SparkRA: Enabling Big Data Scalability for the GATK RNA-seq Pipeline with Apache Spark
Abstract
1. Introduction
- Introducing SparkRA, a Spark-based pipeline to scale up RNA-seq analysis pipelines easily and efficiently, using the Apache Spark in-memory framework.
- Improving the parallelism of the GATK best practices RNA-seq tools by addressing their sequential bottlenecks, and allowing them to take full advantage of the capabilities of SparkRA.
- Comparing the performance of SparkRA with other state-of-the-art pipelines, and measuring an overall speedup of 7.7× as SparkRA scales the GATK RNA-seq pipeline from one node to 16. Compared to Halvade-RNA, our solution is about 1.3× faster on a single node and 1.2× faster on a cluster.
2. Methods
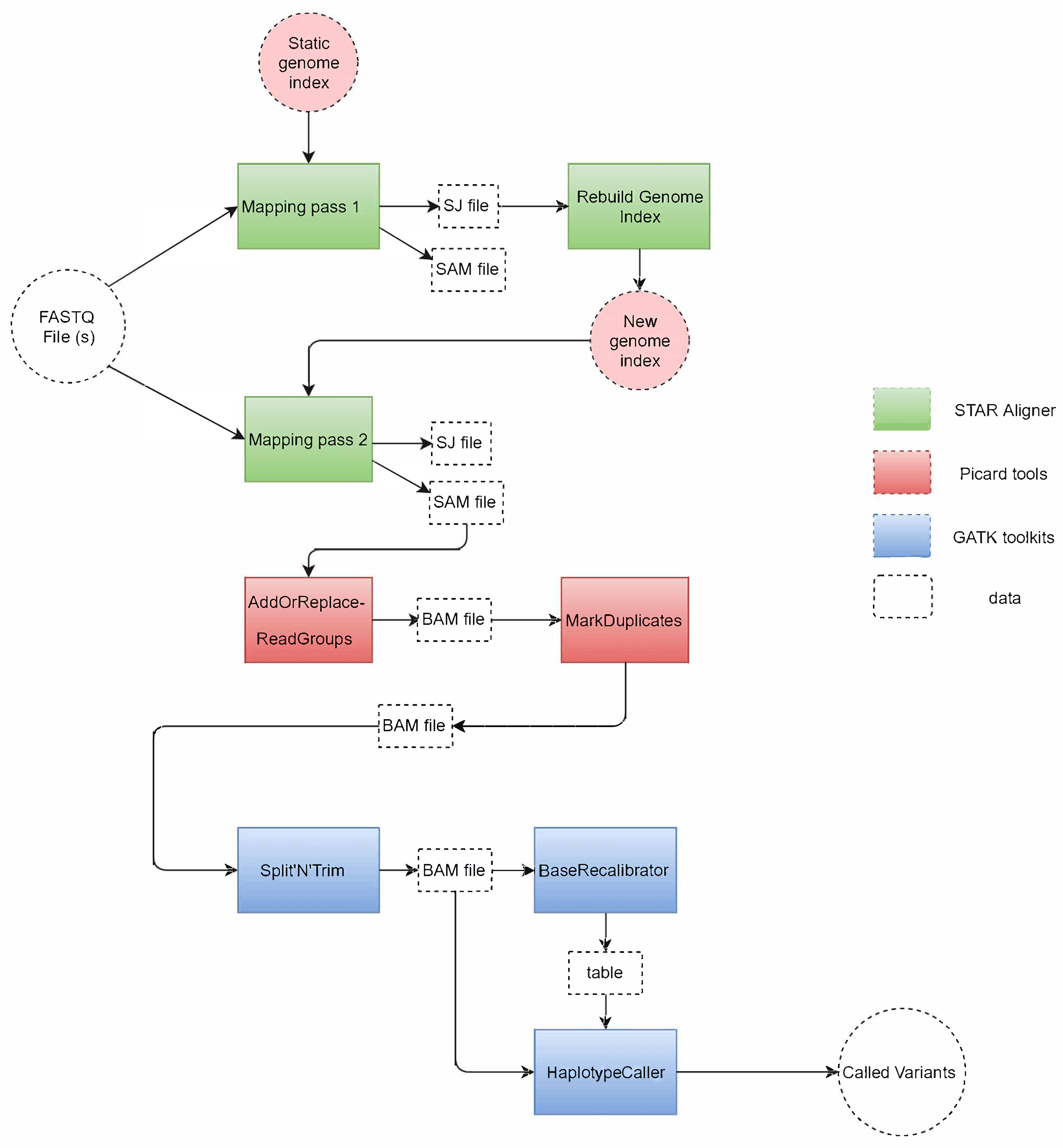
2.1. GATK RNA-seq
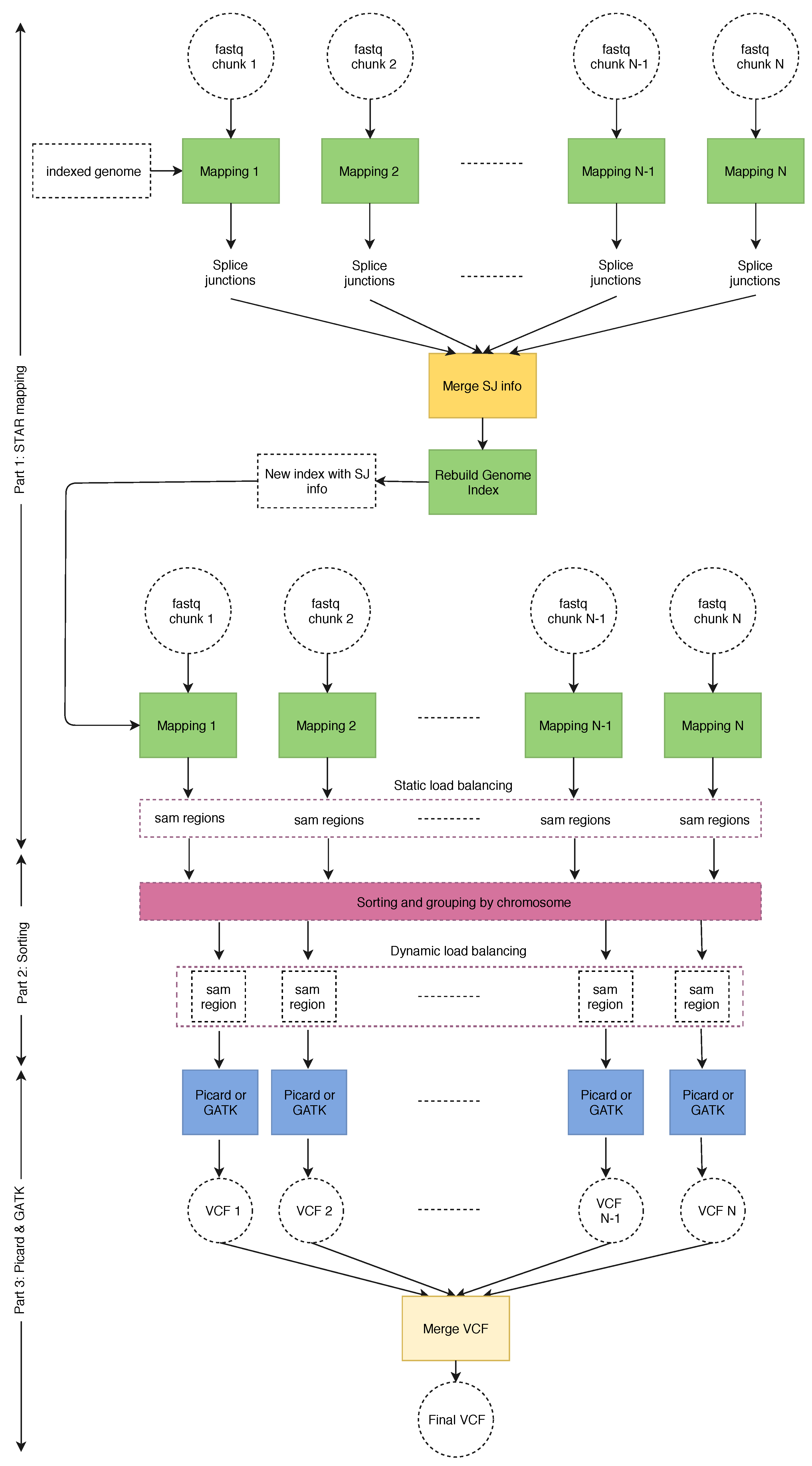
2.2. SparkRA Execution Flow
- Part 1
- does the alignment and performs static load balancing according to the size of each chromosome defined by the reference genome.
- Part 2
- performs dynamic load balancing to divide the SAM files according to the actual number of reads mapped to each chromosome.
- Part 3
- is represented by Picard and GATK.
2.3. Static and Dynamic Load Balancing
2.3.1. Static Load Balancing
2.3.2. Dynamic Load Balancing
3. Results
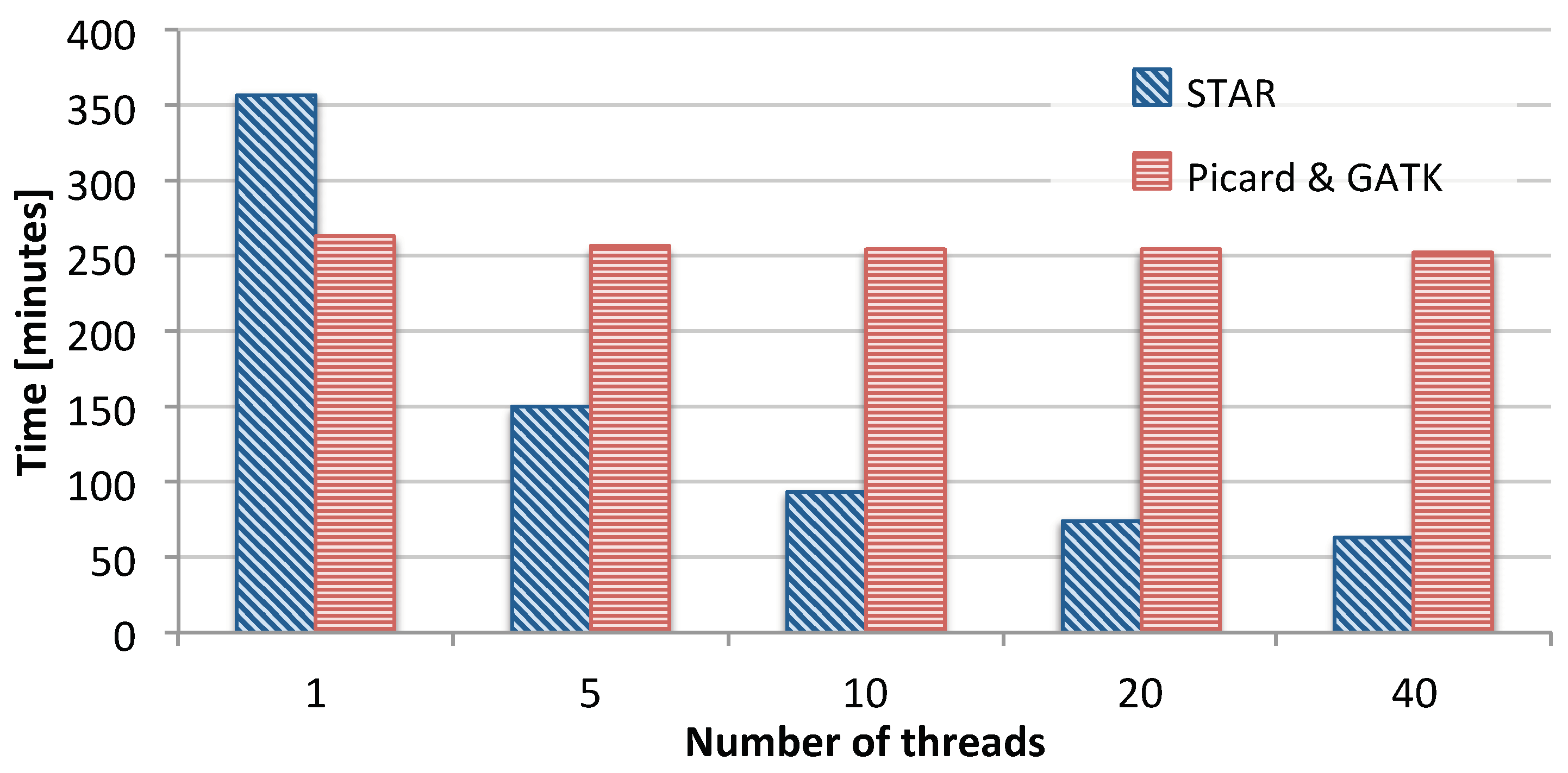
3.1. Single-Node Performance
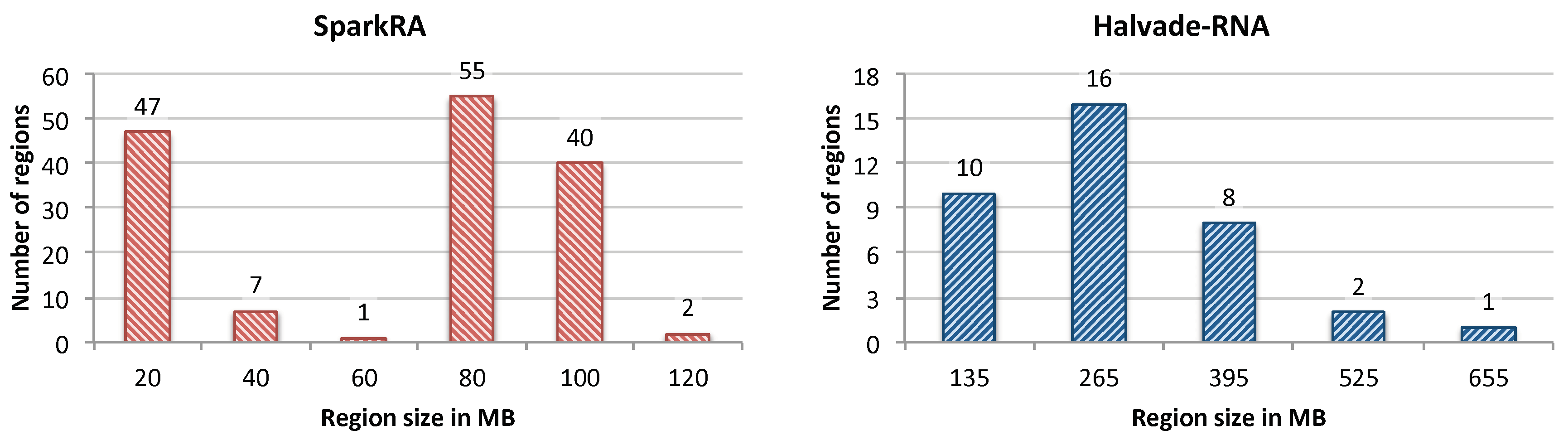
3.1.1. Impact of Load Balancing
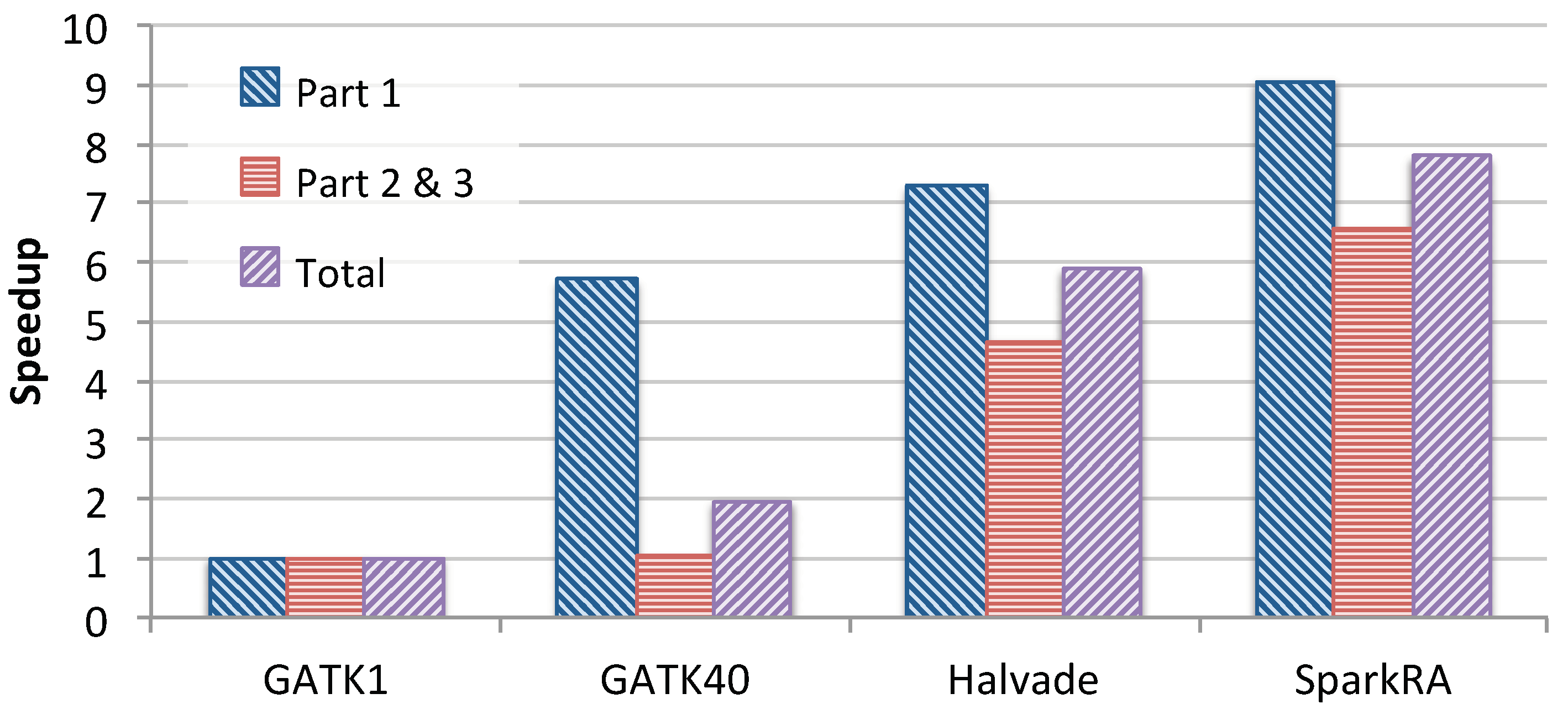
3.1.2. Comparison with Existing Solutions
3.2. Multi-Node Performance
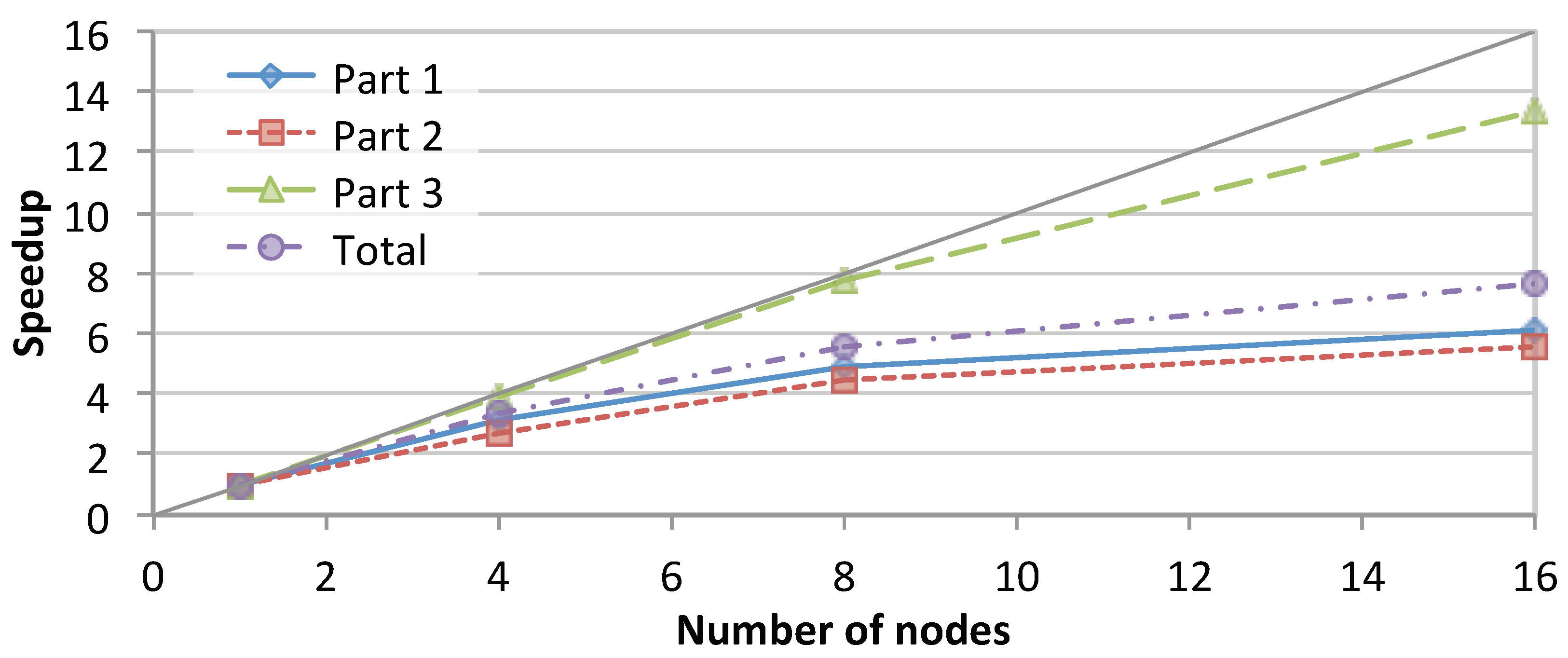
3.2.1. Scalability of SparkRA
3.2.2. Comparison with Existing Solutions
4. Discussion
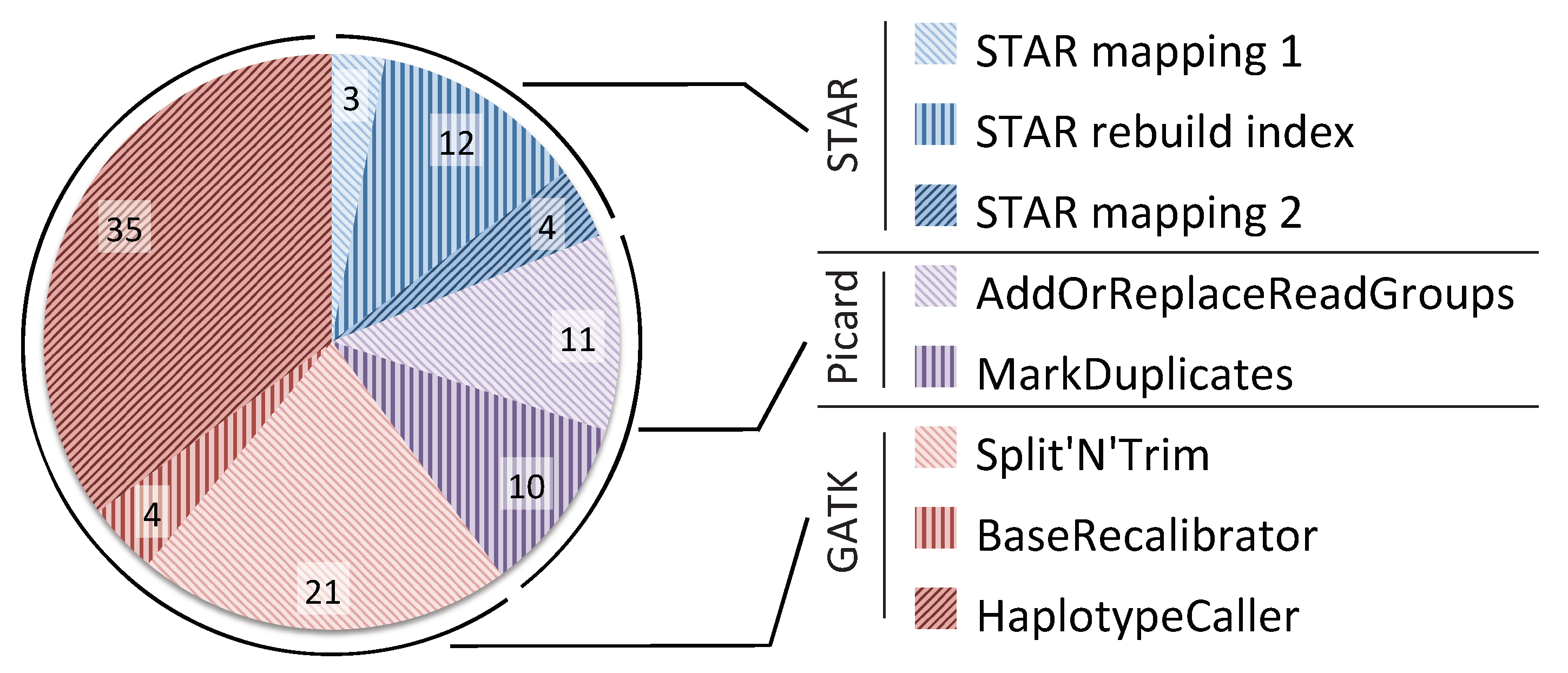
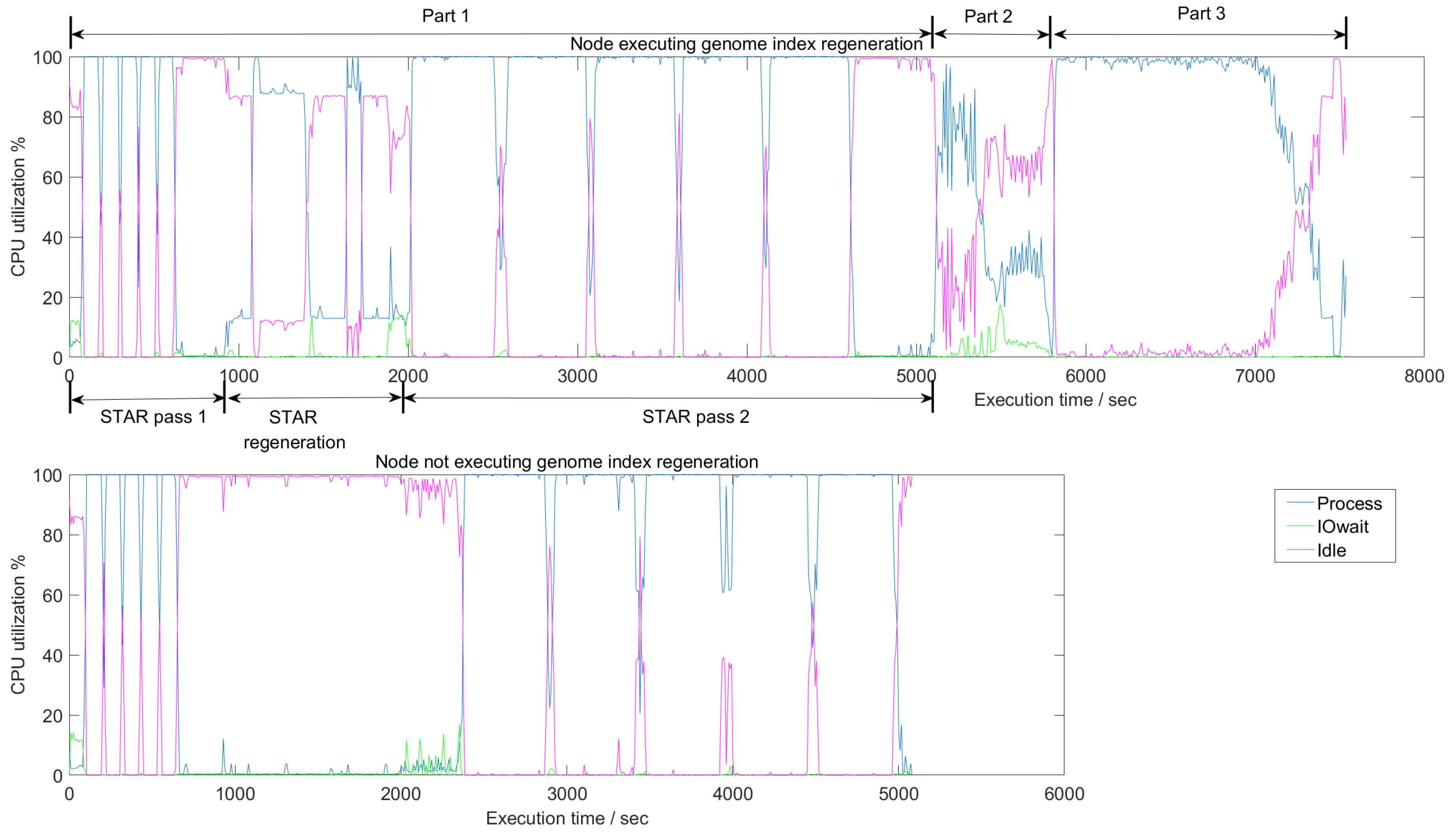
4.1. CPU Utilization
4.2. Pipeline Accuracy
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van der Auwera, G.A.; Carneiro, M.; Hartl, C.; Poplin, R.; del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar]
- Kelly, B.J.; Fitch, J.R.; Hu, Y.; Corsmeier, D.J.; Zhong, H.; Wetzel, A.N.; Nordquist, R.D.; Newsom, D.L.; White, P. Churchill: An ultra-fast, deterministic, highly scalable and balanced parallelization strategy for the discovery of human genetic variation in clinical and population-scale genomics. Genome Biol. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Decap, D.; Reumers, J.; Herzeel, C.; Costanza, P.; Fostier, J. Halvade: Scalable sequence analysis with MapReduce. Bioinformatics 2015, 31, 2482–2488. [Google Scholar] [CrossRef]
- Dean, J.; Ghemawat, S. MapReduce: Simplified Data Processing on Large Clusters. Commun. ACM 2008, 51, 107–113. [Google Scholar] [CrossRef]
- Mushtaq, H.; Liu, F.; Costa, C.; Liu, G.; Hofstee, P.; Al-Ars, Z. SparkGA: A Spark Framework for Cost Effective, Fast and Accurate DNA Analysis at Scale. In Proceedings of the ACM Conference Bioinformatics, Computational Biology and Health Informatics, Boston, MA, USA, 20–23 August 2017. [Google Scholar]
- Mushtaq, H.; Ahmed, N.; Al-Ars, Z. SparkGA2: Production-Quality Memory-Efficient Apache Spark Based Genome Analysis Framework. PLoS ONE 2019, 14, e0224784. [Google Scholar] [CrossRef]
- Mushtaq, H.; Al-Ars, Z. Cluster-based Apache Spark implementation of the GATK DNA analysis pipeline. In Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Washington, DC, USA, 9–12 November 2015; pp. 1471–1477. [Google Scholar]
- Zaharia, M.; Chowdhury, M.; Franklin, M.J.; Shenker, S.; Stoica, I. Spark: Cluster computing with working sets. In Proceedings of the 2nd USENIX Conference on Hot Topics in Cloud Computing (HotCloud’10), Boston, MA, USA, 22–25 June 2010. [Google Scholar]
- Hasan, L.; Al-Ars, Z. An Efficient and High Performance Linear Recursive Variable Expansion Implementation of the Smith-Waterman Algorithm. In Proceedings of the IEEE Engineering in Medicine and Biology Conference, Minneapolis, MN, USA, 3–6 September 2009; pp. 3845–3848. [Google Scholar]
- Hasan, L.; Al-Ars, Z. An Overview of Hardware-based Acceleration of Biological Sequence Alignment. In Computational Biology and Applied Bioinformatics; InTech: Rijeka, Croatia, 2011; pp. 187–202. [Google Scholar]
- Han, Y.; Gao, S.; Muegge, K.; Zhang, W.; Bing, Z. Advanced applications of RNA sequencing and challenges. Bioinform. Biol. Insights 2015. [Google Scholar] [CrossRef] [PubMed]
- Piskol, R.; Ramaswami, G.; Li, J.B. Reliable identification of genomic variants from RNA-seq data. Am. J. Hum. Genet. 2013, 4, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Cummings, B.B.; Marshall, J.L.; Tukiainen, T.; Lek, M.; Donkervoort, S.; Foley, A.R.; Bolduc, V.; dell Wa, L.; Sandaradura, S.; O’Grady, G.L.; et al. Improving genetic diagnosis in mendelian disease with transcriptome sequencing. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.D.; Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010, 26, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Singh, D.; Zeng, Z.; Coleman, S.J.; Huang, Y.; Savich, G.L.; He, X.; Mieczkowski, P.; Grimm, S.A.; Perou, C.M.; et al. Mapsplice: Accurate mapping of rna-seq reads for splice junction discovery. Nucleic Acids Res. 2010, 38, e178. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. Tophat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. Star: Ultrafast universal rna-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- GATK. Calling Variants in RNAseq. Available online: https://software.broadinstitute.org/gatk/documentation/article.php?id=3891 (accessed on 19 October 2019).
- Decap, D.; Reumers, J.; Herzeel, C.; Costanza, P.; Fostier, J. Halvade-rna: Parallel variant calling from transcriptomic data using mapreduce. PLoS ONE 2017, 12, e0174575. [Google Scholar] [CrossRef] [PubMed]
- Engstrom, P.G.; Steijger, T.; Sipos, B.; Grant, G.R.; Kahles, A.; Rätsch, G.; Goldman, N.; Hubbard, T.J.; Harrow, J.; Guigó, R.; et al. Systematic evaluation of spliced alignment programs for rna-seq data. Nat. Methods 2013, 10, 1185. [Google Scholar] [CrossRef] [PubMed]
- Wang, S. Scaling Up the GATK RNA-seq Variant Calling Pipeline with Apache Spark; Delft University of Technology: Delft, The Netherlands, 2018. [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of dna elements in the human genome. Nature 2012, 489, 57. [Google Scholar] [CrossRef] [PubMed]
- SURFsara. Available online: https://www.surf.nl/en/research-ict (accessed on 19 October 2019).
- iostat Linux Man Page. Available online: https://linux.die.net/man/1/iostat (accessed on 19 October 2019).
- RTG Tools, Real Time Genomics. Available online: https://www.realtimegenomics.com/products/rtg-tools (accessed on 19 October 2019).
| Time [minutes] | ||||||
|---|---|---|---|---|---|---|
| Part 1 | Part 2 | Part 3 | Total | |||
| 180 | 224 | 304 | 39.4 | 9.33 | 32.5 | 81.2 |
| 144 | 193 | 262 | 39.7 | 10.3 | 30.8 | 80.8 |
| 108 | 156 | 217 | 40.7 | 11.2 | 29.2 | 81.0 |
| 72 | 123 | 183 | 39.7 | 11.4 | 29.1 | 80.2 |
| 50 | 103 | 161 | 40.0 | 13.8 | 29.4 | 83.2 |
| Pipeline Stage | GATK1 | GATK40 | Halvade | SparkRA | |
|---|---|---|---|---|---|
| Part 1: | STAR (pass 1) | 92.4 | 9.87 | 9.60 | 10.5 |
| Index rebuild | 152 | 39.2 | 6.70 | 6.27 | |
| STAR (pass 2) | 116 | 14.0 | 33.0 | 22.9 | |
| Total Part 1 | 360 | 63.1 | 49.3 | 39.7 | |
| Part 2: | Sorting | n/a | n/a | n/a | 11.4 |
| Part 3: | Picard & GATK | 265 | 255 | 56.8 | 29.1 |
| Total pipeline time | 625 | 318 | 106 | 80.2 | |
| Pipeline Stage | Halvade | SparkRA | Speedup |
|---|---|---|---|
| Part 1: Mapping | 54.7 | 47.2 | 1.16 |
| Parts 2 and 3: Sorting, Picard & GATK | 26.2 | 19.3 | 1.36 |
| Total pipeline time | 80.9 | 66.5 | 1.22 |
| Pipeline | #Regions | TP | FP | FN | Sensitivity | Precision |
|---|---|---|---|---|---|---|
| SparkRA | 183 | 109411 | 6886 | 6363 | 94.50% | 94.08% |
| Halvade | 142 | 109669 | 6850 | 6105 | 94.73% | 94.12% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Ars, Z.; Wang, S.; Mushtaq, H. SparkRA: Enabling Big Data Scalability for the GATK RNA-seq Pipeline with Apache Spark. Genes 2020, 11, 53. https://doi.org/10.3390/genes11010053
Al-Ars Z, Wang S, Mushtaq H. SparkRA: Enabling Big Data Scalability for the GATK RNA-seq Pipeline with Apache Spark. Genes. 2020; 11(1):53. https://doi.org/10.3390/genes11010053
Chicago/Turabian StyleAl-Ars, Zaid, Saiyi Wang, and Hamid Mushtaq. 2020. "SparkRA: Enabling Big Data Scalability for the GATK RNA-seq Pipeline with Apache Spark" Genes 11, no. 1: 53. https://doi.org/10.3390/genes11010053
APA StyleAl-Ars, Z., Wang, S., & Mushtaq, H. (2020). SparkRA: Enabling Big Data Scalability for the GATK RNA-seq Pipeline with Apache Spark. Genes, 11(1), 53. https://doi.org/10.3390/genes11010053




