Breed, Diet, and Interaction Effects on Adipose Tissue Transcriptome in Iberian and Duroc Pigs Fed Different Energy Sources
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Sampling
2.3. RNA Isolation, Library Construction, and Sequencing
2.4. Bioinformatic Analyses
2.5. Results Validation by Quantitative PCR (qPCR)
2.6. Functional Interpretation
3. Results and Discussion
3.1. Breed Effect on Transcriptome
3.2. Diet Effects and Interaction between Breed and Diet on Transcriptome
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Raes, K.; De Smet, S.; Demeyer, D. Effect of dietary fatty acids on incorporation of long chain polyunsaturated fatty acids and conjugated linoleic acid in lamb, beef and pork meat: A review. Anim. Feed Sci. Technol. 2004, 113, 199–221. [Google Scholar] [CrossRef]
- Loor, J.J.; Vailati-Riboni, M.; McCann, J.C.; Zhou, Z.; Bionaz, M. Nutrigenomics in livestock: Systems biology meets nutrition. J. Anim. Sci. 2015, 93, 5554–5574. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Kersten, S. Nutrigenomics: Goals and strategies. Nat. Rev. Genet. 2003, 4, 315–322. [Google Scholar] [CrossRef] [PubMed]
- López-Bote, C. Sustained utilization of the Iberian pig breed. Meat Sci. 1998, 49, S17–S27. [Google Scholar] [CrossRef]
- Ayuso, M.; Fernandez, A.; Nunez, Y.; Benitez, R.; Isabel, B.; Fernandez, A.I.; Lopez-Bote, C.J. Developmental stage, muscle and genetic type modify muscle transcriptome in pigs: Effects on gene expression and regulatory factors involved in growth and metabolism. PLoS ONE 2016, 11, e0167858. [Google Scholar] [CrossRef] [PubMed]
- Pugliese, C.; Sirtori, F. Quality of meat and meat products produced from southern European pig breeds. Meat Sci. 2012, 90, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Cabot, R.A.; Kühholzer, B.; Chan, A.W.; Lai, L.; Park, K.W.; Chong, K.Y.; Schatten, G.; Murphy, C.N.; Abeydeera, L.R.; Day, B.N.; et al. Transgenic pigs produced using in vitro matured oocytes infected with a retroviral vector. Anim. Biotechnol. 2001, 12, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Myers, M.G.; Cowley, M.A.; Münzberg, H. Mechanisms of leptin action and leptin resistance. Annu. Rev. Physiol. 2008, 70, 537–556. [Google Scholar] [CrossRef]
- Walters, E.M.; Wolf, E.; Whyte, J.J.; Mao, J.; Renner, S.; Nagashima, H.; Kobayashi, E.; Zhao, J.; Wells, K.D.; Critser, J.K.; et al. Completion of the swine genome will simplify the production of swine as a large animal biomedical model. BMC Med. Genom. 2012, 5, 55. [Google Scholar] [CrossRef]
- Pérez-Enciso, M.; Ferraz, A.L.; Ojeda, A.; López-Béjar, M. Impact of breed and sex on porcine endocrine transcriptome: A bayesian biometrical analysis. BMC Genom. 2009, 10, 89. [Google Scholar] [CrossRef]
- Ventanas, S.; Ventanas, J.; Jurado, A.; Estevez, M. Quality traits in muscle biceps femoris and back-fat from purebred Iberian and reciprocal Iberian x Duroc crossbred pigs. Meat Sci. 2006, 73, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Óvilo, C.; Benítez, R.; Fernández, A.; Núñez, Y.; Ayuso, M.; Fernández, A.I.; Rodríguez, C.; Isabel, B.; Rey, A.I.; López-Bote, C.; et al. Longissimus dorsi transcriptome analysis of purebred and crossbred Iberian pigs differing in muscle characteristics. BMC Genom. 2014, 15, 413. [Google Scholar] [CrossRef] [PubMed]
- Benítez, R.; Fernández, A.; Isabel, B.; Núñez, Y.; De Mercado, E.; Gómez-Izquierdo, E.; García-Casco, J.; López-Bote, C.; Óvilo, C. Modulatory effects of breed, feeding status, and diet on adipogenic, lipogenic, and lipolytic gene expression in growing iberian and duroc pigs. Int. J. Mol. Sci. 2018, 19, 22. [Google Scholar] [CrossRef] [PubMed]
- FastQC. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 3 August 2019).
- TrimGalore. Available online: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 3 August 2019).
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, 36. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.; Achuthan, P.; Akanni, W.; Amode, R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; García Girón, C.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef] [PubMed]
- Lassmann, T.; Hayashizaki, Y.; Daub, C.O. SAMStat: Monitoring biases in next generation sequencing data. Bioinformatics 2010, 27, 130–131. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2015, 32, 292–294. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Miron, M.; Woody, O.Z.; Marcil, A.; Murie, C.; Sladek, R.; Nadon, R. A methodology for global validation of microarray experiments. BMC Bioinform. 2006, 7, 333. [Google Scholar] [CrossRef]
- Steibel, J.P.; Poletto, R.; Coussens, P.M.; Rosa, G.J.M. A powerful and flexible linear mixed model framework for the analysis of relative quantification RT-PCR data. Genomics 2009, 94, 146–152. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 7. [Google Scholar] [CrossRef]
- Andersen, C.L.; Ledet-Jensen, J.; Orntoft, T. Normalization of real-time quantitative RT-PCR data: A model based variance estimation approach to identify genes suited for normalization—Applied to bladder- and colon-cancer data-sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J.; Tugendreich, S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Olivares, A.; Daza, A.; Rey, A.I.; López-Bote, C.J. Effect of diet saturation on growth performance, carcass characteristics and fat quality of heavy pigs. Food Sci. Technol. Int. 2010, 16, 321–327. [Google Scholar] [CrossRef]
- Fuentes, V.; Ventanas, S.; Ventanas, J.; Estevez, M. The genetic background affects composition, oxidative stability and quality traits of Iberian dry-cured hams: Purebred Iberian versus reciprocal Iberian x Duroc crossbred pigs. Meat Sci. 2014, 96, 737–743. [Google Scholar] [CrossRef]
- Ayuso, M.; Fernández, A.; Núñez, Y.; Benítez, R.; Isabel, B.; Fernández, A.I.; Rey, A.I.; González-Bulnes, A.; Medrano, J.F.; Cánovas, A.; et al. Comparative analysis of muscle transcriptome between pig genotypes identifies genes and regulatory mechanisms associated to growth, fatness and metabolism. PLoS ONE 2015, 10, e0145162. [Google Scholar] [CrossRef]
- Óvilo, C.; Benítez, R.; Fernández, A.; Isabel, B.; Núñez, Y.; Fernández, A.I.; Rodríguez, C.; Daza, A.; Silió, L.; López-Bote, C. Dietary energy source largely affects tissue fatty acid composition but has minor influence on gene transcription in Iberian pigs. J. Anim. Sci. 2014, 92, 939–954. [Google Scholar] [CrossRef]
- Dietert, K.; Reppe, K.; Mundhenk, L.; Witzenrath, M.; Gruber, A.D. mCLCA3 Modulates IL-17 and CXCL-1 induction and leukocyte recruitment in Murine Staphylococcus aureus Pneumonia. PLoS ONE 2014, 9, e102606. [Google Scholar] [CrossRef]
- Cates, M.S.; Berry, M.B.; Ho, E.L.; Li, Q.; Potter, J.D.; Phillips, G.N. Metal-ion affinity and specificity in EF-hand proteins: Coordination geometry and domain plasticity in parvalbumin. Structure 1999, 7, 1269–1278. [Google Scholar] [CrossRef]
- Permyakov, E.A.; Uversky, V.N.; Permyakov, S.E. Parvalbumin as a pleomorphic protein. Curr. Protein Pept. Sci. 2017, 18, 780–794. [Google Scholar] [CrossRef]
- Fernández-Figares, I.; Lachica, M.; Nieto, R.; Rivera-Ferre, M.; Aguilera, J. Serum profile of metabolites and hormones in obese (Iberian) and lean (Landrace) growing gilts fed balanced or lysine deficient diets. Livest. Sci. 2007, 110, 73–81. [Google Scholar] [CrossRef]
- Ceddia, R.B.; William, W.N.; Curi, R. The response of skeletal muscle to leptin. Front Biosci. 2001, 1, 90–97. [Google Scholar] [CrossRef]
- Ahima, R.S. Adipose tissue as an endocrine organ. Obesity 2006, 14, 242–249. [Google Scholar] [CrossRef]
- Hanson, R.W.; Reshef, L. Glyceroneogenesis revisited. Biochimie 2003, 85, 1199–1205. [Google Scholar] [CrossRef]
- Latorre, P.; Burgos, C.; Hidalgo, J.; Varona, L.; Carrodeguas, J.A.; López-Buesa, P.C. A2456C-substitution in Pck1 changes the enzyme kinetic and functional properties modifying fat distribution in pigs. Sci. Rep. 2016, 6, 19617. [Google Scholar] [CrossRef]
- Muñoz, M.; Bozzi, R.; García, F.; Núñez, Y.; Geraci, C.; Crovetti, A.; Martins, J.M. Diversity across major and candidate genes in European local pig breeds. PLoS ONE 2018, 13, e0207475. [Google Scholar] [CrossRef]
- Stelmańska, E. Regulation of extramitochondrial malic enzyme gene expression in lipogenic tissues. Post py Higieny i Medycyny Doświadczalnej 2007, 6, 664–671. [Google Scholar]
- Palma-Granados, P.; Seiquer, I.; Benítez, R.; Óvilo, C.; Nieto, R. Effects of lysine deficiency on carcass composition and activity and gene expression of lipogenic enzymes in muscles and backfat adipose tissue of fatty and lean piglets. Animal 2019, 1–13. [Google Scholar] [CrossRef]
- Van Laere, A.S.; Nguyen, M.; Braunschweig, M.; Nezer, C.; Collette, C.; Moreau, L.; Archibald, A.L.; Haley, C.S.; Buys, N.; Tally, M.; et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 2003, 425, 832–836. [Google Scholar] [CrossRef]
- Burgos, C.; Galve, A.; Moreno, C.; Altarriba, J.; Reina, R.; García, C.; López-Buesa, P. The effects of two alleles of IGF2 on fat content in pig carcasses and pork. Meat Sci. 2012, 90, 309–313. [Google Scholar] [CrossRef]
- Alves, E.; Fernández, A.I.; García-Cortés, L.A.; López, Á.; Benítez, R.; Rodríguez, C.; Silió, L. Is it possible the breed origin traceability of Iberian pigs? In 7th International Symposium on the Mediterranean Pig; 7CICM; CIHEAM: Zaragoza, Spain, 2012. [Google Scholar]
- Murthy, S.; Born, E.; Mathur, S.N.; Field, F.J. LXR/RXR activation enhances basolateral efflux of cholesterol in CaCo2 cells. J. Lipid Res. 2002, 43, 1054–1064. [Google Scholar] [CrossRef]
- Filomeni, G.; Rotilio, G.; Ciriolo, M.R. Cell signalling and the glutathione redox system. Biochem. Pharmacol. 2002, 64, 1057–1064. [Google Scholar] [CrossRef]
- Liu, D.; Xu, Y. p53, Oxidative stress, and aging. Antioxid. Redox. Signal. 2011, 15, 6. [Google Scholar] [CrossRef]
- Wilmann, M.; Gautel, M.; Mayans, O. Activation of calcium/calmodulin regulated kinases. Cell Mol. Biol. 2000, 46, 883–894. [Google Scholar]
- Kanzaki, N.; Treboux, G.; Onuma, K.; Tsutsumi, S.I.A. Calcium phosphate clusters. Biomaterials 2001, 22, 2921–2929. [Google Scholar] [CrossRef]
- Zelcer, N.; Tontonoz, P. Liver X receptors as integrators of metabolic and inflammatory signaling. J. Clin. Investig. 2006, 116, 607–614. [Google Scholar] [CrossRef]
- Laurencikiene, J.; Rydén, M. Liver X receptors and fat cell metabolism. Int. J. Obes. 2012, 36, 1494–1502. [Google Scholar] [CrossRef]
- Kang, M.K.; Mehrazarin, S.; Park, N.H.; Wang, C.Y. Epigenetic gene regulation by histone demethylases: Emerging role in oncogenesis and inflammation. Oral Dis. 2017, 23, 709–720. [Google Scholar] [CrossRef]
- Han, G.; Li, F.; Singh, T.P.; Wolf, P.; Wang, X.J. The Pro-inflammatory Role of TGFβ1: A Paradox? Int. J. Biol. Sci. 2012, 8, 228–235. [Google Scholar] [CrossRef]
- Verrecchia, F.; Mauviel, A. Transforming growth factor-beta signaling through the Smad pathway: Role in extracellular matrix gene expression and regulation. J. Investig. Dermatol. 2002, 118, 211–215. [Google Scholar] [CrossRef]
- Migita, T.; Narita, T.; Asaka, R.; Miyagi, E.; Nagano, H.; Nomura, K.; Matsuura, M.; Satoh, Y.; Okumura, S.; Nakagawa, L.; et al. Role of insulin-like growth factor binding protein 2 in lung adenocarcinoma. Am. J. Pathol. 2010, 176, 4. [Google Scholar] [CrossRef]
- Nieto, R.; Rivera, M.; Garcia, M.A.; Aguilera, J.F. Amino acid availability and energy value of acorn in the Iberian pig. Livest. Prod. Sci. 2002, 77, 227–239. [Google Scholar] [CrossRef]
- Switonski, M.; Stachowiak, M.; Cieslak, J.; Bartz, M.; Grzes, M. Genetics of fat tissue accumulation in pigs: A comparative approach. J. Appl. Genet. 2010, 51, 153–168. [Google Scholar] [CrossRef]
- Fliers, M.D. Obesity wars: Molecular progress confronts an expanding epidemic. Cell 2004, 116, 337–350. [Google Scholar] [CrossRef]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Investig. 2003, 112, 1796–1808. [Google Scholar] [CrossRef]
- Gustafson, B.; Hammarstedt, A.; Andersson, C.X.; Smith, U. Inflamed adipose tissue a culprit underlying the metabolic syndrome and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2007, 2, 2276–2283. [Google Scholar] [CrossRef]
- Pahlavani, M.; Ramalho, T.; Koboziev, I.; LeMieux, M.J.; Jayarathne, S.; Ramalingam, L.; Filgueiras, L.R.; Moustaid-Moussa, N. Adipose tissue inflammation in insulin resistance: Review of mechanisms mediating anti-inflammatory effects of omega-3 polyunsaturated fatty acids. J. Investig. Med. 2017, 65, 1021–1027. [Google Scholar] [CrossRef]
- Gutierrez, D.A.; Puglisi, M.J.; Hasty, A.H. Impact of increased adipose tissue mass on inflammation, insulin resistance, and dyslipidemia. Curr. Diab. Rep. 2009, 9, 26–32. [Google Scholar] [CrossRef]
- Lee, B.C.; Lee, J. Cellular and molecular players in adipose tissue inflammation in the development of obesity-induced insulin resistance. Biochim. Biophys. Acta 2014, 1842, 446–462. [Google Scholar] [CrossRef]
- Torres-Rovira, L.; Astiz, S.; Caro, A.; Lopez-Bote, C.; Ovilo, C.; Pallares, P.; Perez-Solana, M.L.; Sanchez-Sanchez, R.; Gonzalez-Bulnes, A. Diet-induced swine model with obesity/leptin resistance for the study of metabolic syndrome and type 2 diabetes. Sci. World J. 2011, 2012, 510149. [Google Scholar] [CrossRef]
- Xu, H.; Barnes, G.T.; Yang, Q.; Tan, G.; Yang, D.; Chou, C.J.; Sole, J.; Nichols, A.; Ross, J.S.; Tartaglia, L.A.; et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Investig. 2003, 112, 1821–1830. [Google Scholar] [CrossRef]
- Apovian, C.M.; Bigornia, S.; Mott, M.; Meyers, M.R.; Ulloor, J.; Gagua, M.; McDonnell, M.; Hess, D.; Joseph, L.; Gokce, N. Adipose macrophage infiltration is associated with insulin resistance and vascular endothelial dysfunction in obese subjects. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 1654–1659. [Google Scholar] [CrossRef]
- Shepherd, P.R.; Gnudi, L.; Tozzo, E.; Yang, H.; Leach, F.; Kahn, B.B. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J. Biol. Chem. 1993, 268, 22243–22246. [Google Scholar]
- Chung le, T.K.; Hosaka, T.; Harada, N.; Jambaldorj, B.; Fukunaga, K.; Nishiwaki, Y.; Teshigawara, K.; Sakai, T.; Nakaya, Y.; Funaki, M. Myosin IIA participates in docking of Glut4 storage vesicles with the plasma membrane in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 2010, 391, 995–999. [Google Scholar] [CrossRef]
- Stall, R.; Ramos, J.; Fulcher, F.K.; Patel, Y.M. Regulation of myosin IIA and filamentous actin during insulin stimulated glucose uptake in 3T3-L1 adipocytes. Exp. Cell Res. 2014, 10, 81–88. [Google Scholar] [CrossRef]
- Singer, K.; Lumeng, C.N. The initiation of metabolic inflammation in childhood obesity. J. Clin. Investig. 2017, 127, 65–73. [Google Scholar] [CrossRef]
- Elks, C.E.; Heude, B.; de Zegher, F.; Barton, S.J.; Clément, K.; Inskip, H.M.; Koudou, Y.; Cooper, C.; Dunger, D.B.; Ibáñez, L.; et al. Associations between genetic obesity susceptibility and early postnatal fat and lean mass: An individual participant meta-analysis. JAMA Pediatr. 2014, 168, 1122–1130. [Google Scholar] [CrossRef]
- Mariman, E.C.; Wang, P. Adipocyte extracellular matrix composition, dynamics and role in obesity. Cell Mol. Life Sci. 2010, 67, 1277–1292. [Google Scholar] [CrossRef]
- Bresnick, A.R. Molecular mechanisms of nonmuscle myosin-II regulation. Curr. Opin. Cell Biol. 1999, 11, 26–33. [Google Scholar] [CrossRef]
- Shutova, M.S.; Svitkina, T.M. Common and specific functions of nonmuscle myosin II paralogs in cells. Biochemistry 2018, 83, 1459–1468. [Google Scholar] [CrossRef]
- Catalán, V.; Gómez-Ambrosi, J.; Rodríguez, A.; Frühbeck, G. Role of extracellular matrix remodelling in adipose tissue pathophysiology: Relevance in the development of obesity. Histol. Histopathol. 2012, 27, 1515–1528. [Google Scholar]
- Horodyska, J.; Reyer, H.; Wimmers, K.; Trakooljul, N.; Lawlor, P.G.; Hamill, R.M. Transcriptome analysis of adipose tissue from pigs divergent in feed efficiency reveals alteration in gene networks related to adipose growth, lipid metabolism, extracellular matrix, and immune response. Mol. Genet. Genom. 2019, 294, 395–408. [Google Scholar] [CrossRef]
- Bouloumié, A.; Lolmède, K.; Sengenès, C.; Galitzky, J. Angiogenesis in adipose tissue. Ann. Endocrinol. 2002, 63, 91–95. [Google Scholar]
- Wang, P.; Keijer, J.; Bunschoten, A.; Bouwman, F.; Renes, J.; Mariman, E. Insulin modulates the secretion of proteins from mature 3T3-L1 adipocytes: A role for transcriptional regulation of processing. Diabetologia 2006, 49, 2453–2462. [Google Scholar] [CrossRef]
- Vlaicu, S.; Tatomir, A.; Boodhoo, D.; Vesa, S.; Mircea, P.A.; Rus, H. The role of complement system in adipose tissue-related inflammation. Immunol. Res. 2016, 64, 653–664. [Google Scholar] [CrossRef]
- Marissal-Arvy, N.; Langlois, A.; Tridon, C.; Mormede, P. Functional variability in corticosteroid receptors is a major component of strain differences in fat deposition and metabolic consequences of enriched diets in rat. Metabolism 2011, 60, 706–719. [Google Scholar] [CrossRef]
- Peckett, A.J.; Wright, D.C.; Riddell, M.C. The effects of glucocorticoids on adipose tissue lipid metabolism. Metabolism 2011, 60, 1500–1510. [Google Scholar] [CrossRef]
- Björntorp, P.; Rosmond, R. Obesity and cortisol. Nutrition 2000, 16, 924–936. [Google Scholar]
- Boullu-Ciocca, S.; Verger, P.; Bocquier, A.; Oliver, C. Corticotropic axis and chronic stress in abdominal obesity and metabolic syndrome. Presse Med. 2005, 34, 1646–1653. [Google Scholar] [CrossRef]
- Johannsson, G.; Ragnarsson, O. Cardiovascular and metabolic impact of glucocorticoid replacement therapy. Front. Horm. Res. 2014, 43, 33–44. [Google Scholar]
- Gordon, S.; Taylor, P.R. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 2005, 5, 953–964. [Google Scholar] [CrossRef]
- Teng, K.T.; Chang, C.Y.; Chang, L.F.; Nesaretnam, K. Modulation of obesity-induced inflammation by dietary fats: Mechanisms and clinical evidence. Nutr. J. 2014, 13, 12. [Google Scholar] [CrossRef]
- Murray, P.J. Obesity corrupts myelopoiesis. Cell Metab. 2014, 19, 735–736. [Google Scholar] [CrossRef]
- Sehgal, A.; Donaldson, D.; Pridans, C.; Sauter, K.A.; Hume, D.A.; Mabbott, N.A. The role of CSF1R-dependent macrophages in control of the intestinal stem-cell niche. Nat. Commun. 2018, 9, 1272. [Google Scholar] [CrossRef]
- Todoric, J.; Löffler, M.; Huber, J.; Bilban, M.; Reimers, M.; Kadl, A.; Zeyda, M.; Waldhäusl, W.; Stulnig, T.M. Adipose tissue inflammation induced by high-fat diet in obese diabetic mice is prevented by n−3 polyunsaturated fatty acids. Diabetologia 2006, 49, 2109–2119. [Google Scholar] [CrossRef]
- Han, C.Y.; Kargi, A.Y.; Omer, M.; Chan, C.K.; Wabitsch, M.; O’Brien, K.D.; Wight, T.N.; Chait, A. Differential effect of saturated and unsaturated free fatty acids on the generation of monocyte adhesion and chemotactic factors by adipocytes. Diabetes 2010, 59, 386–396. [Google Scholar] [CrossRef]
- Jo, E.K.; Kim, J.K.; Shin, D.M.; Sasakawa, C. Molecular mechanisms regulating NLRP3 inflammasome activation. Cell Mol. Immunol. 2016, 13, 148–159. [Google Scholar] [CrossRef]
- Forrester, S.J.; Kikuchi, D.S.; Hernandes, M.S.; Xu, Q.; Griendling, K.K. Reactive oxygen species in metabolic and inflammatory signaling. Circ. Res. 2018, 122, 877–902. [Google Scholar] [CrossRef]
- Sridhar, J.; Goyal, N.; Liu, J.; Foroozesh, M. Review of ligand specificity factors for CYP1A subfamily enzymes from molecular modeling studies reported to-date. Molecules 2017, 22, 1143. [Google Scholar] [CrossRef]
- Zhang, S.; Hulver, M.W.; McMillan, R.P.; Cline, M.A.; Gilbert, E.R. The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr. Metab. 2014, 11, 10. [Google Scholar] [CrossRef]
- Meager, A.; Das, R.G. Biological standardization of human interferon beta: Establishment of a replacement world health organization international biological standard for human glycosylated interferon beta. J. Immunol. Methods 2005, 306, 1–15. [Google Scholar] [CrossRef]
- Rahman, S.M.; Schroeder-Gloeckler, J.M.; Janssen, R.C.; Jiang, H.; Qadri, I.; Maclean, K.N.; Friedman, J.E. CCAAT/enhancing binding protein beta deletion in mice attenuates inflammation, endoplasmic reticulum stress, and lipid accumulation in diet-induced nonalcoholic steatohepatitis. Hepatology 2007, 45, 1108–1117. [Google Scholar] [CrossRef]
- Rahman, S.M.; Baquero, K.C.; Choudhury, M.; Janssen, R.C.; de la Houssaye, B.A.; Miyazaki-Anzai, S.; Wang, S.; Moustaid-Moussa, N.; Makoto Miyazaki, M.; Friedmanb, J.E. C/EBPβ in bone marrow is essential for diet induced inflammation, cholesterol balance, and atherosclerosis. Atherosclerosis 2016, 250, 172–179. [Google Scholar] [CrossRef]
- Thomas, G.M.; Jacobs, K.B.; Yeager, M.; Kraf, P.; Wacholder, S.; Orr, N.; Yu, K.; Chatterjee, N.; Welch, R.; Hutchinson, A.; et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008, 40, 310–315. [Google Scholar] [CrossRef]
- Zeggini, E.; Scott, L.J.; Saxena, R.; Voight, B.F.; Marchini, J.L.; Hu, T.; de Bakker, P.I.; Abecasis, G.R.; Almgren, P.; Andersen, G.; et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008, 40, 638–645. [Google Scholar] [CrossRef]
- Li, L.; Yang, Y.; Yang, G.; Lu, C.; Yang, M.; Liu, H.; Zong, H. The role of JAZF1 on lipid metabolism and related genes in vitro. Metabolism 2011, 60, 523–530. [Google Scholar] [CrossRef]
- Jang, W.Y.; Bae, K.W.; Kim, S.H.; Yu, D.H.; Kim, H.J.; Ji, Y.R.; Park, S.J.; Park, S.J.; Kang, M.; Jeong, J.I.; et al. Overexpression of Jazf1 reduces body weight gain and regulates lipid metabolism in high fat diet. Biochem. Biophys. Res. Commun. 2014, 444, 296–301. [Google Scholar] [CrossRef]
- Yang, H.; He, J.; Xu, X.L.; Jiang, J.; He, C.Q.; Ma, H.M. Molecular characterization and tissue expression profile analysis of the porcine JAZF1 gene. Genet. Mol. Res. 2015, 14, 542–551. [Google Scholar] [CrossRef]
- Olivares, A.; Daza, A.; Rey, A.I.; Lopez-Bote, C.J. Interactions between genotype, dietary fat saturation and vitamin A concentration on intramuscular fat content and fatty acid composition in pigs. Meat Sci. 2009, 82, 6–12. [Google Scholar] [CrossRef]
- Wood, J.D.; Nute, G.R.; Richardson, R.I.; Whittington, F.M.; Southwood, O.; Plastow, G.; Mansbridge, R.; da Costa, N.; Chang, K.C. Effects of breed, diet and muscle on fat deposition and eating quality in pigs. Meat Sci. 2004, 67, 651–667. [Google Scholar] [CrossRef]
- Godinho, R.; Bergsma, R.; Sevillano, C.A.; Bastiaansen, J.W.M. Genotype by feed interaction in grower-finisher pigs fed different diets. In Proceedings of the World Congress on Genetics Applied to Livestock Production; University of New Zealand: Auckland, New Zealand, 2018. [Google Scholar]
- Zhao, R.; Muehlbauer, E.; Decuypere, E.; Grossmann, R. Effect of genotype—Nutrition interaction on growth and somatotropic gene expression in the chicken. Gen. Comp. Endocrinol. 2004, 136, 2–11. [Google Scholar] [CrossRef]
- Chung, K.Y.; Lunt, D.K.; Kawachi, H.; Yano, H.; Smith, S.B. Lipogenesis and stearoyl-CoA desaturase gene expression and enzyme activity in adipose tissue of short- and long-fed Angus and Wagyu steers fed corn- or hay-based diet. J. Anim. Sci. 2007, 85, 380–387. [Google Scholar] [CrossRef]
| BREED EFFECTS (Ib vs. Du) | DIET EFFECTS (HO vs. CH) | INTERACTION | CORRELATION | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IBERIAN | DUROC | |||||||||||||||
| GENES | RNA Seq (n = 24) | qPCR (n = 48) | RNA Seq (n = 12) | qPCR (n = 29) | RNA Seq (n = 12) | qPCR (n = 19) | DESEQ2 | qPCR | ||||||||
| q-Value | FC | p-Value | FC | q-Value | FC | p-Value | FC | q-Value | FC | p-Value | FC | p-Value | p-Value | Correlation (r) | p-Value (H0: r = 0) | |
| PCK1 | 0.001 | 1.81 | 0.0005 | 2.76 | 0.89 | 1.11 | 0.003 | 1.97 | 0.99 | 1.13 | 0.31 | 1.38 | 0.99 | 0.02 | 0.81 | 3.47 × 10−6 |
| PLIN2 | 0.001 | 1.55 | <0.0001 | 5.02 | 0.99 | 0.92 | 0.63 | 1.08 | 0.49 | 0.91 | 0.51 | 0.86 | 0.99 | 0.42 | 0.64 | 0.001 |
| IGFBP3 | 0.001 | 1.80 | <0.0001 | 5.51 | 0.007 | 2.57 | 0.02 | 1.61 | 0.90 | 1.34 | 0.001 | 1.31 | 0.25 | 0.1 | 0.68 | 4.00 × 10−4 |
| JAZF1 | 0.90 | 1.71 | 0.066 | 1.10 | 0.99 | 0.71 | <0.0001 | 0.01 | 0.90 | 1.02 | 0.01 | 1.03 | 3.51 × 10−6 * | <0.0001 | 0.79 | 1.23 × 10−5 |
| PDLIM3 | 0.001 | 0.11 | 0.001 | 0.13 | 0.85 | 0.82 | 0.02 | 0.44 | 0.02 | 2.78 | 0.02 | 1.58 | 0.19 | 0.76 | 0.63 | 0.002 |
| PYGM | 0.001 | 0.75 | 0.5 | 0.67 | 0.91 | 0.89 | 0.28 | 0.48 | 0.08 | 3.32 | 0.07 | 1.31 | 0.20 | 0.86 | 0.65 | 0.001 |
| RBP7 | 0.001 | 0.46 | <0.0001 | 0.58 | 0.87 | 0.84 | 0.21 | 0.61 | 0.99 | 1.15 | 0.05 | 1.23 | 0.15 | 0.05 | 070 | 0.0002 |
| ASB2 | 0.35 | 0.07 | 0.18 | 0.1 | 0.90 | 0.94 | 0.31 | 0.59 | 0.02 | 3.23 | 0.8 | 1.15 | 0.41 | 0.81 | 0.92 | 5.46 × 10−5 |
| EEF1A2 | 0.02 | 0.01 | 0.05 | 0.10 | 0.02 | 4.49 | 0.001 | 6.66 | 0.02 | 3.86 | 0.11 | 2.14 | 0.80 | 0.04 | 0.96 | 2.42 × 10−6 |
| SERPINE1 | 0.19 | 1.27 | 0.11 | 1.51 | 0.02 | 1.61 | 0.01 | 1.36 | 0.02 | 0.56 | 0.004 | 0.89 | 0.001 | 0.003 | 0.77 | 0.004 |
| CYP1A1 | 0.95 | 0.95 | 0.11 | 0.39 | 0.008 | 2.14 | 0.03 | 2.97 | 0.02 | 3.85 | 0.09 | 1.34 | 0.005 | 0.02 | 0.70 | 0.001 |
| Canonical Pathways | p-Value | Ratio 1 | z-Score 2 | Molecules |
|---|---|---|---|---|
| Glutathione Redox Reactions I | 0.008 | 4/24 | −2 | GPX3,MGST2,GPX1,GSTP1 |
| ILK Signaling | 0.001 | 16/205 | 2 | MYH4,MYL2,ACTN2,DIRAS3,ACTN3,PIK3C2G,MYH7,ITGB8,MYL1,FOS,CDH1,RHOQ,AKT1,MYH2,PTGS2,ACTG2 |
| Actin Cytoskeleton Signaling | 0.002 | 17/234 | 2.111 | MYH4,RASD2,MYL2,ACTN2,MYLPF,MYLK2,ACTN3,PIK3C2G,EGF,MYH7,MYL1,MYH2,FGF18,LBP,ACTG2,NCKAP1L,MATK |
| Opioid Signaling Pathway | 0.01 | 16/250 | 2.138 | CACNG6,ADCY2,CACNA1S,CACNG1,RASD2,CACNB1,RGS3,RGS7,PIK3C2G,GRIN3A,FOSB,FOS,CAMK2A,AKT1,ADCY10,CAMK2B |
| Cardiac Hypertrophy Signaling (Enhanced) | 0.0007 | 31/498 | 2.502 | ADRA2B,IL15RA,CACNA1S,RASD2,LEP,PLCH1,ATP2A1,CAMK2A,NFAT5,AKT1,FGF18,NGFR,WNT4,TNFSF15,CAMK2B,IL11RA,TNFRSF11B,SMPDL3A,HDAC9,ADCY2,IL15,PIK3C2G,IL20RB,ADRA2A,TGFB3,IL2RA,HSPB7,PTGS2,ADCY10,HSPB1,WNT5A |
| Calcium Signaling | 0.00001 | 24/206 | 2.887 | HDAC9,CACNG6,MYH4,TNNT1,CACNB1,CACNG1,CACNA1S,MYL2,TNNI2,TNNT3,TNNC2,GRIA2,MYH7,TPM1,TPM2,MYL1,ATP2A1,ATP2B2,GRIN3A,MYH2,CAMK2A,NFAT5,TNNI1,CAMK2B |
| Upstream Regulator | Expression Log Ratio (Duroc/Iberian) | Molecule Type | Activation Z-Score 1 | p-Value of Overlap | Molecules in Dataset | Related Functions |
|---|---|---|---|---|---|---|
| ACTIVATED IN IBERIAN | ||||||
| KDM5A | transcription regulator | −3.357 | 3.28 × 10−5 | 15 | Epigenetic regulation of inflammation | |
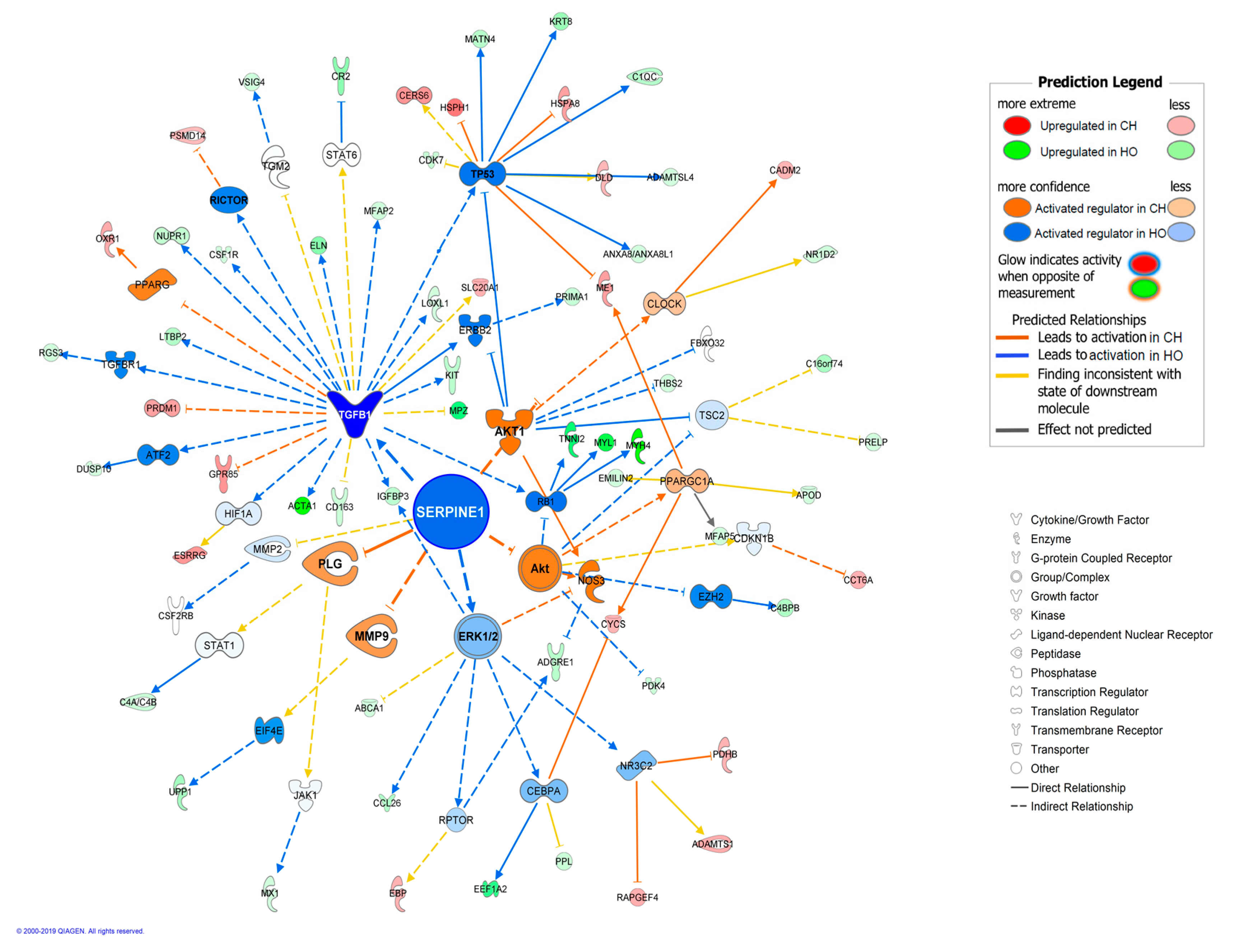
| DNMT3A | enzyme | −3.051 | 2.74 × 10−6 | 16 | Inflammation and lipid metabolism | |
| AHR | ligand-dependent nuclear receptor | −2.702 | 9.15 × 10−10 | 34 | Inflammatory and immune response | |
| SMTNL1 | other | −2.668 | 2.13 × 10−8 | 10 | ||
| PTEN | phosphatase | −2.569 | 0.003 | 21 | Cell migration, survival and proliferation | |
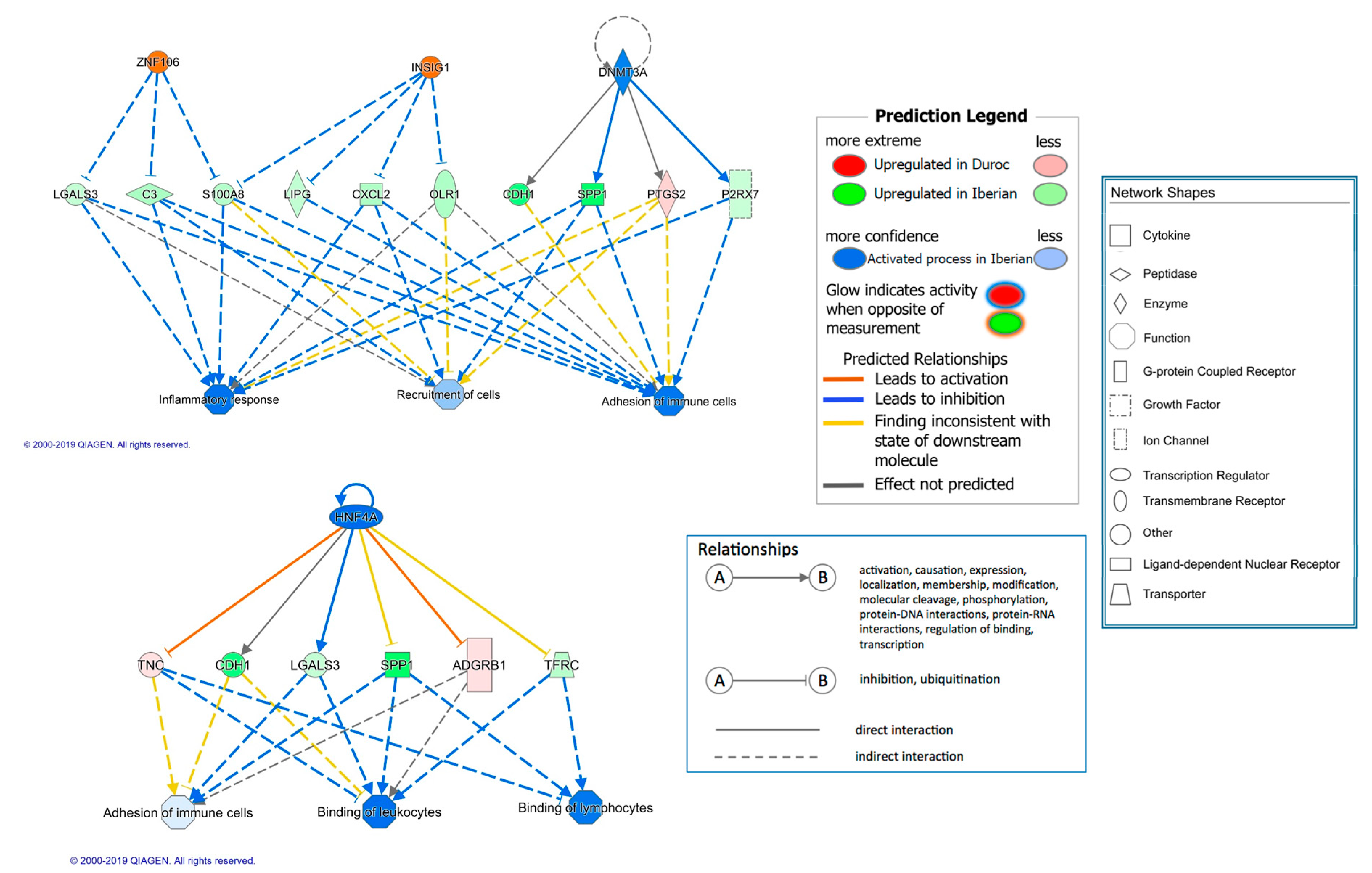
| HNF4A | 1.212 | transcription regulator | −2.53 | 5.71 × 10−5 | 33 | Inflammation |
| NR1H3 | ligand-dependent nuclear receptor | −2.294 | 0.01 | 13 | Inflammation and fat cell metabolism | |
| NOS2 | enzyme | −2.145 | 8.99 × 10−4 | 15 | Inflammation and insulin resistance | |
| MED1 | transcription regulator | −2.138 | 0.001 | 12 | Activation of gene transcription | |
| RBPJ | transcription regulator | −2.101 | 0.0002 | 13 | Polarization of macrophages | |
| ACTIVATED IN DUROC | ||||||
| Bvht | other | 3.148 | 2.16 × 10−4 | 10 | ||
| ZNF106 | other | 3 | 1.68 × 10−5 | 9 | ||
| INSIG1 | other | 3 | 0.004 | 9 | Insulin signaling | |
| ERBB2 | kinase | 2.867 | 0.002 | 30 | ||
| MEF2C | transcription regulator | 2.704 | 1.79 × 10−12 | 17 | Growth | |
| MYOD1 | transcription regulator | 2.53 | 2.19 × 10−9 | 14 | Growth | |
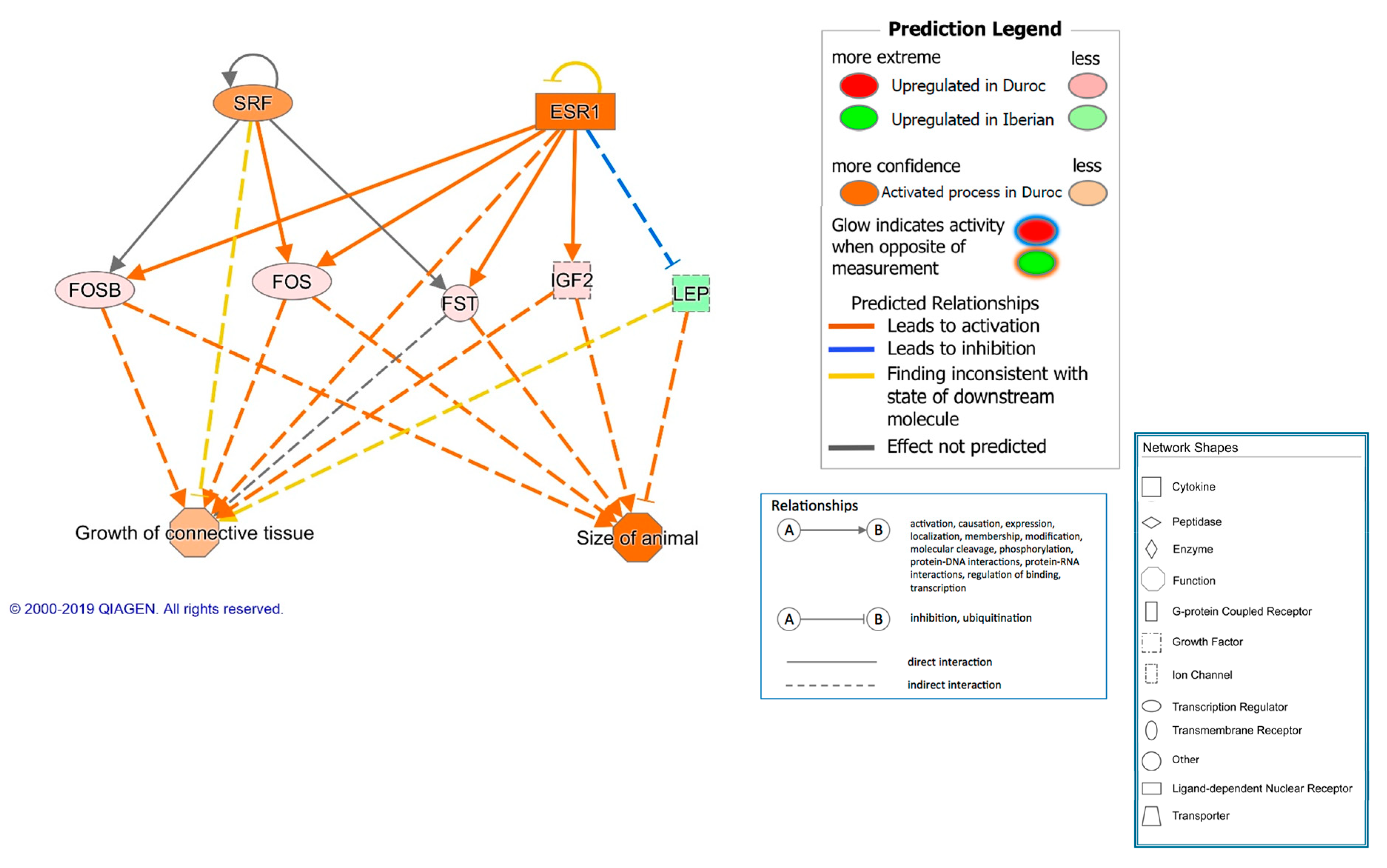
| TGFB1 | growth factor | 2.453 | 3.50 × 10−7 | 20 | Growth and inflammation | |
| HNF1A | transcription regulator | 2.439 | 1.90 × 10−4 | 47 | Cholesterol metabolism | |
| IL1R1 | transmembrane receptor | 2.39 | 7.36 × 10−3 | 22 | Inflammation | |
| SMAD3 | transcription regulator | 2.353 | 3.01 × 10−5 | 6 | Cell differentiation | |
| Akt | group | 2.311 | 1.08 × 10−4 | 6 | signaling of insulin and others receptors | |
| IGFBP2 | other | 2.219 | 0.01 | 17 | Growth | |
| STAT5a/b | group | 2.213 | 0.006 | 15 | ||
| SRF | transcription regulator | 2.204 | 3.01 × 10−7 | 6 | Cell proliferation and differentiation | |
| BDNF | growth factor | 2.202 | 0.001 | 7 | Growth | |
| FSHR | G-protein coupled receptor | 2.177 | 0.008 | 5 | ||
| RB1 | transcription regulator | 2.165 | 1.39 × 10−4 | 5 | ||
| MAPK8 | kinase | 2.138 | 0.002 | 7 | Regulation of development | |
| ESR1 | ligand-dependent nuclear receptor | 2.093 | 1.20 × 10−7 | 26 | Growth | |
| TBX5 | transcription regulator | 2.091 | 3.75 × 10−8 | 11 | Regulation of development | |
| COL6A1 | 0.68 | other | 2.064 | 3.56 × 10−6 | 6 | Growth |
| MET | 0.799 | kinase | 2.039 | 0.001 | 25 | Development and organogenesis |
| PELP1 | other | 2 | 0.003 | 10 | Inflammatory signaling | |
| Canonical Pathways in Iberian Pigs | p-Value | Ratio 1 | Z-Score 2 | Molecules |
| Complement System | 2.45471 × 10−8 | 7/37 | −0.816 | C4A/C4B,C4BPB,C4BPA,C1QC,C1QA,C1QB,CR2 |
| Acetyl-CoA Biosynthesis I (Pyruvate Dehydrogenase Complex) | 2.0893 × 10−5 | 3/7 | DLAT,DLD,PDHB | |
| Agranulocyte Adhesion and Diapedesis | 0.00024 | 8/192 | MYH4,SELE,CXCL14,CCL24,CCL14,CCL26,ACTA1,MYL1 | |
| Mineralocorticoid Biosynthesis | 0.0034 | 2/10 | EBP,HSD3B1 | |
| Interferon Signaling | 0.004 | 3/36 | MX1,IFI6,ISG15 | |
| Glucocorticoid Biosynthesis | 0.004 | 2/11 | EBP,HSD3B1 | |
| Aldosterone Signaling in Epithelial Cells | 0.004 | 6/174 | HSPA8,DNAJB4,HSPH1,HSPA13,DNAJB1,DNAJA1 | |
| Hepatic Fibrosis/Hepatic Stellate Cell Activation | 0.005 | 6/186 | MYH4,IGFBP3,IL10RA,CD14,SERPINE1,MYL1 | |
| Androgen Biosynthesis | 0.006 | 2/14 | EBP,HSD3B1 | |
| Protein Ubiquitination Pathway | 0.009 | 7/271 | HSPA8,DNAJB4,HSPH1,HSPA13,PSMD14,DNAJB1,DNAJA1 | |
| Glucocorticoid Receptor Signaling | 0.01 | 8/350 | HSPA8,SELE,KRT8,CDK7,CDKN1A,PRKAA2,CD163,SERPINE1 | |
| Canonical Pathways in Duroc Pigs | p-Value | Ratio 1 | Z-Score 2 | Molecules |
| Protein Kinase A Signaling | 0.002 | 5/400 | PPP1R14C,CAMK2A,PYGM,TNNI2,MYLK2 | |
| nNOS Signaling in Neurons | 0.005 | 2/47 | CAMK2A,CAPN3 | |
| Calcium Signaling | 0.01 | 3/206 | CAMK2A,TNNI2,MYH8 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benítez, R.; Trakooljul, N.; Núñez, Y.; Isabel, B.; Murani, E.; De Mercado, E.; Gómez-Izquierdo, E.; García-Casco, J.; López-Bote, C.; Wimmers, K.; et al. Breed, Diet, and Interaction Effects on Adipose Tissue Transcriptome in Iberian and Duroc Pigs Fed Different Energy Sources. Genes 2019, 10, 589. https://doi.org/10.3390/genes10080589
Benítez R, Trakooljul N, Núñez Y, Isabel B, Murani E, De Mercado E, Gómez-Izquierdo E, García-Casco J, López-Bote C, Wimmers K, et al. Breed, Diet, and Interaction Effects on Adipose Tissue Transcriptome in Iberian and Duroc Pigs Fed Different Energy Sources. Genes. 2019; 10(8):589. https://doi.org/10.3390/genes10080589
Chicago/Turabian StyleBenítez, Rita, Nares Trakooljul, Yolanda Núñez, Beatriz Isabel, Eduard Murani, Eduardo De Mercado, Emilio Gómez-Izquierdo, Juan García-Casco, Clemente López-Bote, Klaus Wimmers, and et al. 2019. "Breed, Diet, and Interaction Effects on Adipose Tissue Transcriptome in Iberian and Duroc Pigs Fed Different Energy Sources" Genes 10, no. 8: 589. https://doi.org/10.3390/genes10080589
APA StyleBenítez, R., Trakooljul, N., Núñez, Y., Isabel, B., Murani, E., De Mercado, E., Gómez-Izquierdo, E., García-Casco, J., López-Bote, C., Wimmers, K., & Óvilo, C. (2019). Breed, Diet, and Interaction Effects on Adipose Tissue Transcriptome in Iberian and Duroc Pigs Fed Different Energy Sources. Genes, 10(8), 589. https://doi.org/10.3390/genes10080589







