The Molecular Evolution of Circadian Clock Genes in Spotted Gar (Lepisosteus oculatus)
Abstract
1. Introduction
2. Materials and Methods
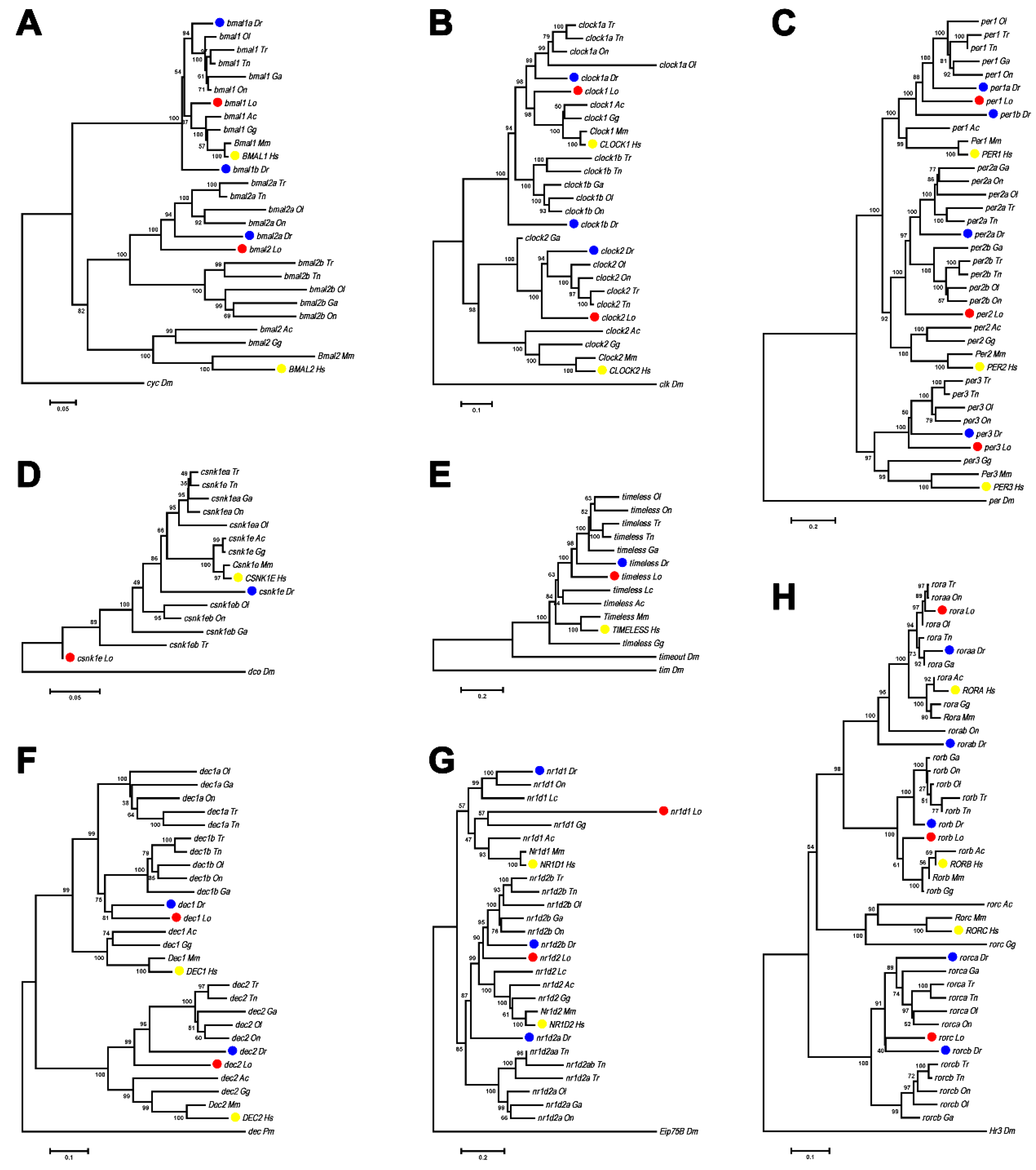
2.1. Data Sets and Phylogenetic Analysis
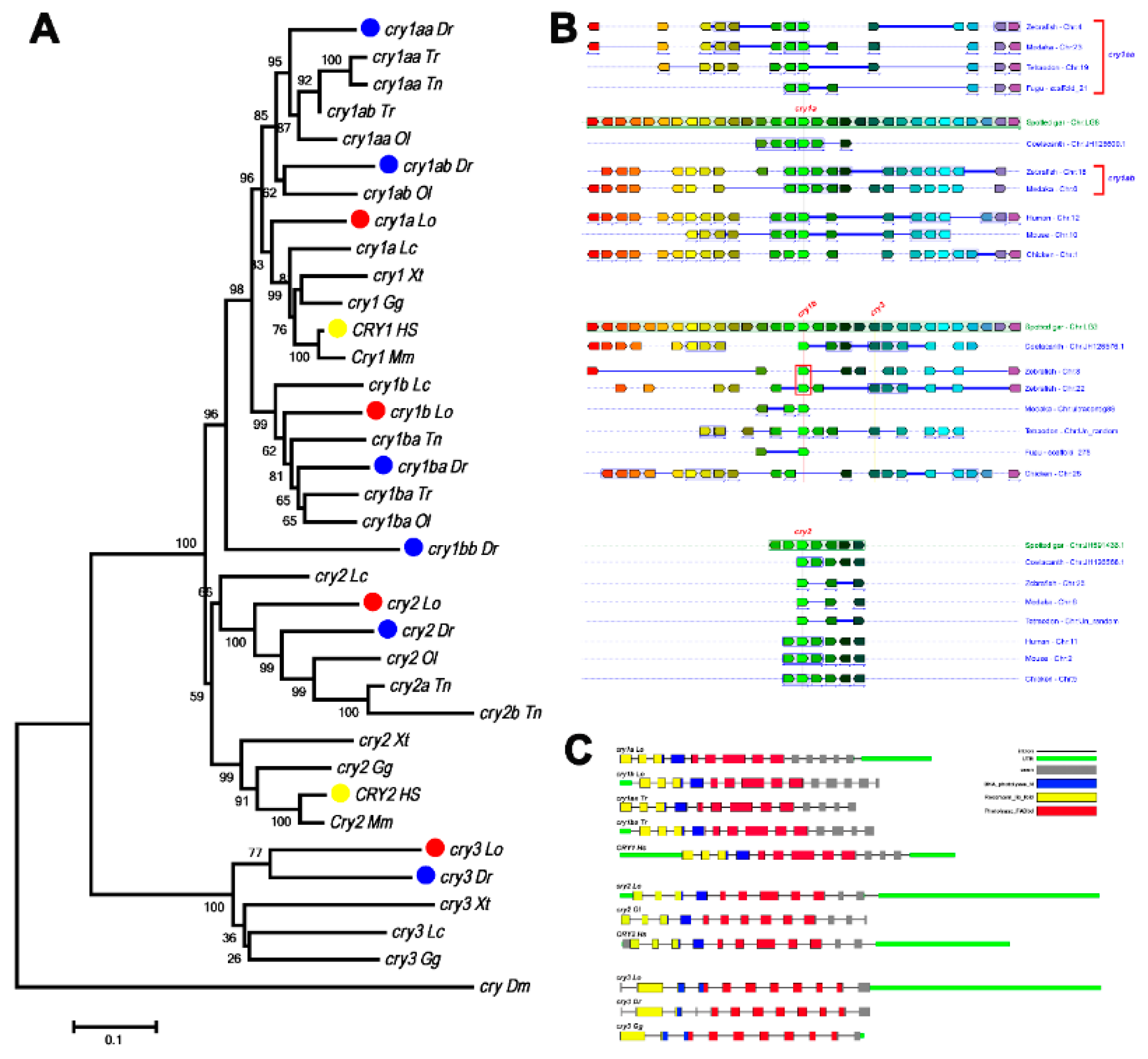
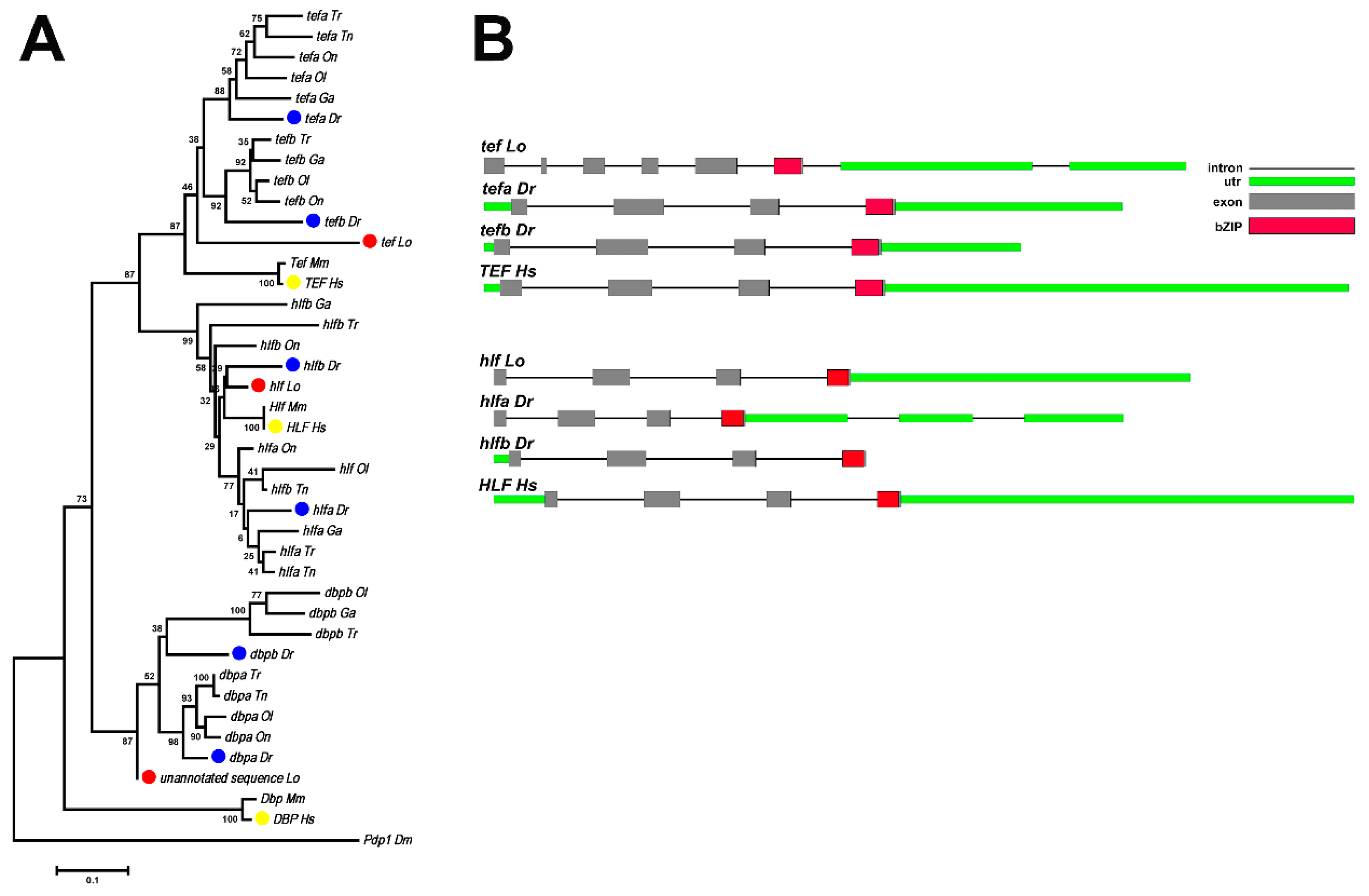
2.2. Exon Structural Analysis
2.3. Conserved Synteny Analysis
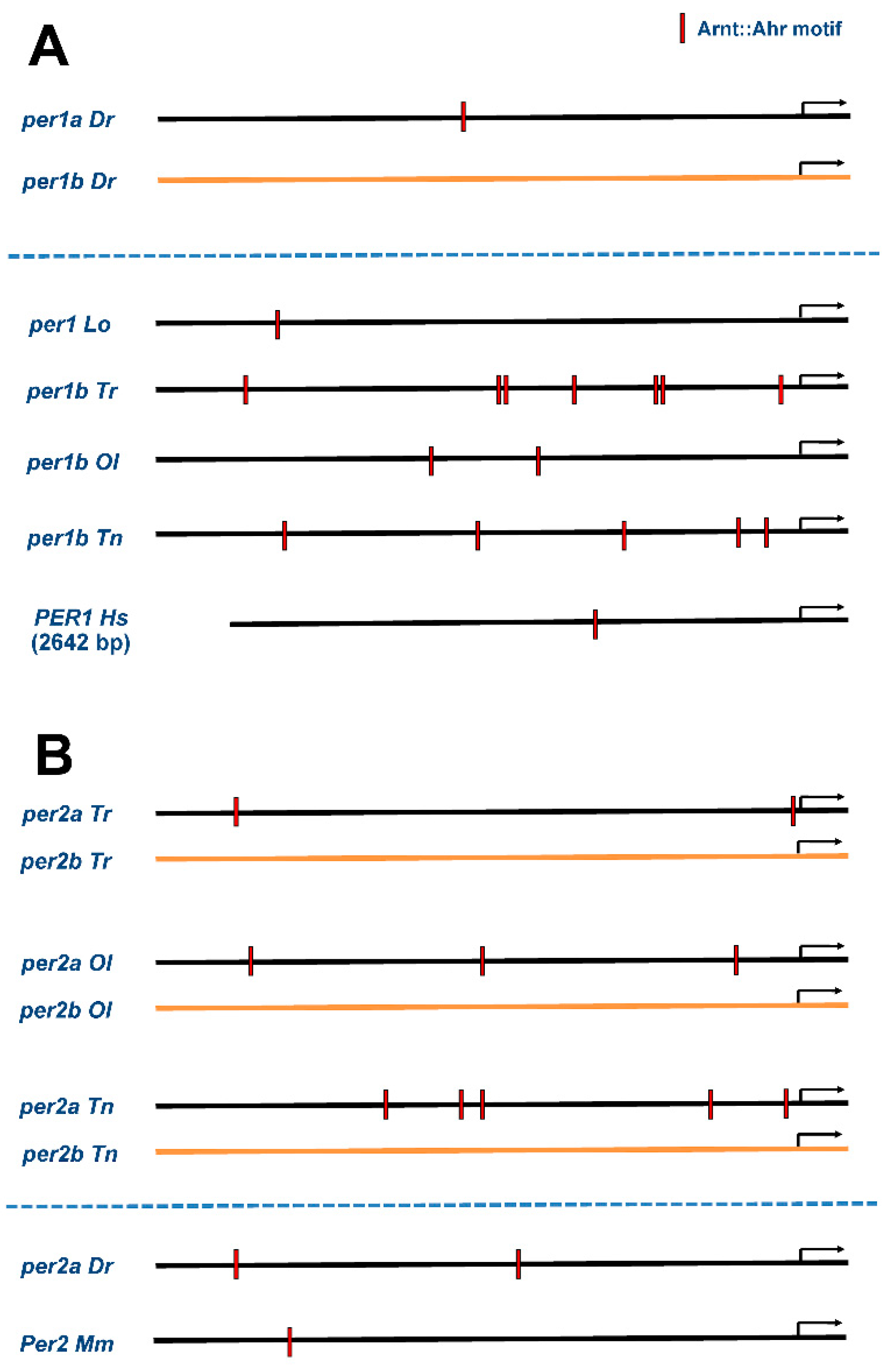
2.4. Regulatory Region Analysis
3. Results
3.1. Phylogenetic Analysis of Spotted Gar Circadian Clock Genes
3.2. Evolution of nfil3 Genes
3.3. Evolution of Cry Genes
3.4. Evolution of the Par Family Genes
3.5. Preservation of Regulatory Elements in Duplicated Circadian Clock Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Panda, S.; Hogenesch, J.B.; Kay, S.A. Circadian rhythms from flies to human. Nature 2002, 417, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Reppert, S.M.; Weaver, D.R. Coordination of circadian timing in mammals. Nature 2002, 418, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shlomo, R.; Kyriacou, C.P. Circadian rhythm entrainment in flies and mammals. Cell Biochem. Biophys. 2002, 37, 141–156. [Google Scholar] [CrossRef]
- Hardin, P.E. The circadian timekeeping system of Drosophila. Curr. Biol. CB 2005, 15, R714–R722. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.H.; Takahashi, J.S. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 2006, 15 (Suppl. 2), R271–R277. [Google Scholar] [CrossRef]
- Bell-Pedersen, D.; Cassone, V.M.; Earnest, D.J.; Golden, S.S.; Hardin, P.E.; Thomas, T.L.; Zoran, M.J. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 2005, 6, 544–556. [Google Scholar] [CrossRef]
- Paibomesai, M.I.; Moghadam, H.K.; Ferguson, M.M.; Danzmann, R.G. Clock genes and their genomic distributions in three species of salmonid fishes: Associations with genes regulating sexual maturation and cell cycling. BMC Res. Notes 2010, 3, 215. [Google Scholar] [CrossRef]
- Dunlap, J.C. Molecular bases for circadian clocks. Cell 1999, 96, 271–290. [Google Scholar] [CrossRef]
- Yu, W.; Hardin, P.E. Circadian oscillators of Drosophila and mammals. J. Cell Sci. 2006, 119, 4793–4795. [Google Scholar] [CrossRef]
- Layeghifard, M.; Rabani, R.; Pirhaji, L.; Yakhchali, B. Evolutionary mechanisms underlying the functional divergence of duplicate genes involved in vertebrates’ circadian rhythm pathway. Gene 2008, 426, 65–71. [Google Scholar] [CrossRef]
- Hughes, A.L. The evolution of functionally novel proteins after gene duplication. Proc. Biol. Sci. R. Soc. 1994, 256, 119–124. [Google Scholar] [CrossRef]
- Aguileta, G.; Bielawski, J.P.; Yang, Z. Gene conversion and functional divergence in the β-globin gene family. J. Mol. Evol. 2004, 59, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. Comparative analysis of teleost fish genomes reveals preservation of different ancient clock duplicates in different fishes. Mar. Genom. 2008, 1, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Volff, J.N. Genome evolution and biodiversity in teleost fish. Heredity 2005, 94, 280–294. [Google Scholar] [CrossRef] [PubMed]
- Amemiya, C.T.; Alfoldi, J.; Lee, A.P.; Fan, S.; Philippe, H.; Maccallum, I.; Braasch, I.; Manousaki, T.; Schneider, I.; Rohner, N.; et al. The African coelacanth genome provides insights into tetrapod evolution. Nature 2013, 496, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Braasch, I.; Gehrke, A.R.; Smith, J.J.; Kawasaki, K.; Manousaki, T.; Pasquier, J.; Amores, A.; Desvignes, T.; Batzel, P.; Catchen, J.; et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 2016, 48, 427–437. [Google Scholar] [CrossRef]
- Xu, P.; Zhang, X.; Wang, X.; Li, J.; Liu, G.; Kuang, Y.; Xu, J.; Zheng, X.; Ren, L.; Wang, G.; et al. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 2014, 46, 1212–1219. [Google Scholar] [CrossRef]
- Amores, A.; Force, A.; Yan, Y.L.; Joly, L.; Amemiya, C.; Fritz, A.; Ho, R.K.; Langeland, J.; Prince, V.; Wang, Y.L.; et al. Zebrafish HOX clusters and vertebrate genome evolution. Science 1998, 282, 1711–1714. [Google Scholar] [CrossRef]
- Postlethwait, J.H.; Yan, Y.L.; Gates, M.A.; Horne, S.; Amores, A.; Brownlie, A.; Donovan, A.; Egan, E.S.; Force, A.; Gong, Z.; et al. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 1998, 18, 345–349. [Google Scholar] [CrossRef]
- Taylor, J.S.; Braasch, I.; Frickey, T.; Meyer, A.; Van de Peer, Y. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003, 13, 382–390. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Brunet, F.; Petit, J.L.; Stange-Thomann, N.; Mauceli, E.; Bouneau, L.; Fischer, C.; Ozouf-Costaz, C.; Bernot, A.; et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946–957. [Google Scholar] [CrossRef]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.L.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar]
- Postlethwait, J.; Amores, A.; Cresko, W.; Singer, A.; Yan, Y.L. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. TIG 2004, 20, 481–490. [Google Scholar] [CrossRef]
- Amores, A.; Catchen, J.; Ferrara, A.; Fontenot, Q.; Postlethwait, J.H. Genome evolution and meiotic maps by massively parallel DNA sequencing: Spotted gar, an outgroup for the teleost genome duplication. Genetics 2011, 188, 799–808. [Google Scholar] [CrossRef]
- Hoegg, S.; Brinkmann, H.; Taylor, J.S.; Meyer, A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J. Mol. Evol. 2004, 59, 190–203. [Google Scholar] [CrossRef]
- Crow, K.D.; Stadler, P.F.; Lynch, V.J.; Amemiya, C.; Wagner, G.P. The “fish-specific” HOX cluster duplication is coincident with the origin of teleosts. Mol. Biol. Evol. 2006, 23, 121–136. [Google Scholar] [CrossRef]
- Toloza-Villalobos, J.; Arroyo, J.I.; Opazo, J.C. The circadian clock of teleost fish: A comparative analysis reveals distinct fates for duplicated genes. J. Mol. Evol. 2015, 80, 57–64. [Google Scholar] [CrossRef]
- Zhang, J.; Rosenberg, H.F.; Nei, M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. USA 1998, 95, 3708–3713. [Google Scholar] [CrossRef]
- Ueda, H.R.; Hayashi, S.; Chen, W.; Sano, M.; Machida, M.; Shigeyoshi, Y.; Iino, M.; Hashimoto, S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005, 37, 187–192. [Google Scholar] [CrossRef]
- Benna, C.; Scannapieco, P.; Piccin, A.; Sandrelli, F.; Zordan, M.; Rosato, E.; Kyriacou, C.P.; Valle, G.; Costa, R. A second timeless gene in Drosophila shares greater sequence similarity with mammalian tim. Curr. Biol. CB 2000, 10, R512–R513. [Google Scholar] [CrossRef][Green Version]
- Kurien, P.; Hsu, P.K.; Leon, J.; Wu, D.; McMahon, T.; Shi, G.; Xu, Y.; Lipzen, A.; Pennacchio, L.A.; Jones, C.R.; et al. TIMELESS mutation alters phase responsiveness and causes advanced sleep phase. Proc. Natl. Acad. Sci. USA 2019, 116, 12045–12053. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Rambaldi, D.; Ciccarelli, F.D. FancyGene: Dynamic visualization of gene structures and protein domain architectures on genomic loci. Bioinformatics 2009, 25, 2281–2282. [Google Scholar] [CrossRef]
- Louis, A.; Muffato, M.; Roest Crollius, H. Genomicus: five genome browsers for comparative genomics in eukaryota. Nucleic Acids Res. 2013, 41, D700–D705. [Google Scholar] [CrossRef]
- Liu, C.; Hu, J.; Qu, C.; Wang, L.; Huang, G.; Niu, P.; Zhong, Z.; Hong, F.; Wang, G.; Postlethwait, J.H.; et al. Molecular evolution and functional divergence of zebrafish (Danio rerio) cryptochrome genes. Sci. Rep. 2015, 5, 8113. [Google Scholar] [CrossRef]
- Ehrlich, J.; Sankoff, D.; Nadeau, J.H. Synteny conservation and chromosome rearrangements during mammalian evolution. Genetics 1997, 147, 289–296. [Google Scholar]
- Woods, I.G.; Wilson, C.; Friedlander, B.; Chang, P.; Reyes, D.K.; Nix, R.; Kelly, P.D.; Chu, F.; Postlethwait, J.H.; Talbot, W.S. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 2005, 15, 1307–1314. [Google Scholar] [CrossRef]
- Venkatesh, B. Evolution and diversity of fish genomes. Curr. Opin. Genet. Dev. 2003, 13, 588–592. [Google Scholar] [CrossRef]
- Glasauer, S.M.; Neuhauss, S.C. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. MGG 2014, 289, 1045–1060. [Google Scholar] [CrossRef]
- Inoue, J.G.; Miya, M.; Venkatesh, B.; Nishida, M. The mitochondrial genome of Indonesian coelacanth Latimeria menadoensis (Sarcopterygii: Coelacanthiformes) and divergence time estimation between the two coelacanths. Gene 2005, 349, 227–235. [Google Scholar] [CrossRef]
- Katsu, Y.; Kohno, S.; Hyodo, S.; Ijiri, S.; Adachi, S.; Hara, A.; Guillette, L.J., Jr.; Iguchi, T. Molecular cloning, characterization, and evolutionary analysis of estrogen receptors from phylogenetically ancient fish. Endocrinology 2008, 149, 6300–6310. [Google Scholar] [CrossRef]
- Salaneck, E.; Larsson, T.A.; Larson, E.T.; Larhammar, D. Birth and death of neuropeptide Y receptor genes in relation to the teleost fish tetraploidization. Gene 2008, 409, 61–71. [Google Scholar] [CrossRef]
- Looby, P.; Loudon, A.S. Gene duplication and complex circadian clocks in mammals. Trends Genet. TIG 2005, 21, 46–53. [Google Scholar] [CrossRef]
- Swanson, H.I. DNA binding and protein interactions of the AHR/ARNT heterodimer that facilitate gene activation. Chem. Biol. Interact. 2002, 141, 63–76. [Google Scholar] [CrossRef]
- Wang, H. Comparative analysis of period genes in teleost fish genomes. J. Mol. Evol. 2008, 67, 29–40. [Google Scholar] [CrossRef]
- Blau, J.; Young, M.W. Cycling vrille expression is required for a functional Drosophila clock. Cell 1999, 99, 661–671. [Google Scholar] [CrossRef]
- Cyran, S.A.; Buchsbaum, A.M.; Reddy, K.L.; Lin, M.C.; Glossop, N.R.; Hardin, P.E.; Young, M.W.; Storti, R.V.; Blau, J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 2003, 112, 329–341. [Google Scholar] [CrossRef]
- Glossop, N.R.; Houl, J.H.; Zheng, H.; Ng, F.S.; Dudek, S.M.; Hardin, P.E. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron 2003, 37, 249–261. [Google Scholar] [CrossRef]
- Doi, M.; Nakajima, Y.; Okano, T.; Fukada, Y. Light-induced phase-delay of the chicken pineal circadian clock is associated with the induction of cE4bp4, a potential transcriptional repressor of cPer2 gene. Proc. Natl. Acad. Sci. USA 2001, 98, 8089–8094. [Google Scholar] [CrossRef]
- Mitsui, S.; Yamaguchi, S.; Matsuo, T.; Ishida, Y.; Okamura, H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 2001, 15, 995–1006. [Google Scholar] [CrossRef]
- Ohno, T.; Onishi, Y.; Ishida, N. The negative transcription factor E4BP4 is associated with circadian clock protein PERIOD2. Biochem. Biophys. Res. Commun. 2007, 354, 1010–1015. [Google Scholar] [CrossRef]
- Ohno, T.; Onishi, Y.; Ishida, N. A novel E4BP4 element drives circadian expression of mPeriod2. Nucleic Acids Res. 2007, 35, 648–655. [Google Scholar] [CrossRef]
- Hogenesch, J.B.; Ueda, H.R. Understanding systems-level properties: Timely stories from the study of clocks. Nat. Rev. Genet. 2011, 12, 407–416. [Google Scholar] [CrossRef]
- Dekens, M.P.; Whitmore, D. Autonomous onset of the circadian clock in the zebrafish embryo. EMBO J. 2008, 27, 2757–2765. [Google Scholar] [CrossRef]
- Vatine, G.; Vallone, D.; Appelbaum, L.; Mracek, P.; Ben-Moshe, Z.; Lahiri, K.; Gothilf, Y.; Foulkes, N.S. Light directs zebrafish period2 expression via conserved D and E boxes. PLoS Biol. 2009, 7, e1000223. [Google Scholar] [CrossRef]


| Gene Names | Ensembl Gene ID | Protein Length (aa) | Genome Location |
|---|---|---|---|
| bmal1 | ENSLOCG00000003999 | 678 | Chromosome LG27: 8,317,763–8,344,549 |
| bmal2 | ENSLOCG00000015224 | 639 | Chromosome LG8: 3,189,570–3,226,867 |
| clock1 | ENSLOCG00000014043 | 744 | Chromosome LG4: 72,323,329–72,339,605 |
| clock2 | ENSLOCG00000014750 | 886 | Chromosome LG7: 42,295,248–42,323,335 |
| cry1a | ENSLOCG00000015272 | 647 | Chromosome LG8: 4,053,475–4,074,670 |
| cry1b | ENSLOCG00000011417 | 675 | Chromosome LG3: 32,901,867–32,938,020 |
| cry2 | ENSLOCG00000014655 | 569 | Scaffold JH591436.1: 96,765–111,323 |
| cry3 | ENSLOCG00000011465 | 586 | Chromosome LG3: 33,014,383–33,053,389 |
| per1 | ENSLOCG00000013344 | 1445 | Chromosome LG2: 58,185,728–58,201,428 |
| per2 | ENSLOCG00000004441 | 1385 | Chromosome LG14: 7,862,125–7,881,568 |
| per3 | ENSLOCG00000002607 | 1165 | Chromosome LG25: 4,785,705–4,800,981 |
| csnk1e | ENSLOCG00000011701 | 273 | Chromosome LG12: 35,099,178-35,103,163 |
| dec1 | ENSLOCG00000010962 | 409 | Chromosome LG5: 27,670,723–27,673,540 |
| dec2 | ENSLOCG00000015327 | 422 | Chromosome LG8: 4,744,466–4,746,682 |
| nfil3-1 | ENSLOCG00000008217 | 443 | Chromosome LG2: 25,281,929–25,283,356 |
| nfil3-2 | ENSLOCG00000018299 | 544 | Chromosome LG6: 17,036,778–17,038,412 |
| nfil3-3 | ENSLOCG00000018298 | 394 | Chromosome LG6: 17,009,772–17,010,956 |
| nr1d1 | ENSLOCG00000006223 | 362 | Chromosome LG4: 15,608,213–15,662,480 |
| nr1d2 | ENSLOCG00000006818 | 604 | Chromosome LG11: 20,445,844–20,461,290 |
| tef | ENSLOCG00000011595 | 323 | Chromosome LG12: 34,848,929–34,860,886 |
| hlf | ENSLOCG00000012233 | 298 | Chromosome LG10: 33,240,269–33,260,238 |
| dbp | 69 | Scaffold JH591448.1:146984–147190 | |
| rora | ENSLOCG00000014779 | 519 | Chromosome LG3: 53,154,612–53,409,505 |
| rorb | ENSLOCG00000009712 | 462 | Chromosome LG2: 34,273,732–34,319,408 |
| rorc | ENSLOCG00000006502 | 467 | Chromosome LG19: 9,503,786–9,519,701 |
| timeless | ENSLOCG00000004180 | 1225 | Chromosome LG4: 11,892,308–11,918,271 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Liu, C.; Huang, M.; Huang, J.; Liu, C.; Zhang, J.; Postlethwait, J.H.; Wang, H. The Molecular Evolution of Circadian Clock Genes in Spotted Gar (Lepisosteus oculatus). Genes 2019, 10, 622. https://doi.org/10.3390/genes10080622
Sun Y, Liu C, Huang M, Huang J, Liu C, Zhang J, Postlethwait JH, Wang H. The Molecular Evolution of Circadian Clock Genes in Spotted Gar (Lepisosteus oculatus). Genes. 2019; 10(8):622. https://doi.org/10.3390/genes10080622
Chicago/Turabian StyleSun, Yi, Chao Liu, Moli Huang, Jian Huang, Changhong Liu, Jiguang Zhang, John H. Postlethwait, and Han Wang. 2019. "The Molecular Evolution of Circadian Clock Genes in Spotted Gar (Lepisosteus oculatus)" Genes 10, no. 8: 622. https://doi.org/10.3390/genes10080622
APA StyleSun, Y., Liu, C., Huang, M., Huang, J., Liu, C., Zhang, J., Postlethwait, J. H., & Wang, H. (2019). The Molecular Evolution of Circadian Clock Genes in Spotted Gar (Lepisosteus oculatus). Genes, 10(8), 622. https://doi.org/10.3390/genes10080622





