Looking for Local Adaptation: Convergent Microevolution in Aleppo Pine (Pinus halepensis)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.1.1. Plant Material and Climatic Data
2.1.2. DNA Extraction, SNP Genotyping, and Gene Annotation
2.2. Statistical Analysis
2.2.1. Population Structure
2.2.2. Looking for Outliers Using PCA
2.2.3. Looking for Outliers Using Bayesian Approaches
- Bayenv
- Baypass
3. Results
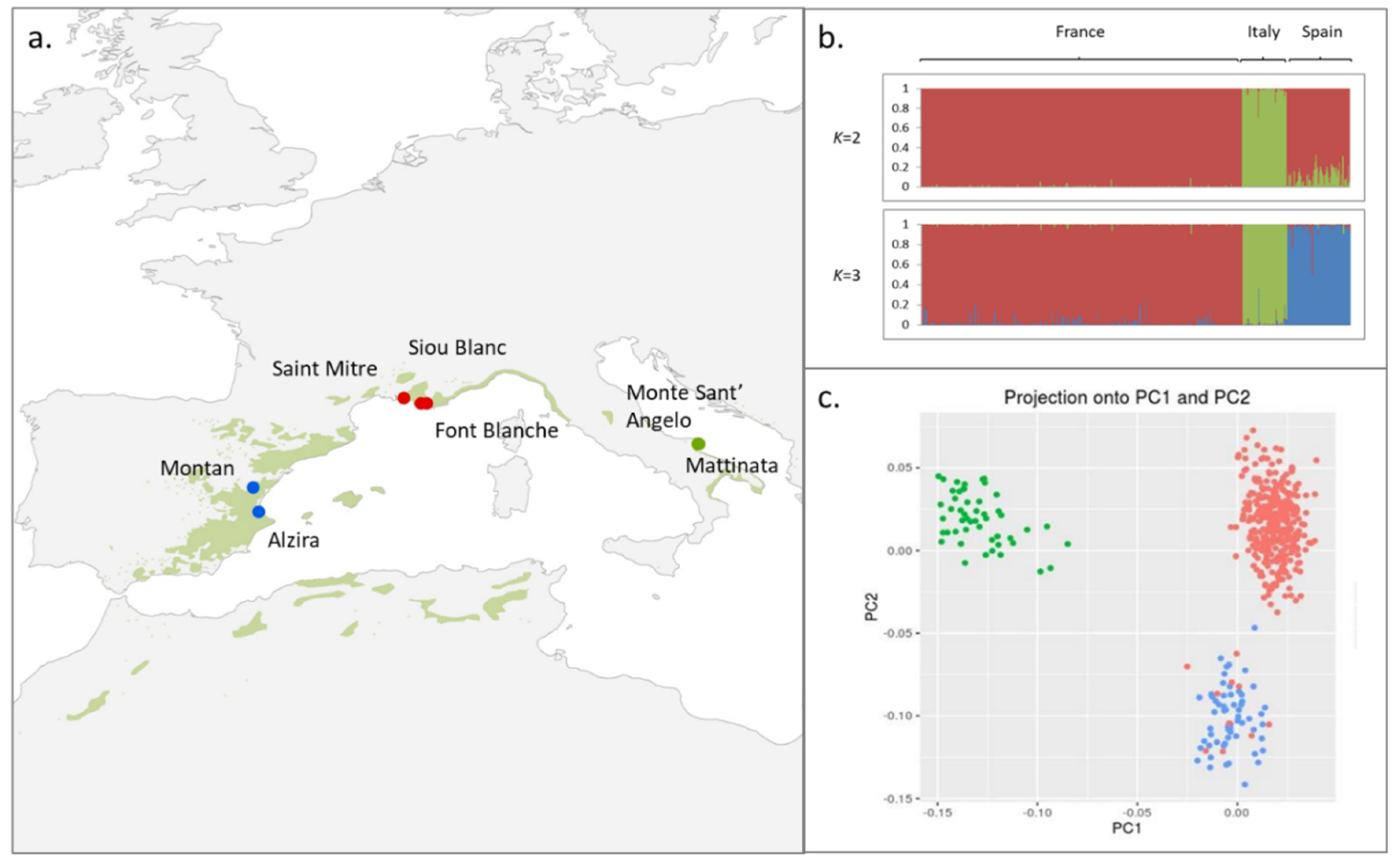
3.1. Population Structure
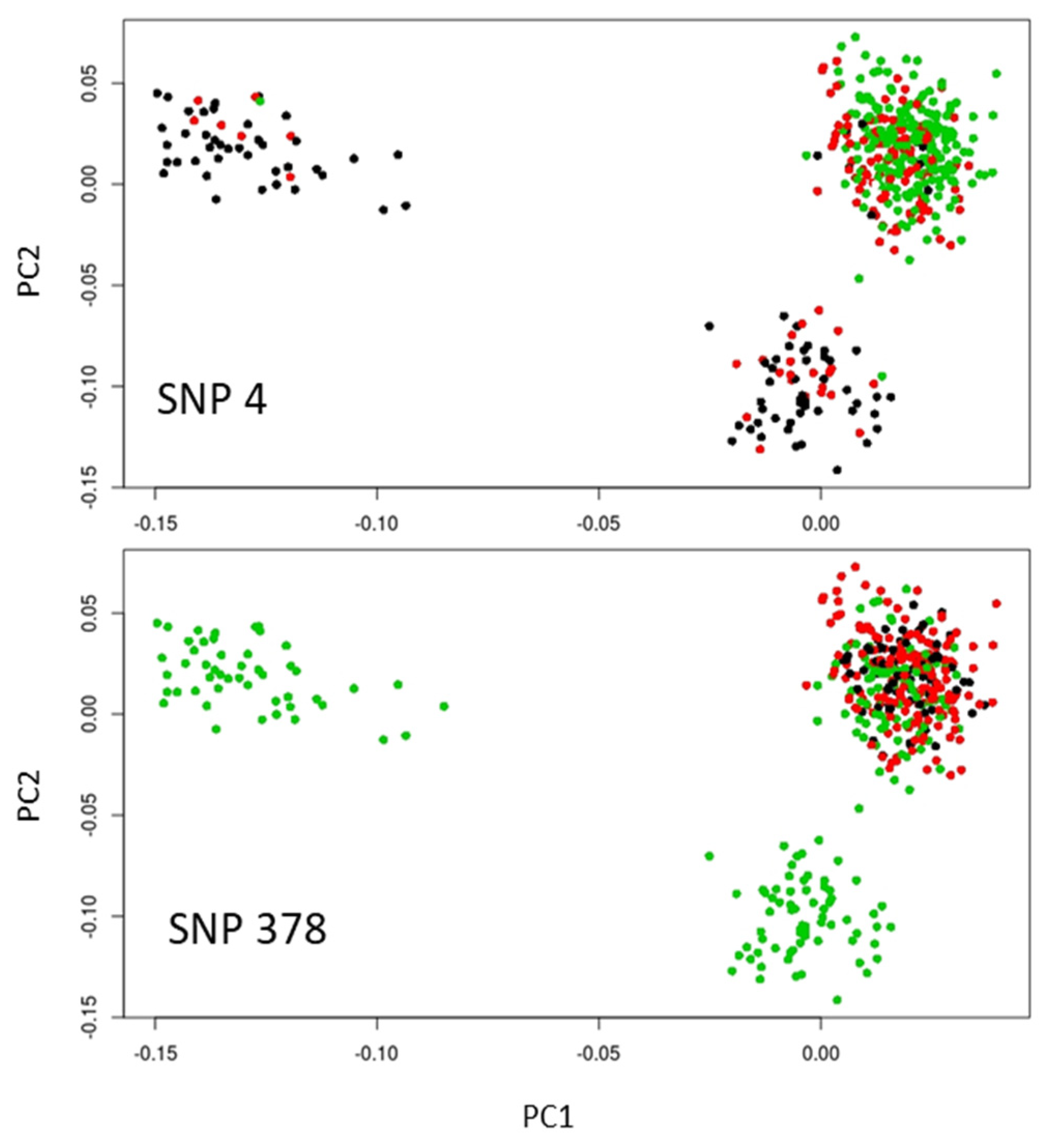
3.2. Looking for Outliers Using PCA
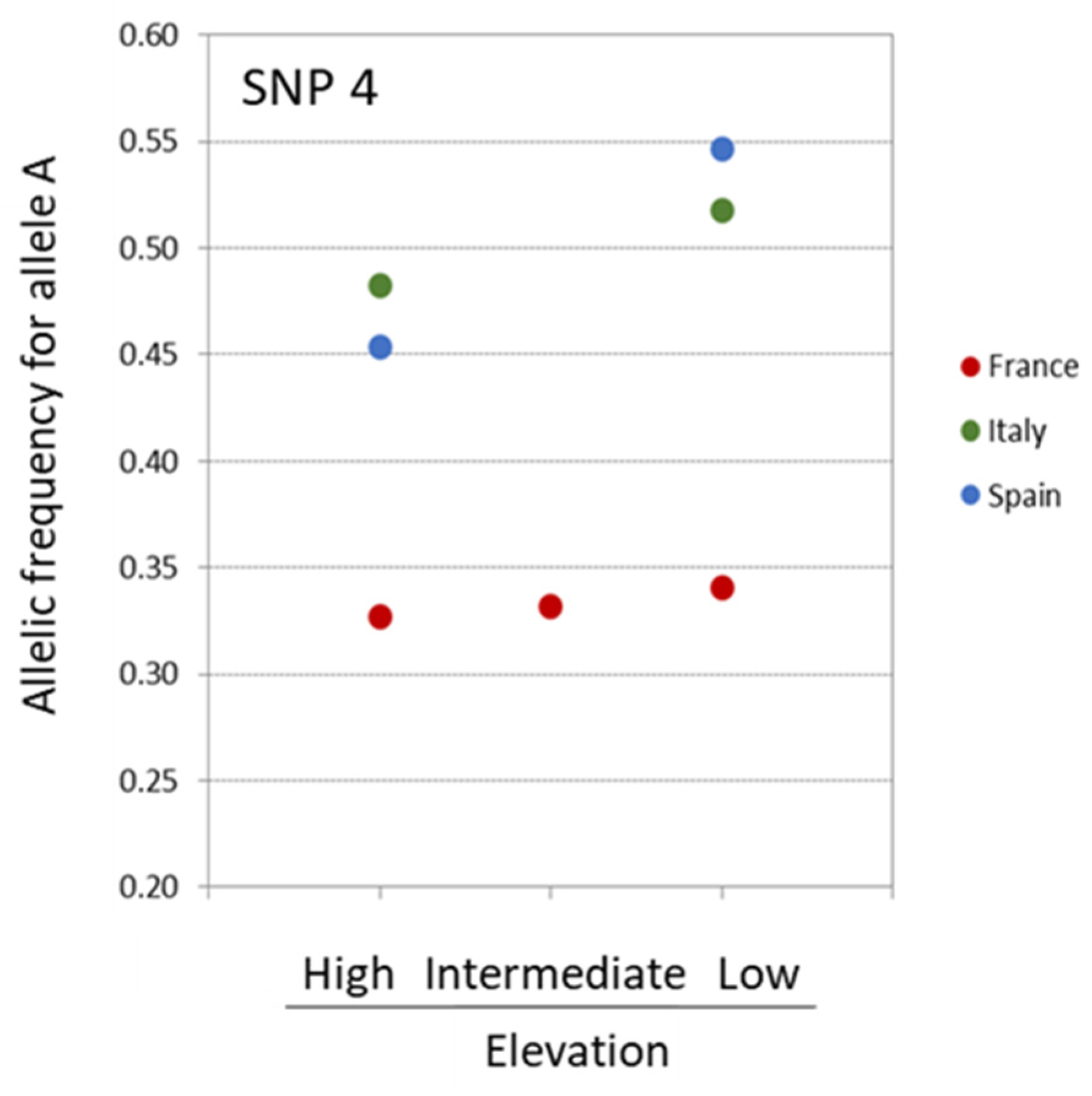
3.3. Looking for Outliers Using Bayesian Approaches
- Bayenv
- Baypass
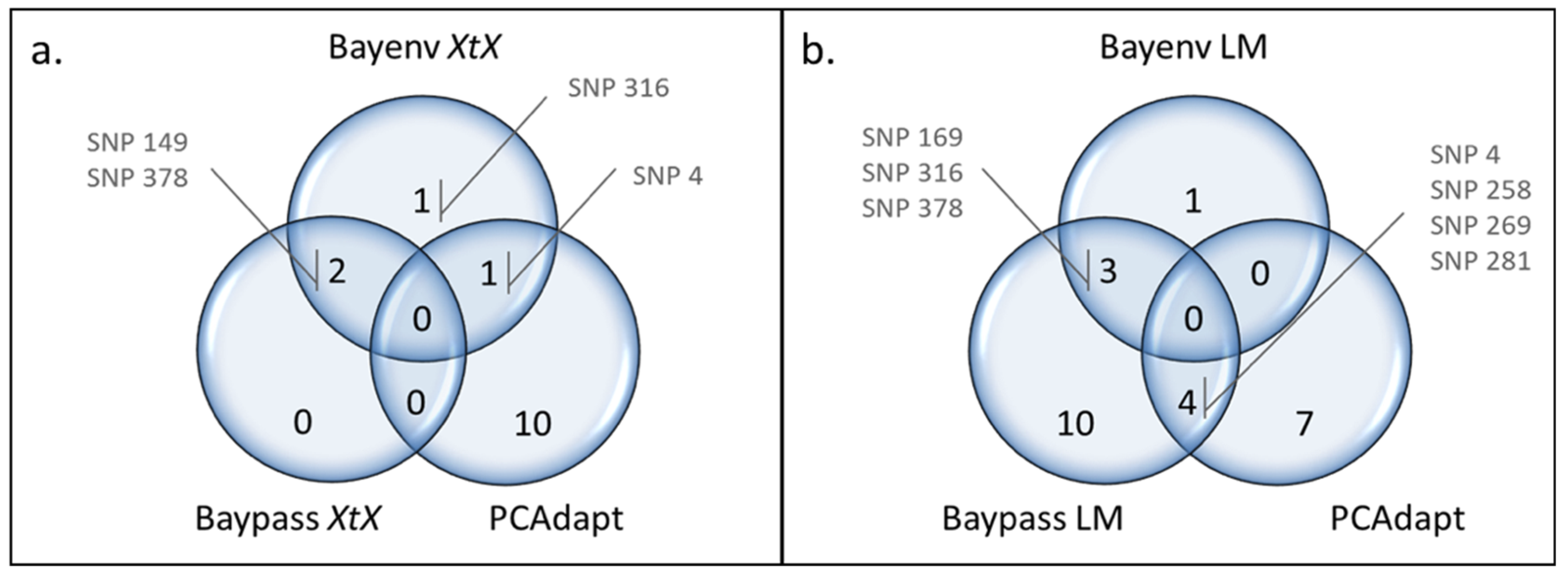
3.4. Combined Results of All Outlier Tests
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Savolainen, O.; Lascoux, M.; Merilä, J. Ecological genomics of local adaptation. Nat. Rev. Genet. 2013, 14, 807–820. [Google Scholar] [CrossRef] [PubMed]
- Hoban, S.; Kelley, J.L.; Lotterhos, K.E.; Antolin, M.F.; Bradburd, G.; Lowry, D.B.; Poss, M.L.; Reed, L.K.; Storfer, A.; Whitlock, M.C. Finding the genomic basis of local adaptation: Pitfalls, practical solutions, and future directions. Am. Nat. 2016, 188, 379–397. [Google Scholar] [CrossRef] [PubMed]
- Leimu, R.; Fischer, M. A meta-analysis of local adaptation in plants. PLoS ONE 2008, 3, e4010. [Google Scholar] [CrossRef] [PubMed]
- Lascoux, M.; Glémin, S.; Savolainen, O. Local Adaptation in Plants; John Wiley & Sons, Ltd.: Chichester, UK, 2016; pp. 1–7. [Google Scholar]
- Savolainen, O.; Pyhäjärvi, T.; Knürr, T. Gene flow and local adaptation in trees. Annu. Rev. Ecol. Evol. Syst. 2007, 38, 595–619. [Google Scholar] [CrossRef]
- Scotti, I.; González-Martínez, S.C.; Budde, K.B.; Lalagüe, H. Fifty years of genetic studies: What to make of the large amounts of variation found within populations? Ann. For. Sci. 2016, 73, 69–75. [Google Scholar] [CrossRef]
- Audigeos, D.; Brousseau, L.; Traissac, S.; Scotti-Saintagne, C.; Scotti, I. Molecular divergence in tropical tree populations occupying environmental mosaics. J. Evol. Biol. 2013, 26, 529–544. [Google Scholar] [CrossRef] [PubMed]
- Brousseau, L.; Foll, M.; Scotti-Saintagne, C.; Scotti, I. Neutral and adaptive drivers of microgeographic genetic divergence within continuous populations: The case of the neotropical tree Eperua falcata (Aubl.). PLoS ONE 2015, 10, e0121394. [Google Scholar] [CrossRef] [PubMed]
- Brousseau, L.; Postolache, D.; Lascoux, M.; Drouzas, A.D.; Källman, T.; Leonarduzzi, C.; Liepelt, S.; Piotti, A.; Popescu, F.; Roschanski, A.M.; et al. Local adaptation in European firs assessed through extensive sampling across altitudinal gradients in southern Europe. PLoS ONE 2016, 11, e0158216. [Google Scholar] [CrossRef]
- Roschanski, A.M.; Csilléry, K.; Liepelt, S.; Oddou-Muratorio, S.; Ziegenhagen, B.; Huard, F.; Ullrich, K.K.; Postolache, D.; Vendramin, G.G.; Fady, B. Evidence of divergent selection for drought and cold tolerance at landscape and local scales in Abies alba Mill. in the French Mediterranean Alps. Mol. Ecol. 2016, 25, 776–794. [Google Scholar] [CrossRef]
- Jaramillo-Correa, J.-P.; Rodríguez-Quilón, I.; Grivet, D.; Lepoittevin, C.; Sebastiani, F.; Heuertz, M.; Garnier-Géré, P.H.; Alía, R.; Plomion, C.; Vendramin, G.G.; et al. Molecular proxies for climate maladaptation in a long-lived tree (Pinus pinaster Aiton, Pinaceae). Genetics 2015, 199, 793–807. [Google Scholar] [CrossRef]
- Nadeau, S.; Meirmans, P.G.; Aitken, S.N.; Ritland, K.; Isabel, N. The challenge of separating signatures of local adaptation from those of isolation by distance and colonization history: The case of two white pines. Ecol. Evol. 2016, 6, 8649–8664. [Google Scholar] [CrossRef] [PubMed]
- Pluess, A.R.; Frank, A.; Heiri, C.; Lalagüe, H.; Vendramin, G.G.; Oddou-Muratorio, S. Genome-environment association study suggests local adaptation to climate at the regional scale in Fagus sylvatica. New Phytol. 2016, 210, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Fady, B. Pinus Halepensis and Pinus Brutia—Technical Guidelines for Genetic Conservation and Use for Aleppo and Brutia Pine: EUFORGEN—European forest genetic resources programme. 2003. Available online: http://www.euforgen.org/publications/publication/ipinus-halepensisi-and-ipinus-brutiai-technical-guidelines-for-genetic-conservation-and/ (accessed on 3 September 2019).
- Rouget, M.; Richardson, D.M.; Milton, S.J.; Polakow, D. Predicting invasion dynamics of four alien Pinus species in a highly fragmented semi-arid shrubland in South Africa. Plant Ecol. 2001, 152, 79–92. [Google Scholar] [CrossRef]
- Lavi, A.; Perevolotsky, A.; Kigel, J.; Noy-Meir, I. Invasion of Pinus halepensis from plantations into adjacent natural habitats. Appl. Veg. Sci. 2005, 8, 85–92. [Google Scholar] [CrossRef]
- Santos-Del-Blanco, L.; Climent, J.; González-Martínez, S.C.; Pannell, J.R. Genetic differentiation for size at first reproduction through male versus female functions in the widespread Mediterranean tree Pinus Pinaster. Ann. Bot. 2012, 110, 1449–1460. [Google Scholar] [CrossRef] [PubMed]
- de Luis, M.; Čufar, K.; Di Filippo, A.; Novak, K.; Papadopoulos, A.; Piovesan, G.; Rathgeber, C.B.K.; Raventós, J.; Saz, M.A.; Smith, K.T. Plasticity in dendroclimatic response across the distribution range of Aleppo pine (Pinus halepensis). PLoS ONE 2013, 8, e83550. [Google Scholar] [CrossRef] [PubMed]
- Grivet, D.; Sebastiani, F.; Alia, R.; Bataillon, T.; Torre, S.; Zabal-Aguirre, M.; Vendramin, G.G.; Gonzalez-Martinez, S.C. Molecular footprints of local adaptation in two Mediterranean conifers. Mol. Biol. Evol. 2011, 28, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Ruiz Daniels, R.; Taylor, R.S.; Serra-Varela, M.J.; Vendramin, G.G.; González-Martínez, S.C.; Grivet, D. Inferring selection in instances of long-range colonization: The Aleppo pine (Pinus halepensis) in the Mediterranean Basin. Mol. Ecol. 2018, 27, 3331–3345. [Google Scholar] [CrossRef]
- Conord, C.; Gurevitch, J.; Fady, B. Large-scale longitudinal gradients of genetic diversity: A meta-analysis across six phyla in the Mediterranean Basin. Ecol. Evol. 2012, 2, 2600–2614. [Google Scholar] [CrossRef]
- Morgante, M.; Felice, N.; Vendramin, G.G. Analysis of hypervariable chloroplast microsatellites in Pinus halepensis reveals a dramatic genetic bottleneck. In Molecular Tools for Screening Biodiversity; Springer: Dordrecht, The Netherlands, 1998; pp. 407–412. [Google Scholar]
- Bucci, G.; Anzidei, M.; Madaghiele, A.; Vendramin, G.G. Detection of haplotypic variation and natural hybridization in halepensis -complex pine species using chloroplast simple sequence repeat (SSR) markers. Mol. Ecol. 1998, 7, 1633–1643. [Google Scholar] [CrossRef]
- Grivet, D.; Sebastiani, F.; González-Martínez, S.C.; Vendramin, G.G. Patterns of polymorphism resulting from long-range colonization in the Mediterranean conifer Aleppo pine. New Phytol. 2009, 184, 1016–1028. [Google Scholar] [CrossRef] [PubMed]
- Serra-Varela, M.J.; Alía, R.; Daniels, R.R.; Zimmermann, N.E.; Gonzalo-Jiménez, J.; Grivet, D. Assessing vulnerability of two Mediterranean conifers to support genetic conservation management in the face of climate change. Divers. Distrib. 2017, 23, 507–516. [Google Scholar] [CrossRef]
- Atzmon, N.; Moshe, Y.; Schiller, G. Ecophysiological response to severe drought in Pinus halepensis Mill. trees of two provenances. Plant Ecol. 2004, 171, 15–22. [Google Scholar] [CrossRef]
- Sathyan, P.; Newton, R.J.; Loopstra, C.A. Genes induced by WDS are differentially expressed in two populations of aleppo pine (Pinus halepensis). Tree Genet. Genomes 2005, 1, 166–173. [Google Scholar] [CrossRef]
- Voltas, J.; Chambel, M.R.; Prada, M.A.; Ferrio, J.P. Climate-related variability in carbon and oxygen stable isotopes among populations of Aleppo pine grown in common-garden tests. Trees 2008, 22, 759–769. [Google Scholar] [CrossRef]
- Santos-del-Blanco, L.; Bonser, S.P.; Valladares, F.; Chambel, M.R.; Climent, J. Plasticity in reproduction and growth among 52 range-wide populations of a Mediterranean conifer: Adaptive responses to environmental stress. J. Evol. Biol. 2013, 26, 1912–1924. [Google Scholar] [CrossRef] [PubMed]
- Vitasse, Y.; Porté, A.J.; Kremer, A.; Michalet, R.; Delzon, S. Responses of canopy duration to temperature changes in four temperate tree species: Relative contributions of spring and autumn leaf phenology. Oecologia 2009, 161, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Bresson, C.C.; Vitasse, Y.; Kremer, A.; Delzon, S. To what extent is altitudinal variation of functional traits driven by genetic adaptation in European oak and beech? Tree Physiol. 2011, 31, 1164–1174. [Google Scholar] [CrossRef] [Green Version]
- Grivet, D.; Avia, K.; Vaattovaara, A.; Eckert, A.J.; Neale, D.B.; Savolainen, O.; González-Martínez, S.C. High rate of adaptive evolution in two widespread European pines. Mol. Ecol. 2017, 26, 6857–6870. [Google Scholar] [CrossRef]
- Luu, K.; Bazin, E.; Blum, M.G.B. Pcadapt: An R package to perform genome scans for selection based on principal component analysis. Mol. Ecol. Resour. 2017, 17, 67–77. [Google Scholar] [CrossRef]
- Coop, G.; Witonsky, D.; Di Rienzo, A.; Pritchard, J.K. Using environmental correlations to identify loci underlying local adaptation. Genetics 2010, 185, 1411–1423. [Google Scholar] [CrossRef] [PubMed]
- Günther, T.; Coop, G. Robust identification of local adaptation from allele frequencies. Genetics 2013, 195, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Gautier, M. Genome-wide scan for adaptive divergence and association with population-specific covariates. Genetics 2015, 201, 1555–1579. [Google Scholar] [CrossRef] [PubMed]
- De Mita, S.; Thuillet, A.C.; Gay, L.; Ahmadi, N.; Manel, S.; Ronfort, J.; Vigouroux, Y. Detecting selection along environmental gradients: Analysis of eight methods and their effectiveness for outbreeding and selfing populations. Mol. Ecol. 2013, 22, 1383–1399. [Google Scholar] [CrossRef] [PubMed]
- Blair, L.M.; Granka, J.M.; Feldman, M.W. On the stability of the Bayenv method in assessing human SNP-environment associations. Hum. Genom. 2014, 8, 1. [Google Scholar] [CrossRef]
- Lotterhos, K.E.; Whitlock, M.C. The relative power of genome scans to detect local adaptation depends on sampling design and statistical method. Mol. Ecol. 2015, 24, 1031–1046. [Google Scholar] [CrossRef] [PubMed]
- Rellstab, C.; Gugerli, F.; Eckert, A.J.; Hancock, A.M.; Holderegger, R. A practical guide to environmental association analysis in landscape genomics. Mol. Ecol. 2015, 24, 4348–4370. [Google Scholar] [CrossRef] [PubMed]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces from global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Gonzalo, J. Phytoclimatic analysis of the Spanish Peninsula. Update and geostatistical analysis. Ph.D. Thesis, Universidad de Valladolid, Valladolid, Spain, 2007. [Google Scholar]
- Körner, C. The use of ‘altitude’ in ecological research. Trends Ecol. Evol. 2007, 22, 569–574. [Google Scholar] [CrossRef]
- Pinosio, S.; González-Martínez, S.C.; Bagnoli, F.; Cattonaro, F.; Grivet, D.; Marroni, F.; Lorenzo, Z.; Pausas, J.G.; Verdú, M.; Vendramin, G.G. First insights into the transcriptome and development of new genomic tools of a widespread circum-Mediterranean tree species, Pinus halepensis Mill. Mol. Ecol. Resour. 2014, 14, 846–856. [Google Scholar] [CrossRef]
- Ruiz Daniels, R.; Taylor, R.S.; González-Martínez, S.C.; Vendramin, G.G.; Fady, B.; Oddou-Muratorio, S.; Piotti, A.; Simioni, G.; Grivet, D.; Beaumont, M.A. Looking for local adaptation: convergent microevolution in Aleppo pine (Pinus halepensis). Zenodo 2018. [Google Scholar] [CrossRef]
- Budde, K.B.; Heuertz, M.; Hernández-Serrano, A.; Pausas, J.G.; Vendramin, G.G.; Verdú, M.; González-Martínez, S.C. In situ genetic association for serotiny, a fire-related trait, in Mediterranean maritime pine (Pinus pinaster). New Phytol. 2014, 201, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Eckert, A.J.; Wegrzyn, J.L.; Liechty, J.D.; Lee, J.M.; Cumbie, W.P.; Davis, J.M.; Goldfarb, B.; Loopstra, C.A.; Palle, S.R.; Quesada, T.; et al. The evolutionary genetics of the genes underlying phenotypic associations for Loblolly pine (Pinus taeda, Pinaceae). Genetics 2013, 195, 1353–1372. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.geneious.com/ (accessed on 3 September 2019).
- François, O.; Martins, H.; Caye, K.; Schoville, S.D. Controlling false discoveries in genome scans for selection. Mol. Ecol. 2016, 25, 454–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rousset, F. Genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- McVean, G. A genealogical interpretation of principal components analysis. PLoS Genet. 2009, 5, e1000686. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.B.; Lee, S.H.; Zhu, Z.X.; Benyamin, B.; Robinson, M.R. EigenGWAS: Finding loci under selection through genome-wide association studies of eigenvectors in structured populations. Heredity 2016, 117, 51–61. [Google Scholar] [CrossRef]
- Duforet-Frebourg, N.; Luu, K.; Laval, G.; Bazin, E.; Blum, M.G.B. Detecting genomic signatures of natural selection with principal component analysis: Application to the 1000 genomes data. Mol. Biol. Evol. 2016, 33, 1082–1093. [Google Scholar] [CrossRef]
- Galinsky, K.J.; Bhatia, G.; Loh, P.R.; Georgiev, S.; Mukherjee, S.; Patterson, N.J.; Price, A.L. Fast Principal-Component Analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am. J. Hum. Genet. 2016, 98, 456–472. [Google Scholar] [CrossRef]
- Hao, W.; Song, M.; Storey, J.D. Probabilistic models of genetic variation in structured populations applied to global human studies. Bioinformatics 2016, 32, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Lotterhos, K.E.; Whitlock, M.C. Evaluation of demographic history and neutral parameterization on the performance of FST outlier tests. Mol. Ecol. 2014, 23, 2178–2192. [Google Scholar] [CrossRef] [PubMed]
- Kass, R.E.; Raftery, A.E. Bayes Factors. J. Am. Stat. Assoc. 1995, 90, 773–795. [Google Scholar] [CrossRef]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, R.M.; VonHoldt, B.M.; Harrigan, R.; Knowles, J.C.; Musiani, M.; Coltman, D.; Novembre, J.; Wayne, R.K. Genetic subdivision and candidate genes under selection in North American grey wolves. Mol. Ecol. 2016, 25, 380–402. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, S.; Hodgins, K.A.; Lotterhos, K.E.; Suren, H.; Nadeau, S.; Degner, J.C.; Nurkowski, K.A.; Smets, P.; Wang, T.; Gray, L.K.; et al. Convergent local adaptation to climate in distantly related conifers. Science 2016, 353, 1431–1433. [Google Scholar] [CrossRef] [Green Version]
- Turchin, M.C.; Chiang, C.W.K.; Palmer, C.D.; Sankararaman, S.; Reich, D.; Hirschhorn, J.N. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat. Genet. 2012, 44, 1015–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daub, J.T.; Hofer, T.; Cutivet, E.; Dupanloup, I.; Quintana-Murci, L.; Robinson-Rechavi, M.; Excoffier, L. Evidence for polygenic adaptation to pathogens in the human genome. Mol. Biol. Evol. 2013, 30, 1544–1558. [Google Scholar] [CrossRef]
- Berg, J.J.; Coop, G. A Population genetic signal of polygenic adaptation. PLoS Genet. 2014, 10, e1004412. [Google Scholar] [CrossRef]
- Robinson, M.R.; Hemani, G.; Medina-Gomez, C.; Mezzavilla, M.; Esko, T.; Shakhbazov, K.; Powell, J.E.; Vinkhuyzen, A.; Berndt, S.I.; Gustafsson, S.; et al. Population genetic differentiation of height and body mass index across Europe. Nat. Genet. 2015, 47, 1357–1361. [Google Scholar] [CrossRef]
- Racimo, F.; Berg, J.J.; Pickrell, J.K. Detecting polygenic adaptation in admixture graphs. Genetics 2018, 208, 1565–1584. [Google Scholar] [CrossRef] [PubMed]
- Křeček, P.; Skůpa, P.; Libus, J.; Naramoto, S.; Tejos, R.; Friml, J.; Zažímalová, E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009, 10, 249. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.; Camarero, J.J.; Carrer, M. Shifts of irrigation in Aleppo pine under semi-arid conditions reveal uncoupled growth and carbon storage and legacy effects on wood anatomy. Agric. For. Meteorol. 2018, 253–254, 225–232. [Google Scholar] [CrossRef]
- Tiffin, P.; Ross-Ibarra, J. Advances and limits of using population genetics to understand local adaptation. Trends Ecol. Evol. 2014, 29, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Coop, G. Distinguishing among modes of convergent adaptation using population genomic data. Genetics 2017, 207, 1591–1619. [Google Scholar] [CrossRef] [PubMed]
- IPCC. Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Core Writing Team, Pachauri, R.K., Meyer, L.A., Eds.; IPCC: Geneva, Switzerland; p. 151. Available online: http://www.ipcc.ch/report/ar5/syr/ (accessed on 3 September 2019).
- Ruiz-Labourdette, D.; Nogués-Bravo, D.; Ollero, H.S.; Schmitz, M.F.; Pineda, F.D. Forest composition in Mediterranean mountains is projected to shift along the entire elevational gradient under climate change. J. Biogeogr. 2012, 39, 162–176. [Google Scholar] [CrossRef]
- Benito-Garzón, M.; Fernández-Manjarrés, J.F. Testing scenarios for assisted migration of forest trees in Europe. New For. 2015, 46, 979–994. [Google Scholar] [CrossRef]
| Bayenv2 | SNP | Sequence | XtX Value |
| 149 | seq-8671-529 | 20.42 | |
| 4 | seq-9882-801 | 20.83 | |
| 316 | seq-10373-2483 | 21.52 | |
| 378 | seq-2_3941_01-381 | 23.76 | |
| Baypass | SNP | Sequence | XtX value |
| 378 | seq-2_3941_01-381 | 13.44 | |
| 149 | seq-8671-529 | 14.14 | |
| PCAdapt | SNP | Sequence | p-value |
| 335 | seq-1_6493_01-100 | 2.64E-009 | |
| 94 | seq-55383-900 | 1.74E-007 | |
| 19 | seq-55383-1485 | 2.05E-007 | |
| 331 | seq-55383-141 | 2.95E-007 | |
| 258 | seq-9882-2209 | 4.74E-004 | |
| 4 | seq-9882-801 | 7.23E-004 | |
| 384 | seq-0_3073_01-92 | 1.21E-003 | |
| 281 | seq-16094-410 | 3.36E-003 | |
| 10 | seq-44358-1615 | 3.51E-003 | |
| 113 | seq-44358-2515 | 3.80E-003 | |
| 269 | seq-16094-1379 | 5.90E-003 |
| SNP | Sequence | Env. | BF Bayenv2 | eBPis Baypass |
|---|---|---|---|---|
| 169 | seq-0_10162_01-244 | BIO9 | 41.97 | 5.48 |
| 316 | seq-10373-2483 | Elevation | 20.90 | 3.77 |
| 378 | seq-2_3941_01-381 | BIO12 | 47.38 | 3.65 |
| SNP | Sequence | Description | E-Value |
|---|---|---|---|
| 4, 258 | seq-9882 | PIN-like protein in various conifers | 1e−96 |
| 149 | seq-8671 | No significant similarity found | |
| 169 | seq-0_10162_01 | Anonymous locus in Pinus taeda | 1e−86 |
| 269, 281 | seq-16094 | Anonymous locus in Picea glauca | 1e−51 |
| 316 | seq-10373 | Putative alpha-xylosidase (XYL1) in Pinus pinaster | 2e−99 |
| 378 | seq-2_3941_01 | Anonymous locus in Pinus taeda | 5e−91 |
| SNP | Contig | p-Value PCAdapt | eBPis Baypass | Env. |
|---|---|---|---|---|
| 4 | seq-9882-801 | 7.23E−004 | 5.38 | BIO12 |
| 258 | seq-9882-2209 | 4.74E−004 | 5.06 | Elevation |
| 258 | seq-9882-2209 | 4.74E−004 | 6.34 | BIO12 |
| 258 | seq-9882-2209 | 4.74E−004 | 3.02 | BIO19 |
| 269 | seq-16094-1379 | 5.90E−003 | 4.86 | BIO12 |
| 269 | seq-16094-1379 | 5.90E−003 | 3.77 | BIO16 |
| 269 | seq-16094-1379 | 5.90E−003 | 4.72 | BIO19 |
| 281 | seq-16094-410 | 3.36E−003 | 3.21 | Elevation |
| 281 | seq-16094-410 | 3.36E−003 | 5.21 | BIO11 |
| 281 | seq-16094-410 | 3.36E−003 | 3.03 | BIO12 |
| 281 | seq-16094-410 | 3.36E−003 | 4.64 | BIO16 |
| 281 | seq-16094-410 | 3.36E−003 | 4.92 | BIO19 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz Daniels, R.; Taylor, R.S.; González-Martínez, S.C.; Vendramin, G.G.; Fady, B.; Oddou-Muratorio, S.; Piotti, A.; Simioni, G.; Grivet, D.; Beaumont, M.A. Looking for Local Adaptation: Convergent Microevolution in Aleppo Pine (Pinus halepensis). Genes 2019, 10, 673. https://doi.org/10.3390/genes10090673
Ruiz Daniels R, Taylor RS, González-Martínez SC, Vendramin GG, Fady B, Oddou-Muratorio S, Piotti A, Simioni G, Grivet D, Beaumont MA. Looking for Local Adaptation: Convergent Microevolution in Aleppo Pine (Pinus halepensis). Genes. 2019; 10(9):673. https://doi.org/10.3390/genes10090673
Chicago/Turabian StyleRuiz Daniels, Rose, Richard S. Taylor, Santiago C. González-Martínez, Giovanni G. Vendramin, Bruno Fady, Sylvie Oddou-Muratorio, Andrea Piotti, Guillaume Simioni, Delphine Grivet, and Mark A. Beaumont. 2019. "Looking for Local Adaptation: Convergent Microevolution in Aleppo Pine (Pinus halepensis)" Genes 10, no. 9: 673. https://doi.org/10.3390/genes10090673
APA StyleRuiz Daniels, R., Taylor, R. S., González-Martínez, S. C., Vendramin, G. G., Fady, B., Oddou-Muratorio, S., Piotti, A., Simioni, G., Grivet, D., & Beaumont, M. A. (2019). Looking for Local Adaptation: Convergent Microevolution in Aleppo Pine (Pinus halepensis). Genes, 10(9), 673. https://doi.org/10.3390/genes10090673






