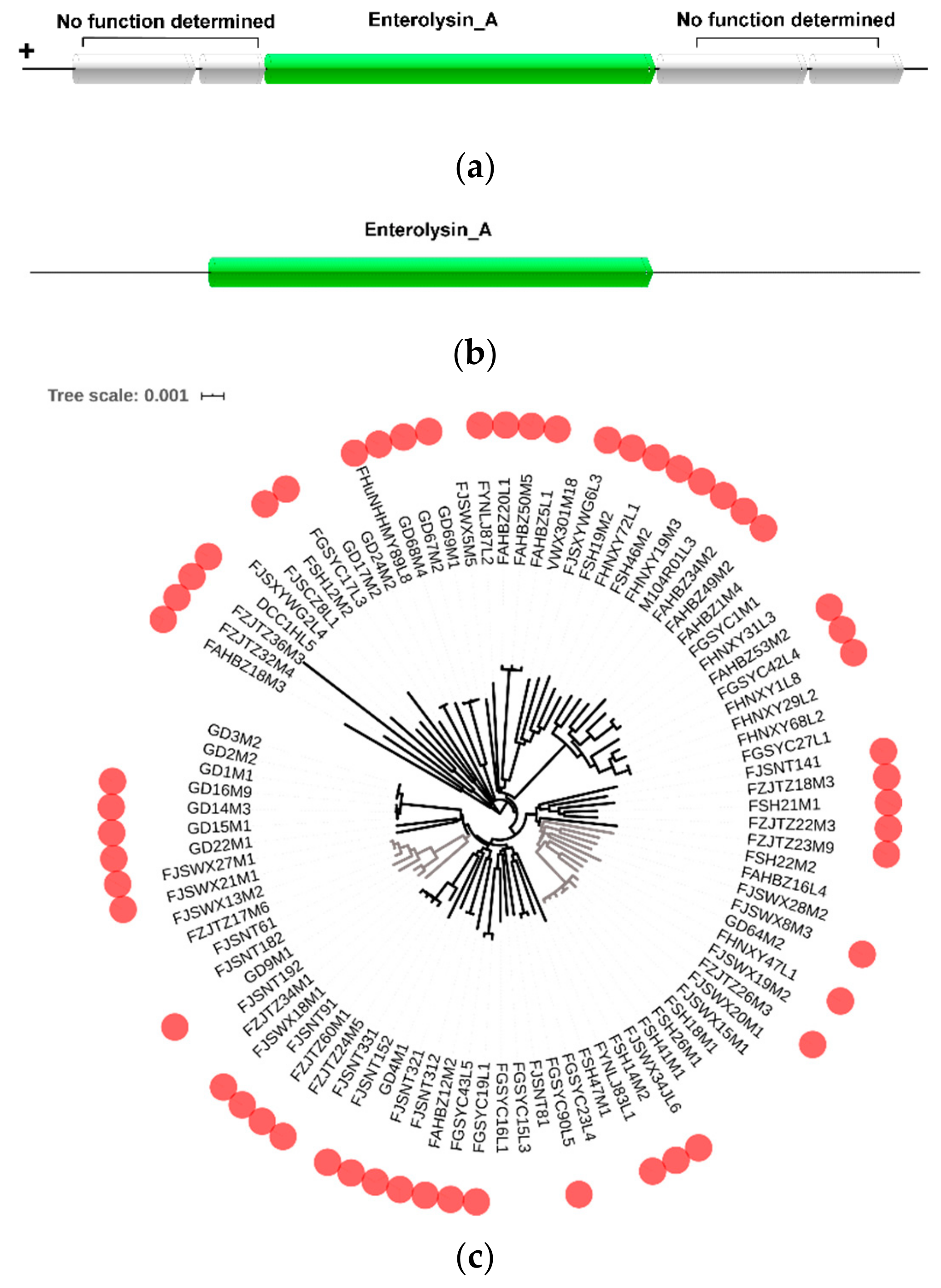
2.6. Prediction of Bacteriocin Operon
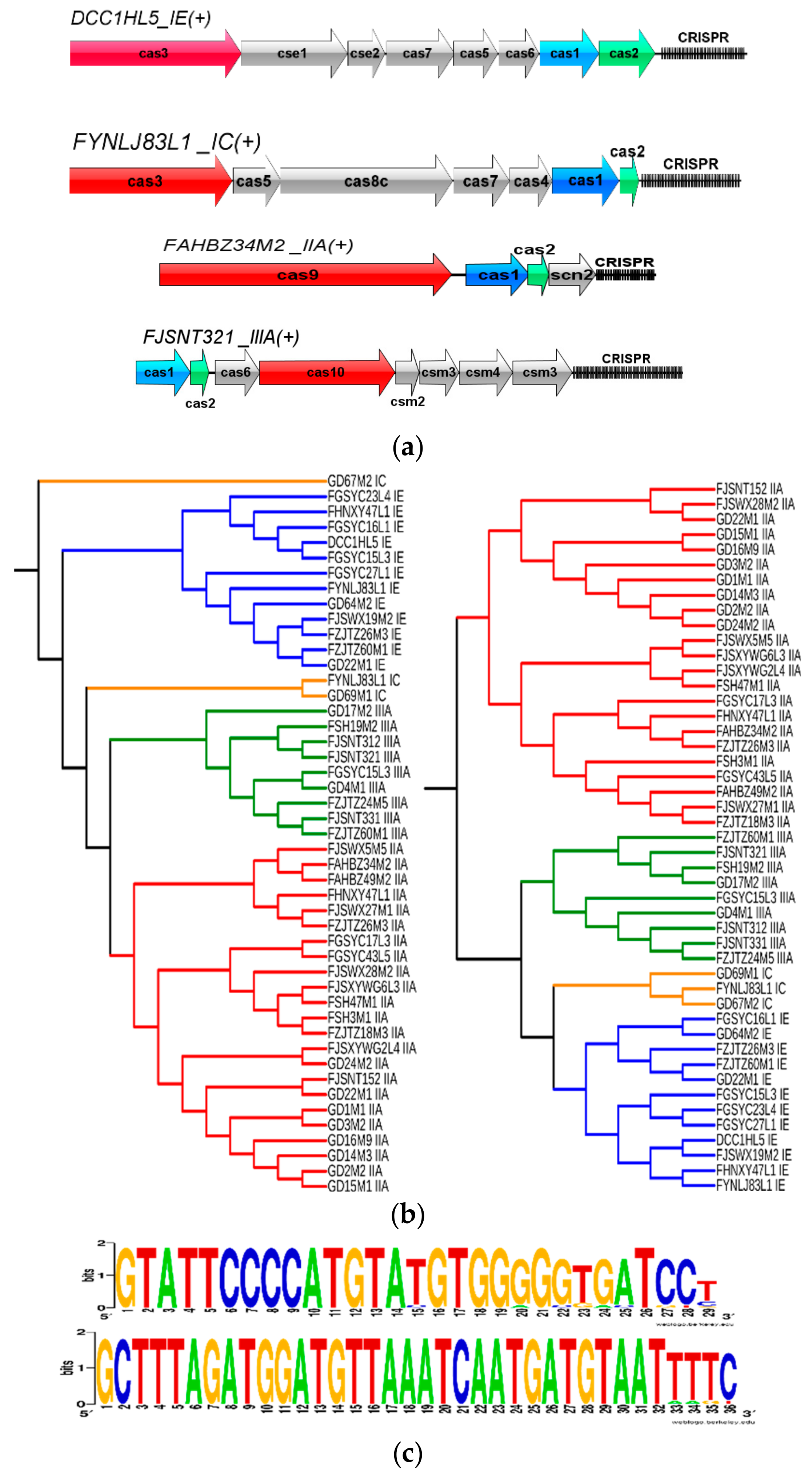
2.7. CRISPR-Cas Identification and Characterization
3. Results
3.1. General Genome Features of L. mucosae
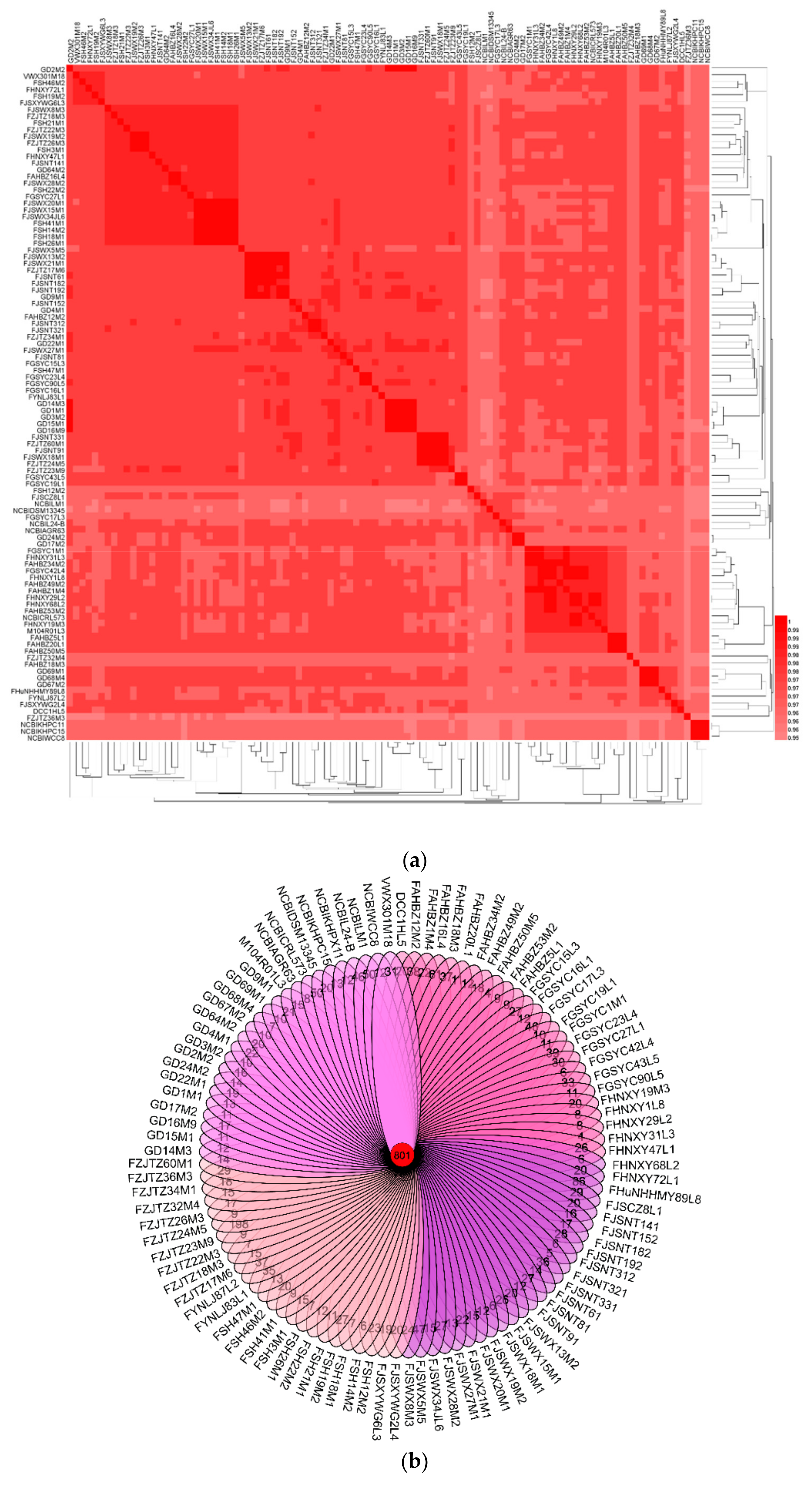
3.2. ANI and Phylogenetic Analyses of L. mucosae
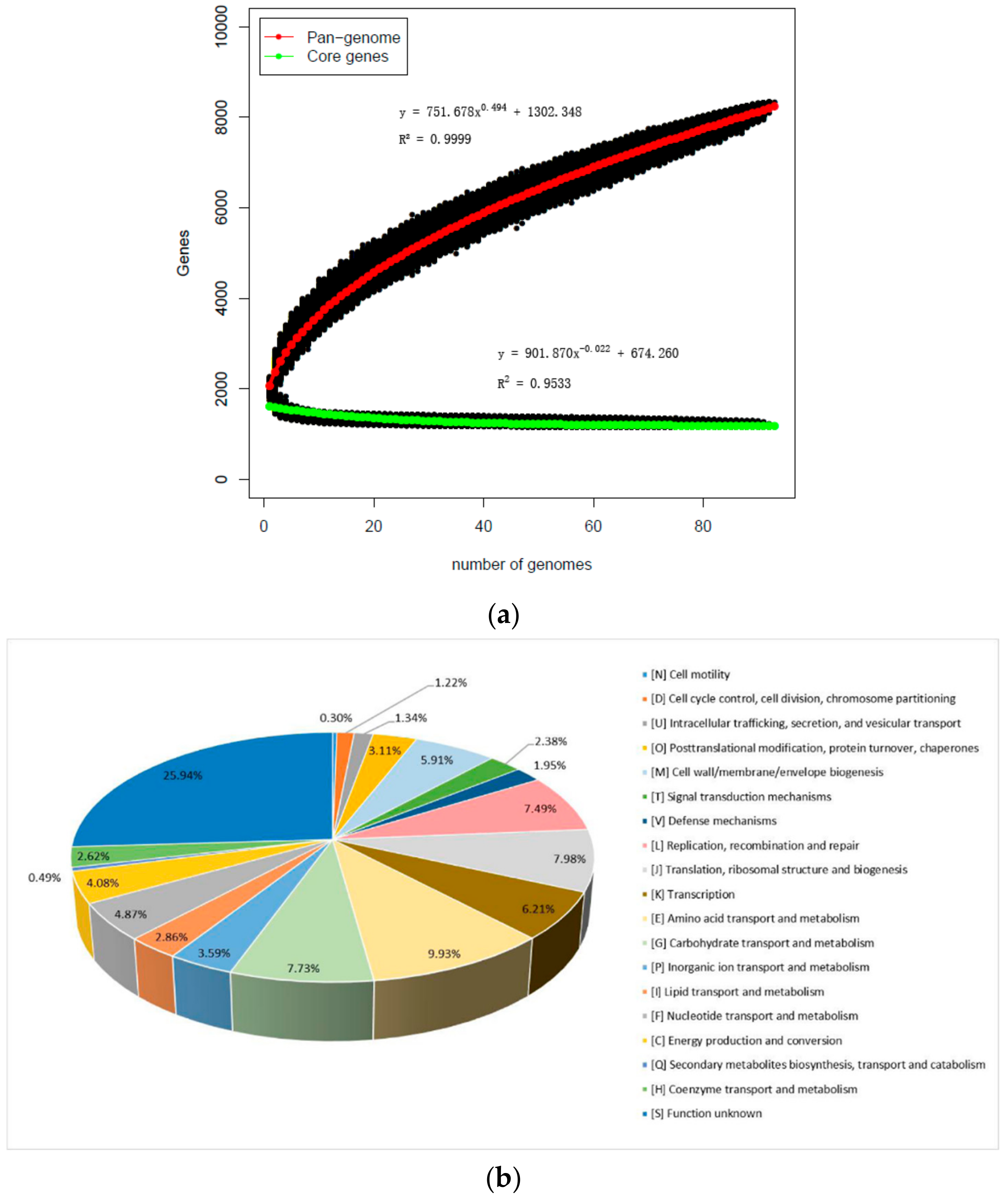
3.3. Pan-Genome and Core Genes of L. mucosae
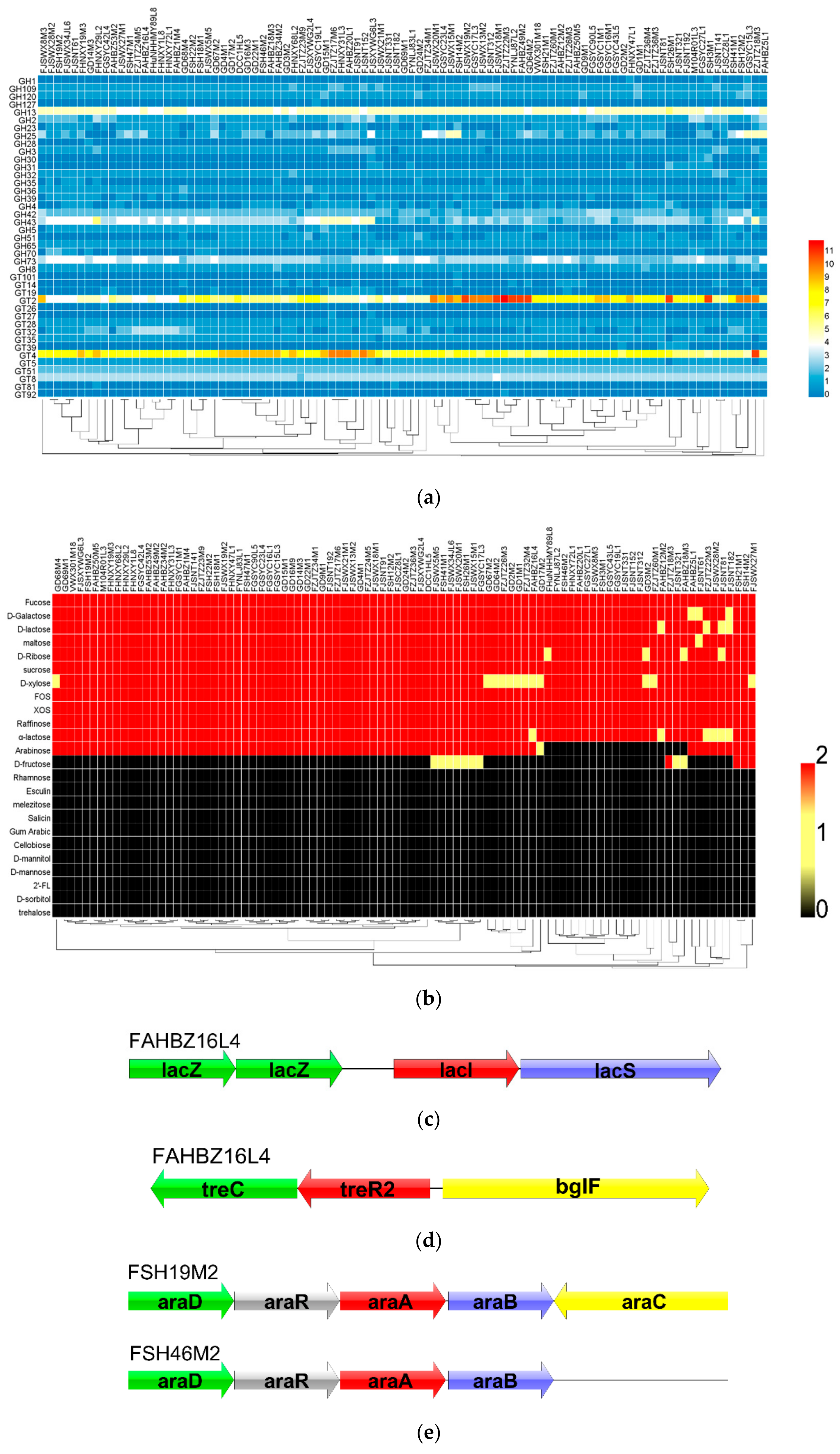
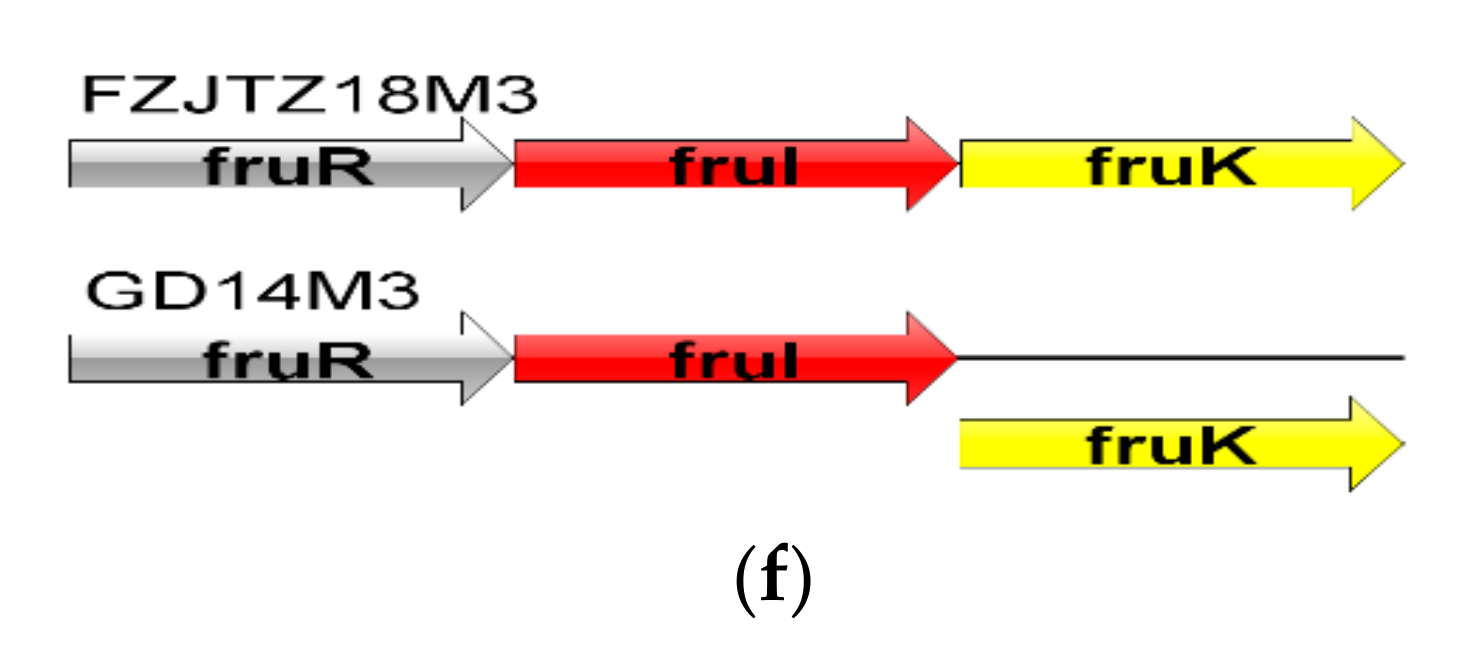
3.4. In Silico Gene–Trait Matching for Carbohydrate Utilization
3.5. Prediction of the EPS Operon in L. mucosae
3.6. Prediction of Bacteriocin Production in L. mucosae
3.7. CRISPR-Cas Systems in L. mucosae
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mamuad, L.L.; Kim, S.H.; Choi, Y.J.; Soriano, A.P.; Cho, K.K.; Lee, K.; Bae, G.S.; Lee, S.S. Increased propionate concentration in Lactobacillus mucosae-fermented wet brewers grains and during in vitro rumen fermentation. J. Appl. Microbiol. 2017, 123, 29–40. [Google Scholar] [CrossRef] [PubMed]
- London, L.E.E.; Chaurin, V.; Auty, M.A.E.; Fenelon, M.A.; Fitzgerald, G.F.; Ross, R.P.; Stanton, C. Use of Lactobacillus mucosae DPC 6426, an exopolysaccharide-producing strain, positively influences the techno-functional properties of yoghurt. Int. Dairy J. 2015, 40, 33–38. [Google Scholar] [CrossRef]
- Ryan, P.M.; Burdikova, Z.; Beresford, T.; Auty, M.A.; Fitzgerald, G.F.; Ross, R.P.; Sheehan, J.J.; Stanton, C. Reduced-fat Cheddar and Swiss-type cheeses harboring exopolysaccharide-producing probiotic Lactobacillus mucosae DPC 6426. J. Dairy Sci. 2015, 98, 8531–8544. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.M.; Stolte, E.H.; London, L.E.E.; Wells, J.M.; Long, S.L.; Joyce, S.A.; Gahan, C.G.M.; Fitzgerald, G.F.; Ross, R.P.; Caplice, N.M.; et al. Lactobacillus mucosae DPC 6426 as a bile-modifying and immunomodulatory microbe. BMC Microbiol. 2019, 19, 33. [Google Scholar] [CrossRef]
- Long, M.; Zang, J.; Peng, L.I.; Zhang, W.K.; Liu, G.W.; Wang, Z. The removal capability of zearalenone by Lactobacillus mucosae in vitro. Chin. J. Vet. Sci. 2013, 33, 458–461. [Google Scholar]
- Valeriano, V.D.V.; Oh, J.K.; Bagon, B.B.; Kim, H.; Kang, D.K. Comparative genomic analysis of Lactobacillus mucosae LM1 identifies potential niche-specific genes and pathways for gastrointestinal adaptation. Genomics 2017, 111, 24–33. [Google Scholar] [CrossRef]
- Valeriano, V.D.; Parungao-Balolong, M.M.; Kang, D.K. In vitro evaluation of the mucin-adhesion ability and probiotic potential of Lactobacillus mucosae LM1. J. Appl. Microbiol. 2014, 117, 485–497. [Google Scholar] [CrossRef]
- Soriano, A.P.; Mamuad, L.L.; Kim, S.H.; Choi, Y.J.; Jeong, C.D.; Bae, G.S.; Chang, M.B.; Lee, S.S. Effect of Lactobacillus mucosae on in vitro rumen fermentation characteristics of dried brewers grain, methane production and bacterial diversity. Asian-Australas. J. Anim. Sci. 2014, 27, 1562–1570. [Google Scholar] [CrossRef]
- Roos, S.; Karner, F.; Axelsson, L.; Jonsson, H. Lactobacillus mucosae sp. nov., a new species with in vitro mucus-binding activity isolated from pig intestine. Int. J. Syst. Evol. Microbiol. 2000, 50(Pt. 1), 251–258. [Google Scholar] [CrossRef]
- London, L.E.; Price, N.P.; Ryan, P.; Wang, L.; Auty, M.A.; Fitzgerald, G.F.; Stanton, C.; Ross, R.P. Characterization of a bovine isolate Lactobacillus mucosae DPC 6426 which produces an exopolysaccharide composed predominantly of mannose residues. J. Appl. Microbiol. 2014, 117, 509–517. [Google Scholar] [CrossRef]
- De Moraes, G.M.D.; de Abreu, L.R.; do Egito, A.S.; Salles, H.O.; da Silva, L.M.F.; Nero, L.A.; Todorov, S.D.; dos Santos, K.M.O. Functional properties of lactobacillus mucosae strains isolated from Brazilian goat milk. Probiotics Antimicrob. Proteins 2017, 9, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Drissi, F.; Labas, N.; Merhej, V.; Raoult, D. Draft genome sequence of the Lactobacillus agilis strain marseille. Genome Announc. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Bleckwedel, J.; Teran, L.C.; Bonacina, J.; Saavedra, L.; Mozzi, F.; Raya, R.R. Draft genome sequence of the mannitol-producing strain Lactobacillus mucosae CRL573. Genome Announc. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Kant, R.; Blom, J.; Palva, A.; Siezen, R.J.; de Vos, W.M. Comparative genomics of Lactobacillus. Microb. Biotechnol. 2011, 4, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, P.; Bottacini, F.; Mahony, J.; Kilcawley, K.N.; van Sinderen, D. Comparative and functional genomics of the Lactococcus lactis taxon; insights into evolution and niche adaptation. BMC Genomics 2017, 18, 267. [Google Scholar] [CrossRef] [PubMed]
- Ruibang, L.; Binghang, L.; Yinlong, X.; Zhenyu, L.; Weihua, H.; Jianying, Y.; Guangzhu, H.; Yanxiang, C.; Qi, P.; Yunjie, L. SOAPdenovo2: An empirically improved memory-efficient short-readde novoassembler. GigaScience 2012, 1, 18. [Google Scholar]
- Li, L.; Stoeckert, C.J., Jr.; Roos, D.S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef]
- Chen, F.; Mackey, A.J.; Stoeckert, C.J.; Roos, D.S. OrthoMCL-DB: Querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006, 34, D363–D368. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Zhao, Y.B.; Wu, J.Y.; Yang, J.H.; Sun, S.X.; Xiao, J.F.; Yu, J. PGAP: pan-genomes analysis pipeline. Bioinformatics 2012, 28, 416–418. [Google Scholar] [CrossRef]
- Jensen, L.J.; Julien, P.; Kuhn, M.; von Mering, C.; Muller, J.; Doerks, T.; Bork, P. eggNOG: Automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008, 36, D250–D254. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, H.P.; Motherway, M.O.; Lakshminarayanan, B.; Stanton, C.; Ross, R.P.; Brulc, J.; Menon, R.; O’Toole, P.W.; van Sinderen, D. Carbohydrate catabolic diversity of bifidobacteria and lactobacilli of human origin. Int. J. Food Microbiol. 2015, 203, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.R.; Tang, W.; Zheng, Y.N.; Xing, Z.Q.; Wang, Y.P. Functional and bioinformatics analysis of an exopolysaccharide-related gene (epsN) from Lactobacillus kefiranofaciens ZW3. Arch. Microbiol. 2016, 198, 611–618. [Google Scholar] [CrossRef]
- Petrova, M.I.; Macklaim, J.M.; Wuyts, S.; Verhoeven, T.; Vanderleyden, J.; Gloor, G.B.; Lebeer, S.; Reid, G. Comparative Genomic and Phenotypic Analysis of the Vaginal Probiotic Lactobacillus rhamnosus GR-1. Front. Microbiol. 2018, 9, 1278. [Google Scholar] [CrossRef]
- Van Heel, A.J.; de Jong, A.; Montalban-Lopez, M.; Kok, J.; Kuipers, O.P. BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 2013, 41, W448–W453. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Neron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Hidalgo-Cantabrana, C.; Crawley, A.B.; Sanchez, B.; Barrangou, R. Characterization and exploitation of crispr loci in Bifidobacterium longum. Front. Microbiol. 2017, 8, 1851. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Lee, J.H.; Valeriano, V.D.; Shin, Y.R.; Chae, J.P.; Kim, G.B.; Ham, J.S.; Chun, J.; Kang, D.K. Genome sequence of Lactobacillus mucosae LM1, isolated from piglet feces. J. Bacteriol. 2012, 194, 4766. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.H.; Harris, H.M.B.; McCann, A.; Guo, C.Y.; Argimon, S.; Zhang, W.Y.; Yang, X.W.; Jeffery, I.B.; Cooney, J.C.; Kagawa, T.F.; et al. Expanding the biotechnology potential of Lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 2015, 6, 8322. [Google Scholar] [CrossRef] [PubMed]
- Seshadri, R.; Leahy, S.C.; Attwood, G.T.; Teh, K.H.; Lambie, S.C.; Cookson, A.L.; Eloe-Fadrosh, E.A.; Pavlopoulos, G.A.; Hadjithomas, M.; Varghese, N.J.; et al. Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection. Nat. Biotechnol. 2018, 36, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.M.; Guinane, C.M.; London, L.E.E.; Kelleher, P.R.; Fitzgerald, G.F.; Caplice, N.M.; Ross, R.P.; Stanton, C. Genome sequence of the heteropolysaccharide-producing strain Lactobacillus mucosae DPC 6426. Microbiol. Resour. Ann. 2015, 3. [Google Scholar] [CrossRef]
- Ayala, D.I.; Cook, P.W.; Franco, J.G.; Bugarel, M.; Kottapalli, K.R.; Loneragan, G.H.; Brashears, M.M.; Nightingale, K.K. A systematic approach to identify and characterize the effectiveness and safety of novel probiotic strains to control foodborne pathogens. Front. Microbiol. 2019, 10, 1108. [Google Scholar] [CrossRef]
- Tzortzis, G.; Baillon, M.L.; Gibson, G.R.; Rastall, R.A. Modulation of anti-pathogenic activity in canine-derived Lactobacillus species by carbohydrate growth substrate. J. Appl. Microbiol. 2004, 96, 552–559. [Google Scholar] [CrossRef]
- Zhuang, X.R.; Zhu, Y.Z. Research progress of bacterial pan-genome. J. Shanghai Jiaotong Univ. (Medical Science) 2012, 32, 1440–1443. [Google Scholar]
- Riley, M.A.; Lizotte-Waniewski, M. Population genomics and the bacterial species concept. Methods Mol. Biol. 2009, 532, 367–377. [Google Scholar] [CrossRef]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef]
- Chan, Z.M.; Halachev, M.R.; Loman, N.J.; Constantinidou, C.; Pallen, M.J. Defining bacterial species in the genomic era: Insights from the genusAcinetobacter. BMC Microbiol. 2012, 12, 302. [Google Scholar] [CrossRef]
- Odamaki, T.; Bottacini, F.; Kato, K.; Mitsuyama, E.; Yoshida, K.; Horigome, A.; Xiao, J.Z.; van Sinderen, D. Genomic diversity and distribution of Bifidobacterium longum subsp. longum across the human lifespan. Sci. Rep. 2018, 8, 85. [Google Scholar] [CrossRef] [PubMed]
- Hammes, W.P.; Hertel, C. Lactobacillus. In Bergey’s Manual of Systematics of Archaea and Bacteria; Whitman, W.B., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 1–76. [Google Scholar] [CrossRef]
- Wheatley, R.W.; Lo, S.; Jancewicz, L.J.; Dugdale, M.L.; Huber, R.E. Structural explanation for allolactose (lac operon inducer) synthesis by lacz beta-galactosidase and the evolutionary relationship between allolactose synthesis and the lac repressor. J. Biol. Chem. 2013, 288, 12993–13005. [Google Scholar] [CrossRef] [PubMed]
- Odamaki, T.; Horigome, A.; Sugahara, H.; Hashikura, N.; Minami, J.; Xiao, J.Z.; Abe, F. Comparative genomics revealed genetic diversity and species/strain-level differences in carbohydrate metabolism of three probiotic Bifidobacterial species. Int. J. Genomics 2015, 2015, 12. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.; Slesarev, A.; Wolf, Y.; Sorokin, A.; Mirkin, B.; Koonin, E.; Pavlov, A.; Pavlova, N.; Karamychev, V.; Polouchine, N.; et al. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 2006, 103, 15611–15616. [Google Scholar] [CrossRef]
- Arboleya, S.; Bottacini, F.; O’Connell-Motherway, M.; Ryan, C.A.; Ross, R.P.; van Sinderen, D.; Stanton, C. Gene-trait matching across the Bifidobacterium longum pan-genome reveals considerable diversity in carbohydrate catabolism among human infant strains. BMC Genomics 2018, 19, 33. [Google Scholar] [CrossRef]
- Schluepmann, H.; Pellny, T.; van Dijken, A.; Smeekens, S.; Paul, M. Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 6849–6854. [Google Scholar] [CrossRef]
- Helanto, M.; Aarnikunnas, J.; Palva, A.; Leisola, M.; Nyyssola, A. Characterization of genes involved in fructose utilization by Lactobacillus fermentum. Arch. Microbiol. 2006, 186, 51–59. [Google Scholar] [CrossRef]
- Okano, K.; Yoshida, S.; Tanaka, T.; Ogino, C.; Fukuda, H.; Kondo, A. Homo-d-Lactic acid fermentation from arabinose by redirection of the phosphoketolase pathway to the pentose phosphate pathway in L-Lactate dehydrogenase gene-deficient Lactobacillus plantarum. Appl. Environ. Microbiol. 2009, 75, 5175. [Google Scholar] [CrossRef]
- Franco, I.S.; Mota, L.J.; Soares, C.M.; de Sa-Nogueira, I. Functional domains of the bacillus subtilis transcription factor arar and identification of amino acids important for nucleoprotein complex assembly and effector binding. J. Bacteriol. 2006, 188, 3024–3036. [Google Scholar] [CrossRef]
- Prechtl, R.M.; Janssen, D.; Behr, J.; Ludwig, C.; Kuster, B.; Vogel, R.F.; Jakob, F. Sucrose-induced proteomic response and carbohydrate utilization of Lactobacillus sakei TWM 1.411 during dextran formation. Front. Microbiol. 2018, 9, 2796. [Google Scholar] [CrossRef]
- Hendrickson, W.; Schleif, R.F. Regulation of the Escherichia coli L-arabinose operon studied by gel electrophoresis DNA binding assay. J. Mol. Biol. 1984, 178, 611–628. [Google Scholar] [CrossRef]
- Dan, T.; Fukuda, K.; Sugai-Bannai, M.; Takakuwa, N.; Motoshima, H.; Urashima, T. Characterization and expression analysis of the exopolysaccharide gene cluster in Lactobacillus fermentum TDS030603. Biosci. Biotech. Biochem. 2009, 73, 2656–2664. [Google Scholar] [CrossRef] [PubMed]
- Lebeer, S.; Verhoeven, T.L.A.; Francius, G.; Schoofs, G.; Lambrichts, I.; Dufrene, Y.; Vanderleyden, J.; De Keersmaecker, S.C.J. Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Appl. Environ. Microbiol. 2009, 75, 3554–3563. [Google Scholar] [CrossRef] [PubMed]
- Vastano, V.; Perrone, F.; Marasco, R.; Sacco, M.; Muscariello, L. Transcriptional analysis of exopolysaccharides biosynthesis gene clusters in Lactobacillus plantarum. Arch. Microbiol. 2016, 198, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Dertli, E.; Mayer, M.J.; Colquhoun, I.J.; Narbad, A. EpsA is an essential gene in exopolysaccharide production in Lactobacillus johnsonii FI9785. Microb. Biotechnol. 2016, 9, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Xiong, Z.Q.; Kong, L.H.; Wang, G.Q.; Ai, L.Z. Relationship between putative eps genes and production of exopolysaccharide in Lactobacillus casei LC2W. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Azevedo, A.C.; Bento, C.B.; Ruiz, J.C.; Queiroz, M.V.; Mantovani, H.C. Distribution and genetic diversity of bacteriocin gene clusters in rumen microbial genomes. Appl. Environ. Microbiol. 2015, 81, 7290–7304. [Google Scholar] [CrossRef]
- Nilsen, T.; Nes, I.F.; Holo, H. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 2003, 69, 2975–2984. [Google Scholar] [CrossRef]
- Veljovic, K.; Terzic-Vidojevic, A.; Tolinacki, M.; Kojic, M.; Topisirovic, L. Molecular analysis of enterolysin a and entl gene cluster from natural isolate Enterococcus faecalis bgpt1-10p. Genetika-Belgrade 2013, 45, 479–492. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ross, R.P.; Hill, C.; Dobson, A. Bacteriocin production: A probiotic trait? Appl. Environ. Microbiol. 2011, 78, 1–6. [Google Scholar] [CrossRef]
- Walsh, M.C.; Gardiner, G.E.; Hart, O.M.; Lawlor, P.G.; Daly, M.; Lynch, B.; Richert, B.T.; Radcliffe, S.; Giblin, L.; Hill, C.; et al. Predominance of a bacteriocin-producing Lactobacillus salivarius component of a five-strain probiotic in the porcine ileum and effects on host immune phenotype. FEMS Microbiol. Ecol. 2008, 64, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Gillor, O.; Giladi, I.; Riley, M.A. Persistence of colicinogenic Escherichia coli in the mouse gastrointestinal tract. BMC Microbiol. 2009, 9, 165. [Google Scholar] [CrossRef] [PubMed]
- Haft, D.H.; Selengut, J.; Mongodin, E.F.; Nelson, K.E. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005, 1, e60. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Grishin, N.V.; Shabalina, S.A.; Wolf, Y.I.; Koonin, E.V. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 2006, 1, 7. [Google Scholar] [CrossRef]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- Chylinski, K.; Makarova, K.S.; Charpentier, E.; Koonin, E.V. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014, 42, 6091–6105. [Google Scholar] [CrossRef]
- Horvath, P.; Romero, D.A.; Coute-Monvoisin, A.C.; Richards, M.; Deveau, H.; Moineau, S.; Boyaval, P.; Fremaux, C.; Barrangou, R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 2008, 190, 1401–1412. [Google Scholar] [CrossRef]
- Boto, L. Horizontal gene transfer in evolution: Facts and challenges. Proc. Biol. Sci. 2010, 277, 819–827. [Google Scholar] [CrossRef]

| Strain | Region (City/Province) | Origin/Age | Gene Size | GC (%) | tRNA | Accession No. | Reference |
|---|---|---|---|---|---|---|---|
| (Mb) | |||||||
| DCC1HL5 | Dali, Yunnan | Bovine Feces | 1.96 | 48.17 | 57 | SAMN13220078 | This work |
| FAHBZ1M4 | Bozhou, Anhui | Human feces, 24y | 2.22 | 48.14 | 66 | SAMN13220079 | This work |
| FAHBZ5L1 | Bozhou, Anhui | Human feces, 77y | 2.05 | 48.17 | 62 | SAMN13220080 | This work |
| FAHBZ12M2 | Bozhou, Anhui | Human feces, 6m | 2.21 | 47.95 | 62 | SAMN13220081 | This work |
| FAHBZ16L4 | Bozhou, Anhui | Human feces, 63y | 2.02 | 47.85 | 52 | SAMN13220082 | This work |
| FAHBZ18M3 | Bozhou, Anhui | Human feces, 26y | 2.07 | 48.09 | 48 | SAMN13220083 | This work |
| FAHBZ20L1 | Bozhou, Anhui | Human feces, 52y | 2.00 | 47.84 | 54 | SAMN13220084 | This work |
| FAHBZ34M2 | Bozhou, Anhui | Human feces, 51y | 2.11 | 48.12 | 61 | SAMN13220085 | This work |
| FAHBZ49M2 | Bozhou, Anhui | Human feces, 53y | 2.01 | 48.21 | 53 | SAMN13220086 | This work |
| FAHBZ50M5 | Bozhou, Anhui | Human feces, 14y | 1.97 | 47.89 | 59 | SAMN13220087 | This work |
| FAHBZ53M2 | Bozhou, Anhui | Human feces, 54y | 2.01 | 48.08 | 47 | SAMN13220088 | This work |
| FGSYC1M1 | Yongchang, Gansu | Human feces, 77y | 2.13 | 48.07 | 50 | SAMN13220089 | This work |
| FGSYC15L3 | Yongchang, Gansu | Human feces, 78y | 2.07 | 47.84 | 52 | SAMN13220090 | This work |
| FGSYC16M1 | Yongchang, Gansu | Human feces, 77y | 2.07 | 47.68 | 55 | SAMN13220091 | This work |
| FGSYC17L3 | Yongchang, Gansu | Human feces, 53y | 2.11 | 47.73 | 48 | SAMN13220092 | This work |
| FGSYC19L1 | Yongchang, Gansu | Human feces, 66y | 2.17 | 47.98 | 54 | SAMN13220093 | This work |
| FGSYC23L4 | Yongchang, Gansu | Human feces, 67y | 2.14 | 47.98 | 54 | SAMN13220094 | This work |
| FGSYC27L1 | Yongchang, Gansu | Human feces, 54y | 2.07 | 48.23 | 50 | SAMN13220095 | This work |
| FGSYC42L4 | Yongchang, Gansu | Human feces, 57y | 2.09 | 47.96 | 48 | SAMN13220096 | This work |
| FGSYC43L5 | Yongchang, Gansu | Human feces, 55y | 2.33 | 48.08 | 58 | SAMN13220097 | This work |
| FGSYC90L5 | Yongchang, Gansu | Human feces, 67y | 2.22 | 48.1 | 52 | SAMN13220098 | This work |
| FHNXY1L8 | Xiayi, Henan | Human feces, 65y | 2.06 | 48.06 | 60 | SAMN13220099 | This work |
| FHNXY19M3 | Xiayi, Henan | Human feces, 85y | 2.14 | 47.98 | 49 | SAMN13220100 | This work |
| FHNXY29L2 | Xiayi, Henan | Human feces, 66y | 2.07 | 47.56 | 65 | SAMN13220101 | This work |
| FHNXY31L3 | Xiayi, Henan | Human feces, 33y | 2.06 | 48.08 | 44 | SAMN13220102 | This work |
| FHNXY47L1 | Xiayi, Henan | Human feces, 86y | 2.01 | 48.12 | 46 | SAMN13220103 | This work |
| FHNXY68L2 | Xiayi, Henan | Dog feces | 2.06 | 48.06 | 47 | SAMN13220104 | This work |
| FHNXY72L1 | Xiayi, Henan | Dog feces | 2.01 | 48.15 | 53 | SAMN13220105 | This work |
| FHuNHHMY89L8 | Mayang, Hunan | Pig feces | 2.25 | 47.84 | 53 | SAMN13220106 | This work |
| FJSCZ8L1 | Changzhou, Jiangsu | Dog feces | 1.99 | 48.23 | 42 | SAMN13220107 | This work |
| FJSNT61 | Nantong, Jiangsu | Human feces, 82y | 2.13 | 48.38 | 48 | SAMN13220108 | This work |
| FJSNT81 | Nantong, Jiangsu | Human feces, 60y | 2.20 | 48.1 | 47 | SAMN13220109 | This work |
| FJSNT91 | Nantong, Jiangsu | Human feces, 79y | 2.14 | 48.36 | 55 | SAMN13220110 | This work |
| FJSNT141 | Nantong, Jiangsu | Human feces, 82y | 2.05 | 48.26 | 61 | SAMN13220111 | This work |
| FJSNT152 | Nantong, Jiangsu | Human feces, 95y | 2.08 | 47.91 | 48 | SAMN13220112 | This work |
| FJSNT182 | Nantong, Jiangsu | Human feces, 85y | 2.27 | 47.65 | 56 | SAMN13220113 | This work |
| FJSNT192 | Nantong, Jiangsu | Human feces, 84y | 2.23 | 48.01 | 56 | SAMN13220114 | This work |
| FJSNT312 | Nantong, Jiangsu | Human feces, 64y | 2.04 | 48.04 | 59 | SAMN13220115 | This work |
| FJSNT321 | Nantong, Jiangsu | Human feces, 63y | 2.04 | 48.04 | 45 | SAMN13220116 | This work |
| FJSNT331 | Nantong, Jiangsu | Human feces, 92y | 2.18 | 48.1 | 66 | SAMN13220117 | This work |
| FJSWX5M5 | Wuxi, Jiangsu | Human feces, 81y | 2.12 | 48.22 | 50 | SAMN13220118 | This work |
| FJSWX8M3 | Wuxi, Jiangsu | Human feces, 88y | 2.08 | 48.07 | 50 | SAMN13220119 | This work |
| FJSWX13M2 | Wuxi, Jiangsu | Human feces, 79y | 2.17 | 48.21 | 56 | SAMN13220120 | This work |
| FJSWX15M1 | Wuxi, Jiangsu | Human feces, 82y | 2.08 | 47.81 | 59 | SAMN13220121 | This work |
| FJSWX18M1 | Wuxi, Jiangsu | Human feces, 81y | 2.2 | 48.15 | 56 | SAMN13220122 | This work |
| FJSWX19M2 | Wuxi, Jiangsu | Human feces, 78y | 2.12 | 47.83 | 62 | SAMN13220123 | This work |
| FJSWX20M1 | Wuxi, Jiangsu | Human feces, 90y | 2.16 | 47.84 | 57 | SAMN13220124 | This work |
| FJSWX21M1 | Wuxi, Jiangsu | Human feces, 90y | 2.17 | 48.08 | 49 | SAMN13220125 | This work |
| FJSWX27M1 | Wuxi, Jiangsu | Human feces, 88y | 2.05 | 47.68 | 60 | SAMN13220126 | This work |
| FJSWX28M2 | Wuxi, Jiangsu | Human feces, 83y | 2.05 | 47.84 | 62 | SAMN13220127 | This work |
| FJSWX34JL6 | Wuxi, Jiangsu | Human feces, 3y | 2.19 | 48.27 | 48 | SAMN13220128 | This work |
| FJSXYWG2L4 | Xuyi, Jiangsu | Silage | 1.97 | 48.33 | 41 | SAMN13220129 | This work |
| FJSXYWG6L3 | Xuyi, Jiangsu | Cow feces | 1.94 | 48.1 | 54 | SAMN13220130 | This work |
| FSH3M1 | Shanghai | Human feces, 83y | 1.99 | 48.37 | 41 | SAMN13220131 | This work |
| FSH12M2 | Shanghai | Human feces, 84y | 2.02 | 48.25 | 57 | SAMN13220132 | This work |
| FSH14M2 | Shanghai | Human feces, 85y | 2.10 | 48.17 | 54 | SAMN13220133 | This work |
| FSH18M1 | Shanghai | Human feces, 85y | 2.03 | 48.04 | 60 | SAMN13220134 | This work |
| FSH19M2 | Shanghai | Human feces, 86y | 2.04 | 48.23 | 59 | SAMN13220135 | This work |
| FSH21M1 | Shanghai | Human feces, 87y | 1.94 | 48.15 | 60 | SAMN13220136 | This work |
| FSH22M2 | Shanghai | Human feces, 78y | 2.02 | 48.16 | 60 | SAMN13220137 | This work |
| FSH26M1 | Shanghai | Human feces, 71y | 2.03 | 47.88 | 59 | SAMN13220138 | This work |
| FSH41M1 | Shanghai | Human feces, 86y | 2.09 | 48.24 | 59 | SAMN13220139 | This work |
| FSH46M2 | Shanghai | Human feces, 84y | 1.99 | 47.87 | 59 | SAMN13220140 | This work |
| FSH47M1 | Shanghai | Human feces, 85y | 2.08 | 47.88 | 58 | SAMN13220141 | This work |
| FYNLJ83L1 | Lijiang, Yunnan | Pig feces | 2.11 | 48.15 | 56 | SAMN13220142 | This work |
| FYNLJ87L2 | Lijiang, Yunnan | Pig feces | 2.10 | 47.97 | 49 | SAMN13220143 | This work |
| FZJTZ17M6 | Taizhou, Zhejiang | Human feces, 67y | 2.2 | 48.1 | 59 | SAMN13220144 | This work |
| FZJTZ18M3 | Taizhou, Zhejiang | Human feces, 79y | 1.86 | 48.03 | 64 | SAMN13220145 | This work |
| FZJTZ22M3 | Taizhou, Zhejiang | Human feces, 55y | 1.93 | 47.84 | 64 | SAMN13220146 | This work |
| FZJTZ23M9 | Taizhou, Zhejiang | Human feces, 76y | 2.45 | 47.71 | 46 | SAMN13220147 | This work |
| FZJTZ24M5 | Taizhou, Zhejiang | Human feces, 62y | 2.28 | 48.29 | 64 | SAMN13220148 | This work |
| FZJTZ26M3 | Taizhou, Zhejiang | Human feces, 40y | 1.97 | 48.59 | 44 | SAMN13220149 | This work |
| FZJTZ32M4 | Taizhou, Zhejiang | Human feces, 58y | 1.99 | 47.95 | 60 | SAMN13220150 | This work |
| FZJTZ34M1 | Taizhou, Zhejiang | Human feces, 76y | 2.20 | 48.16 | 50 | SAMN13220151 | This work |
| FZJTZ36M3 | Taizhou, Zhejiang | Human feces, 80y | 2.05 | 48.37 | 58 | SAMN13220152 | This work |
| FZJTZ60M1 | Taizhou, Zhejiang | Human feces, 78y | 2.25 | 48.3 | 53 | SAMN13220153 | This work |
| GD1M1 | Lianzhou, Guangdong | Human feces, 75y | 2.33 | 48.28 | 51 | SAMN13220154 | This work |
| GD2M2 | Lianzhou, Guangdong | Human feces, 84y | 2.33 | 48.15 | 58 | SAMN13220155 | This work |
| GD3M2 | Lianzhou, Guangdong | Human feces, 76y | 2.32 | 47.97 | 79 | SAMN13220156 | This work |
| GD4M1 | Lianzhou, Guangdong | Human feces, 5y | 2.13 | 48.06 | 38 | SAMN13220157 | This work |
| GD9M1 | Lianzhou, Guangdong | Human feces, 76y | 2.21 | 48.17 | 52 | SAMN13220158 | This work |
| GD14M3 | Lianzhou, Guangdong | Human feces, 10y | 2.36 | 48.18 | 55 | SAMN13220159 | This work |
| GD15M1 | Lianzhou, Guangdong | Human feces, 10y | 2.36 | 48.3 | 67 | SAMN13220160 | This work |
| GD16M9 | Lianzhou, Guangdong | Human feces, 81y | 2.36 | 48.23 | 56 | SAMN13220161 | This work |
| GD17M2 | Lianzhou, Guangdong | Human feces, 8y | 2.14 | 48.36 | 66 | SAMN13220162 | This work |
| GD22M1 | Lianzhou, Guangdong | Human feces, 73y | 2.15 | 48.23 | 57 | SAMN13220163 | This work |
| GD24M2 | Lianzhou, Guangdong | Human feces, 58y | 2.13 | 48.25 | 58 | SAMN13220164 | This work |
| GD64M2 | Lianzhou, Guangdong | Human feces, 64y | 2.05 | 48.19 | 55 | SAMN13220165 | This work |
| GD67M2 | Lianzhou, Guangdong | Human feces, 9y | 2.01 | 48.19 | 56 | SAMN13220166 | This work |
| GD68M4 | Lianzhou, Guangdong | Human feces, 10y | 2.00 | 48.31 | 53 | SAMN13220167 | This work |
| GD69M1 | Lianzhou, Guangdong | Human feces, 63y | 2.03 | 47.87 | 67 | SAMN13220168 | This work |
| M104R01L3 | Dangxiong, Tibet | Yak milk | 2.07 | 47.92 | 57 | SAMN13220169 | This work |
| VWX301M18 | Wuxi, Jiangsu | Human vagina | 2.01 | 47.84 | 61 | SAMN13220170 | This work |
| LM1 | Brazil | Pig small intestine | 2.43 | 46.13 | 91 | SAMN02470226 | [31] |
| DSM13345 | Sweden | Pig small intestine | 2.25 | 46.40 | - | SAMN02369406 | [32] |
| KHPC15 | United States | Bovine rumen | 1.88 | 46.70 | 46 | SAMN05216545 | [33] |
| KHPX11 | United States | Bovine rumen | 1.88 | 46.70 | 64 | SAMN05216461 | [33] |
| WCC8 | United States | Bovine rumen | 1.88 | 46.70 | 59 | SAMN05216430 | [33] |
| DPC6426 | Ireland | Bovine rumen | 2.80 | 47.00 | 89 | SAMN03145820 | [34] |
| AGR63 | United States | Bovine rumen | 1.94 | 47.00 | 94 | SAMN02744693 | [33] |
| L24-B | United States | Bovine rumen | 2.00 | 46.90 | 88 | SAMN10744154 | [35] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, Y.; Yang, B.; Ross, P.; Stanton, C.; Zhang, H.; Zhao, J.; Chen, W. Comparative Genomics Analysis of Lactobacillus mucosae from Different Niches. Genes 2020, 11, 95. https://doi.org/10.3390/genes11010095
Jia Y, Yang B, Ross P, Stanton C, Zhang H, Zhao J, Chen W. Comparative Genomics Analysis of Lactobacillus mucosae from Different Niches. Genes. 2020; 11(1):95. https://doi.org/10.3390/genes11010095
Chicago/Turabian StyleJia, Yan, Bo Yang, Paul Ross, Catherine Stanton, Hao Zhang, Jianxin Zhao, and Wei Chen. 2020. "Comparative Genomics Analysis of Lactobacillus mucosae from Different Niches" Genes 11, no. 1: 95. https://doi.org/10.3390/genes11010095
APA StyleJia, Y., Yang, B., Ross, P., Stanton, C., Zhang, H., Zhao, J., & Chen, W. (2020). Comparative Genomics Analysis of Lactobacillus mucosae from Different Niches. Genes, 11(1), 95. https://doi.org/10.3390/genes11010095





